ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2011; 7(8):1145-1160. doi:10.7150/ijbs.7.1145 This issue Cite
Research Paper
Stability of a Long Noncoding Viral RNA Depends on a 9-nt Core Element at the RNA 5' End to Interact with Viral ORF57 and Cellular PABPC1
1. Tumor Virus RNA Biology Laboratory, HIV and AIDS Malignancy Branch, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA
2. Nanobiology Program, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA
3. Biostatistics & Data Management Section, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA
Abstract
Kaposi sarcoma-associated herpesvirus (KSHV) ORF57, also known as Mta (mRNA transcript accumulation), enhances viral intron-less transcript accumulation and promotes splicing of intron-containing viral RNA transcripts. In this study, we identified KSHV PAN, a long non-coding polyadenylated nuclear RNA as a main target of ORF57 by a genome-wide CLIP (cross-linking and immunoprecipitation) approach. KSHV genome lacking ORF57 expresses only a minimal amount of PAN. In cotransfection experiments, ORF57 alone increased PAN expression by 20-30-fold when compared to vector control. This accumulation function of ORF57 was dependent on a structured RNA element in the 5' PAN, named MRE (Mta responsive element), but not much so on an ENE (expression and nuclear retention element) in the 3' PAN previously reported by other studies. We showed that the major function of the 5' PAN MRE is increasing the RNA half-life of PAN in the presence of ORF57. Further mutational analyses revealed a core motif consisting of 9 nucleotides in the MRE-II , which is responsible for ORF57 interaction and function. The 9-nt core in the MRE-II also binds cellular PABPC1, but not the E1B-AP5 which binds another region of the MRE-II. In addition, we found that PAN RNA is partially exportable in the presence of ORF57. Together, our data provide compelling evidence as to how ORF57 functions to accumulate a non-coding viral RNA in the course of virus lytic infection.
Keywords: KSHV, long non-coding RNA, ORF57, PAN, RNA stability, RNA accumulation, PABPC1, E1B-AP5
Introduction
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV8), belongs to the gamma herpesvirus family and is the causative agent for Kaposi's sarcoma, the most common AIDS-associated cancer [1-3]. KSHV is also associated with other lymphotropic malignancies, such as primary effusion lymphoma (PEL) and multicentric Castleman's disease [4,5]. Similar to other herpesviruses, KSHV undergoes two distinguishable viral life cycles: latency and a lytic replication. However, and unlike other viruses, the KSHV latent program does not efficiently immortalize virus infected cells [6,7]. Experimental evidence also indicates that viral factors encoded during the lytic cycle, such as paracrine signals [8], or viral proteins, like the G protein-coupled receptor (vGPCR) [9,10], might supply the inflammatory and angiogenic components required for cell transformation [7].
Given the importance of the KSHV lytic cycle, we are interested in the mechanisms underlying the regulation of viral lytic gene expression. During the lytic cycle, KSHV expresses two main regulatory proteins: ORF50 (Rta, replication and transcription activator) which initiates transcription of viral lytic genes [11,12] and ORF57 (Mta, mRNA transcript accumulation), a multifunctional protein that promotes the accumulation of viral intron-less transcripts [13-17] and stimulates intron removal of viral intron-containing RNAs [18,19]. The intron removal function of ORF57 is especially important for the virus, as pre-mRNA lacking appropriate posttranscriptional processing is less stable than its properly processed functional counterpart [20]. Lytic KSHV replication also promotes widespread shutoff of cellular gene expression by hyperadenylation and nuclear retention of host mRNA which leads to enhancement of cellular mRNA turnover [21-23]. Thus, it is no surprise that KSHV has evolved mechanisms for regulating the ORF57-mediated post-transcriptional viral RNA processing and accumulation. To understand how ORF57 functions in host cells is a key step to determine how KSHV ensures the expression of its own genes, while also shutting-off host gene expression during virus lytic infection. Previously, we have described a mechanism as to how ORF57 stimulates the expression of intron-containing viral RNA transcripts in [18]. However, it remains unclear how ORF57 accumulates intron-less viral RNAs, and whether the accumulation is mediated by interfering with nuclear export [13,16,24-26].
Recently, we conducted a genome-wide CLIP (UV-crosslinking and immunoprecipitation) assay from B cells with KSHV lytic infection and identified 11 viral transcripts as putative ORF57 targets [27]. One of those targets was vIL-6, which stimulates cell proliferation during KSHV infection [8]. We found that vIL-6 RNA contains an ORF57 responsive element. ORF57 binding to this element in vIL-6 RNA stabilizes vIL-6 RNA and promotes vIL-6 translation by interfering with miR-1293-mediated vIL-6 mRNA repression [27].
Subsequently, we identified PAN RNA (polyadenylated nuclear RNA) as a major target of ORF57 and, therefore, became interested in whether ORF57 is also involved in modulating PAN RNA metabolism. PAN is the most abundant viral transcript, constituting ~80% of all polyadenylated transcripts in a cell with lytic KSHV infection [28,29]. PAN is a long non-coding, polyadenylated nuclear RNA transcribed by RNA polymerase II [28-30] which differs from EBV non-coding, small nuclear RNAs, EBER-1 and EBER-2 transcribed by RNA polymerase III [31]. In the nucleus, PAN forms a speckled pattern typical of U snRNAs and colocalizes with Sm protein [28,32]. We, along with other groups, have shown that ORF57 promotes PAN RNA accumulation [14,33]. Previous studies showed that PAN RNA stability in the absence of ORF57 seems to be dependent on the activity of a cis-acting 79-nucleotide (nt) RNA element located in the 3' PAN, called ENE (expression and nuclear retention element)[34-37]. In the absence of the ENE, PAN's stability appears to rely on ORF57 binding to a 5' motif [38]. In this report, we identified a 9-nt core motif in the 5' PAN region that is responsible for ORF57-mediated PAN accumulation. The element containing the core motif is designated as MRE (Mta responsive element) and its major function is increasing the half-life of PAN RNA in the presence of ORF57. In contrast, the ENE plays only a small role in ORF57-mediated accumulation of PAN. We found that the PAN MRE interacts with ORF57 and cellular proteins PABPC1 and E1B-AP5.
Materials and methods
Cells
Human HEK293 and HeLa cells were cultivated in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, HyClone, Logan, UT), 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). HEK293 cell lines stably harboring a KSHV wt genome (Bac36-wt) or an ORF57-null KSHV genome (Bac36-Δ57) [33] were induced with 1 mM valproate (VA) for lytic infection. JSC-1 cells (KSHV+/EBV+) and BCBL1 cells (KSHV+) were grown in RPMI 1640 containing 10% FBS and were induced, respectively, with sodium butyrate (Bu, 3 mM) and VA (1 mM) for lytic infection. TREx BCBL1-vector and TREx BCBL1-Rta cells [39] were cultivated in RPMI 1640 supplemented with 10% FBS and hygromycin B (50 μg/ml). To induce the expression of KSHV lytic genes, TREx BCBL1-Rta or TREx BCBL1-vector (a negative control) cells at 5 x 105 cells/ml were seeded and cultivated for 24 h in the presence of 1μg/ml of doxycycline (Dox).
Mammalian expression vectors
Mammalian expression vectors pVM7 (for FLAG-tagged ORF57), pVM8 (for GFP-tagged ORF57) and pVM36 (for GFP-tagged inactive ORF57 with point mutations in NLS2+3 or ORF57 mtNLS2+3) were described in our previous report [13]. KSHV ORF50 (Rta) expression vector has been used in our laboratory for various studies [33,40]. Other plasmids used in this study and the oligos and strategy used for their construction are listed in Supplementary Material: Table S1. See oligo sequences in Supplementary Material: Table S2. Overlapped PCR for mutant constructions was performed as described [41].
Transient transfection assay
Cotransfection of expression vectors PAN (wt and mutants), vGPCR, or K5 was performed in HEK293 or HeLa cells (5 x105 cells/well) in a six-well plate, with 1 μg of individual vector together with 0.2 μg (5:1 ratio) of pVM7 (ORF57-FLAG fusion) [13] or an empty pFLAG-CMV-5.1 vector using Lipofectamine 2000 (Invitrogen).
Western blot
Protein samples for Western blotting were prepared by direct lysis of the cells in 0.5 ml of 2X SDS sample buffer (Quality Biological, Inc., Gathersburg, MD) plus 10% 2-mercaptoethanol. Samples were boiled and resolved in a 4%-12% SDS-polyacrylamide gel by electrophoresis. The following antibodies were used in Western blot analyses: a rabbit polyclonal anti-ORF57 antibody [33](1:3,000 dilution), mouse monoclonal anti-ORF57 antibody (unpublished data, used at a dilution of 1:1,000), rabbit polyclonal anti-PABPC1 (1:700, Abcam ab21060), and rabbit polyclonal anti-E1B-AP5 (1:1,000, ProteinTech Group 0578-1-AP, Chicago, IL), together with corresponding peroxidase-conjugated secondary antibodies (1:10,000, Sigma). The signal on the western blot was detected with West Pico chemiluminiscence substrate (Pierce, Rockford, IL).
RNA preparation and Northern blot
Total cell RNA samples were prepared 24 or 48 h after transfection (HEK293, HeLa) or after induction (B cells) by the addition of 1 ml of TRIzol reagent according to the TRIzol protocol (Invitrogen). Cytoplasmic and nuclear total RNAs were fractionated as described [13]. RNA (~5 μg) was separated in a 1% agarose gel and analyzed by Northern blotting [13]. PAN, K5 and vGPCR RNAs expressed from individual plasmids were detected with 2 × 106 cpm of a γ-32P-labeled T7 probe oZMZ243 (see Supplementary Material: Table S2 for sequence details). PAN was also detected with a PAN-specific probe oJM7 (Supplementary Material: Table S2). ORF57 was detected with an oligo probe oVM11 (Supplementary Material: Table S2). The hybridization signal was captured using a Molecular Dynamics PhosphorImager Storm 860 and analyzed with ImageQuant software.
CLIP and RT-PCR
CLIP assays were conducted as described [27]. RT-PCR was performed by using a PAN-specific primer pair oVM52 and oJM7 on DNase-treated total RNA extracted from the CLIP complexes obtained with an anti-ORF57 antibody or rabbit IgG (a negative control).
Luciferase Assays
To evaluate luciferase activity in HEK293 cells, 2.5 x105 cells were grown in 24-well plates and transfected with 50 ng of each firefly luciferase reporter plasmid (pMIR-REPORT-Luciferase reporter or its derived pJM19, pJM43 and pJM44, see Supplementary Material: Table S1 for details) together with 10 ng of Renilla luciferase reporter (pRL-TS) [42] using Lipofectamine 2000. Cells were harvested in dual luciferase assay 1x lysis buffer (Promega, Madison, WI). To evaluate activity in TREx BCBL1 cells, 2.5 x105 TREx BCBL1-Rta cells were grown in 24-well plates for 24 h and the reactivated by addition of Dox (1 μg/ml). One hour after Dox addition, the cells were transfected with 100 ng of each firefly and Renilla luciferase reporter plasmid (psiCHECK2 or its derived pJM47, pJM48, pJM49, pJM50 and pJM51, see Supplementary Material: Table S1 for details) using FugeneHD transfection agent (Roche Diagnostics, Indianapolis, IN). 24 h after transfection, the cells were harvested in dual luciferase assay 1x lysis buffer. The volume of the buffer added was adjusted according to cell number of each testing condition in order to obtain an equal amount of protein levels of induced vs un-induced conditions. Luciferase activity measured from cell lysate supernatant using a GloMax®-Multi Microplate Multimode Reader (Promega) was expressed in ratios of firefly to Renilla (F/R for HEK293 cells) or Renilla to firefly (R/F for TREx BCBL1-Rta cells).
We performed a mixed model ANOVA on luciferase activities (Fig. 5D). An eight level treatment group (4 plasmids x 2 treatments) was used as the fixed effect. Two-tailed p-values were determined after adjusting for the number of comparisons performed, using a standard method (due to Holm). Residuals were examined for normality and homogeneity and were partitioned if found to be heterogeneous.
RNA-protein pulldown assay
Total cell extracts used in RNA pulldown assays were prepared from ~5 x 106 of TREx BCBL1-Rta or -vector cells which were reactivated by Dox (1 μg/ml) for 24 h. The cells were harvested by centrifugation at 800 x g at 4oC for 5 min, washed twice with 1 x PBS and resuspended in 500 μl (Rta cells) or 1000 μl (vector cells) of 1x RIPA buffer containing protease inhibitor (Complete Mini EDTA-free Protease Inhibitor Cocktail, 1x, Roche). The samples were incubated for 10 min on ice, briefly sonicated (10 times at level 4), and centrifuged for 25 min at 10,000 x g at 4oC. The extract supernatant ready for RNA pulldown was aliquoted and stored at -20oC.
RNA pulldown assay using biotinylated RNA oligomers was performed as described [27,43]. The following RNA oligos used in the pulldowns and schematized in Fig. 6A were oJM32, oJM33, oJM34, oJM35 and oJM68 (see sequence details in Supplementary Material: Table S2). RNA oligos oNP41 and oNP42 derived from vIL-6 RNA served, respectively, as a negative and positive control for ORF57 binding [27]. The proteins in the pulldowns were analyzed by Western blot. Alternatively, the proteins resolved in a 4%-12% SDS-PAGE gel were silver-stained using SilverQuest™ Silver Staining Kit (Invitrogen) and the corresponding proteins in a separate gel without silver staining were gel-purified and identified using Nano LC-MS/MS peptide sequencing technology (ProtTech, Inc., Norristown, PA).
RNA stability assay
PAN decay was analyzed in HEK293 cells transiently transfected with individual PAN expression plasmids in the presence or absence of ORF57. Since PAN is expressed at a very low level in the absence of ORF57, HEK293 cells (1x 106) in 60-mm dishes were cotransfected using FuGene HD (Roche) with 4 μg of a wt PAN expression plasmid plus 800 ng of an empty pFLAG-CMV-5.1 vector, 20-times more PAN expression vector in the absence of ORF57 than its transfection in a 24-well plate in the presence of ORF57. Real-time PCR for PAN and GAPDH RNA was carried out on the cDNA in duplicates as described previously [26]. GAPDH RNA levels remained constant in the course of the experiments by normalization with β-actin RNA. The following PAN TaqMan primers from IDT (Coralville, IA) were used: PAN Probe; PAN primer 1; PAN primer 2 (Supplementary Material: Table S2). The relative expression of PAN was determined using 2-ΔΔCT method [27,44,45] and converted to percentage, with the amount at time zero of actinommycin D (act. D) addition as 100 %. The RNA decay represents remaining RNA (%) over the time zero in the presence or absence of ORF57. A non-linear regression analysis on the raw data was performed and applied to an exponential decay model [Fold Percent = α*exp(β*time)], where α (alpha) is the intercept when time = 0, and β (beta) is the decay rate. Alpha was constrained to be equal to 100% (α=100%), and beta to be less than or equal to zero (β<=0). Comparison of beta parameter estimates between the treatments was performed by using a model with a dummy variable (model based two-tailed p-value via t-test). The half-life for each treatment was estimated by letting the fold percent equal 50% and solving for time.
Results
PAN is associated with ORF57 in B cells with KSHV lytic infection and responds to ORF57 function
Among 11 potential RNA targets of ORF57 identified in B cells with KSHV lytic infection by an anti-ORF57 CLIP assay [27], PAN RNA sequences were pulled down more than 90 times, listing the PAN as one of the most prominent targets in the ORF57 CLIP assays. Considering PAN is the most abundant KSHV RNA constituting ~80% of all polyadenylated transcripts in a cell with lytic KSHV infection [28,29], the high frequency of PAN sequences identified in the ORF57 CLIP assays might simply reflect the abundance of PAN in an RNA pool of the KSHV-infected B cells. However, when aligned against the KSHV genome, the isolated PAN sequences were clustered mainly at the 5'-half (nt 28667-28750 with 16 sequence tags) and 3'-half (nt 29544-29658 with 43 sequence tags) of PAN (NCBI accession no.: U75698) [46], along with a fewer sequence tags clustering in the middle regions of PAN [Supplementary Material: Fig. S1, also see Supplementary Material: Table S1 in the reference [27], indicating a sequence-specific PAN association of ORF57 in B cells with KSHV lytic infection. Interestingly, the PAN 3' region interacting with ORF57 encompasses a structured ENE (expression and nuclear retention element) previously identified for the nuclear abundance of PAN [34] through its U-rich internal loop interacting with the PAN poly (A) tail [36].
The interaction between PAN RNA and ORF57 in B cells with KSHV lytic infection was then verified using CLIP/RT-PCR, by which PAN RNA was specifically pulled down with an anti-ORF57 antibody, but not an IgG control (Fig. 1A). Previous studies have demonstrated a role of ORF57 in PAN expression [14,33]. Consistent with these findings, we observed that PAN expression was increased during lytic KSHV induction in two B cell lines BCBL-1 (Fig. 1B, compare lane 1 to lane 2) and JSC-1 (Fig 1B, compare lane 5 to lane 6).
PAN expression depends on viral ORF57. (A) ORF57 binds PAN RNA during lytic KSHV infection. RT-PCR was performed by using a PAN-specific primer pair on total RNA isolated from the CLIP complexes obtained with an anti-ORF57 antibody from butyrate-treated JSC-1 cells. (B) PAN expression depends on ORF57 during lytic KSHV infection. Total RNA obtained from BCBL-1 and JSC-1 cells induced with butyrate (Bu, 3 mM) or from Bac36 cells with a wild-type KSHV genome (Bac36 wt) and Bac36-Δ57 cells with an ORF57-null KSHV genome [33] induced with VA (1 mM) was examined by Northern blot for PAN RNA with a PAN-specific probe oJM7. GAPDH RNA served as a loading control. (C) ORF57 enhances PAN accumulation at the posttranscriptional level. Total RNA from HEK293 cells transfected with an expression vector of PAN, K5 or vGPCR (see diagrams) or an empty pcDNA3 vector, together with a FLAG-tagged ORF57 (+) or an empty FLAG vector (-) was examined by Northern blot with a T7 probe for expression of PAN, K5 or vGPCR. (D) ORF57 inactive mutant (mtNLS2+3) is unable to enhance PAN expression. Total RNA from HEK293 cells transfected with a PAN expression vector together with a GFP-tagged ORF57 wt (ORF57 wild type), GFP-tagged ORF57 inactive mutant (mtNLS2+3) [13] or GFP empty vector pEGFP-N1 was examined by Northern blot for PAN RNA.
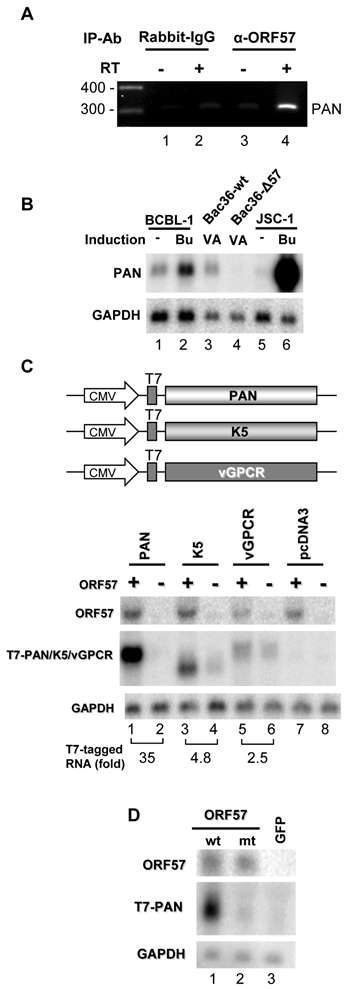
This increase of PAN expression during lytic KSHV infection, in the context of KSHV genome, is dependent on ORF57 expression. As shown in Fig. 1B, Bac36-wt cells with stable transfection of a wt KSHV genome responded to viral lytic induction by VA for PAN expression, but Bac36-Δ57 cells with stable transfection of an ORF57-null KSHV genome did not (compare lane 3 to lane 4). By cotransfection analyses of HEK293 cells, we further demonstrated that PAN was the most prominent responder (~35-fold increase) to ORF57 in the absence of other viral proteins when comparing with KSHV K5 and vGPCR (Fig. 1C), although expression of all three tested genes in the vector were driven by the same cytomegalovirus (CMV) immediate-early promoter. Both KSHV K5 and vGPCR did not interact with ORF57 in our CLIP assays and were previously shown as a non-responder to ORF57 [14]. Consistently, we found only a slight increase of K5 (~5 fold) and vGPCR (~3 fold) in the presence of ORF57 in our cotransfection assays (Fig. 1C). Together, these data suggest that ORF57 regulates PAN expression at the posttranscriptional level. When an ORF57 inactive mutant (a NLS2+3 mutant) [13] was included for comparison, increased PAN expression was found in the cotransfection with wt ORF57, but not with the mt ORF57 (Fig. 1D). Our data further indicate the specificity of ORF57 in posttranscriptional regulation of PAN expression.
An MRE at the 5' end of PAN is required for ORF57 to accumulate PAN
Given ORF57-specific association with PAN sequences that could be clustered at three major regions, a 5'-end region (nt 28667-28750), an internal region (nt 28971-29114), and a 3'-half region (nt 29544-29658) (Supplementary Material: Fig. S1), we constructed a series of PAN deletion mutants (Fig. 2A) and evaluated the relevance of these ORF57-binding sequences on PAN accumulation in the presence or absence of ORF57 by cotransfection of HEK293 cells. As shown in Fig. 2B, we demonstrated that wt PAN, Δ2 mt with deletion of the internal region, and Δ3b mt with partial deletion of the 3'-half region were all higher responders to ORF57, despite a minor reduction of PAN expression observed in two mutants in the presence of ORF57. Deletion of the 5'-end region was detrimental and greatly reduced the response of PAN to ORF57 function. Interestingly, deletion of the entire 3'-half region (Δ3 mt) including the ENE only had a small (~ 2 fold) effect on PAN to ORF57. From this analysis, we concluded that the 5'-end region is essential for PAN RNA accumulation and it contributed at least 25-fold enhancement of PAN expression in the presence of ORF57 (Figure 2B). The ENE is a much weaker element in response to ORF57 and the internal region examined in the presence of ORF57 did not play a significant role in PAN expression. As a result, we designated the 5'-end region of PAN as a functional MRE.
ORF57-mediated enhancement of PAN expression depends on a 5' PAN element. Shown in (A) are schematic diagrams of wt PAN and its deletion mutants. Shaded and numbered boxes represent the positions of ORF57 interacting regions identified by CLIP in PAN: box 1 (black), a PAN 5' region from nt 28667-28750; box 2 (dark grey), a PAN internal region from nt 28971-29114; box 3 (hatched and solid grey), a PAN 3' region from nt 29544-29658 including the entire ENE (hatched grey, nt 29554-29632). Total RNA from HEK293 cells transfected with each of the PAN constructs in the presence (+) or absence (-) of ORF57 was examined by Northern blot with a T7 probe for PAN expression (B). The same membrane in each panel was stripped and reprobed separately with a 32P-labeled ORF57 or GAPDH probe for ORF57 expression and sample loading. Relative levels of PAN RNA in each sample were shown at the bottom of each Northern blot panel after normalization to the level of the corresponding GAPDH for sample loading.
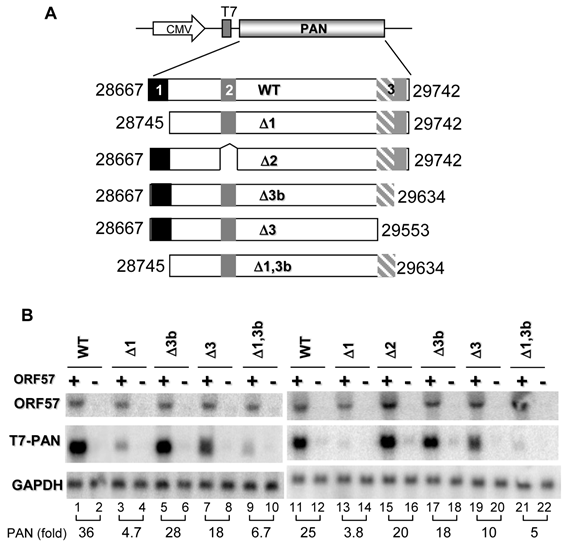
PAN stability depends on both ORF57 and MRE
To further analyze the function of the MRE, we first evaluated the stability of PAN with or without the identified MRE in the presence or absence of ORF57. After inhibiting Pol II transcription in HEK293 cells by treating with actinomycin D, we measured the remaining PAN RNA level over the time by RT-qPCR and the PAN RNA level was normalized to the corresponding GAPDH RNA level for sample loading. As shown in Fig. 3, PAN, under our experimental conditions, was relatively unstable, with a half-life of 3.87 h in the absence of ORF57. However, in the presence of ORF57, PAN became a significantly more stable transcript, with a half-life of 12.2 h, equating to a ~3-times longer half-life compared to its absence of ORF57. Interestingly, this significant (P=0.005) increase in decay rate of PAN by ORF57 was greatly decreased when the MRE was deleted from PAN. In the presence of ORF57, PANΔ1 with deletion of the MRE (Fig. 3) had a half-life of only 4.78 h, with the decay rates being significantly different than wt PAN under the same condition (P=0.042). The determined half-life (½ = 4.78 h) of the mt PAN in the presence of ORF57 was comparable with the half life (½ = 3.87 h) of wt PAN in the absence of ORF57, indicating that ORF57 enhances PAN expression by stabilizing PAN RNA through interaction with the MRE in the 5' region of PAN.
Stability of PAN depends on MRE and ORF57. (A) Calculation of half life of wt PAN and mt PANΔ1 lacking the 5' MRE in the presence or absence of ORF57. HEK293 cells were transfected with a vector expressing wt or mt PAN together with ORF57 or a vector control. 24 h after transfection, transcription was stopped by addition of 10 μg/ml of actinomycin D and total RNA was extracted from each sample collected over the time as indicated. PAN and GAPDH (for normalization) were quantified by qRT-PCR. The remaining level of PAN RNA in each time point relative to time zero was determined as described in Methods, after normalization to GAPDH RNA for sample loading. Results are presented as mean, minimum and maximum. A non-linear regression analysis on the raw data was performed, choosing an exponential decay model [Fold Percent = α*exp(β*time) ] where α (alpha) is the intercept when time = 0, and β (beta) is the decay rate. (B) Summary of the estimated betas (Est. β) and its standard errors (SE) for each study group. Also included are the adjusted R-squares and calculated half-life for each study group as well as two-tailed p-values for the indicated pair-wise comparisons between the estimated parameters. The p-values for the estimated betas were adjusted using a step-down Bonferroni method.
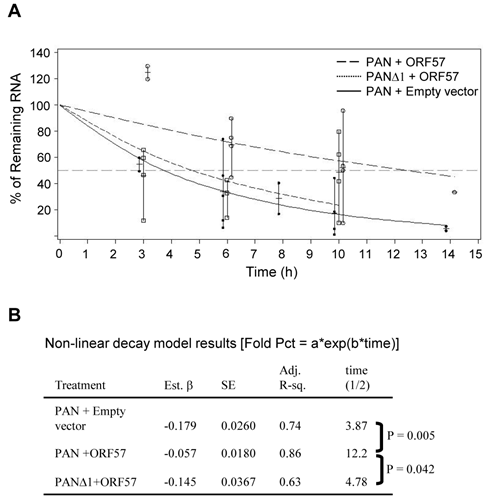
A 9-nt core in the MRE motif II (MRE-II) is necessary for ORF57-mediated PAN accumulation
By alignment of all MRE sequence tags identified by ORF57 CLIP assays (Supplementary Material: Fig. S1A) in combination with RNA folding analysis, we found that the 84-nt MRE is composed of three stem-loop structures which were designated as MRE-I, -II and III in the 5' to 3' order (See Fig. 4A for underlines and 4B for labels). Subsequently, we constructed a series of successive 5' to 3' deletions of the MRE into the MRE-I (ΔMRE-a), the MRE-I plus the MRE-II (ΔMRE-c) and an internal deletion of the entire MRE-II (ΔMRE-d) in the context of PAN (Fig. 4A). By cotransfection of HEK293 cells in the presence or absence of ORF57, as shown in Fig. 4C and 4D, wt PAN expression increased almost 30-fold by ORF57 when compared to its absence of ORF57, as expected. The expression of ΔMRE-a, with complete deletion of the MRE-I, also exhibited 34-fold increased PAN expression in the presence of ORF57 (Fig. 4C), whereas the expression of ΔMRE-c, with deletion of both MRE-I and entire MRE-II, showed a significant reduction in PAN expression when ORF57 was present (Fig. 4D). The expression level of PAN from the ΔMRE-c (Fig. 4D) was similar to that observed in the MREΔ1 mutant in the presence of ORF57 (Fig. 2). Collectively, these data indicate that the PAN MRE-II is the element through which ORF57 functionally modulates PAN expression. The importance of the MRE-II in ORF57-mediated PAN expression was further verified using an additional construct ΔMRE-d, containing an internal deletion of the entire MRE-II (Fig. 4A and 4D). The level of PAN ΔMRE-d expression observed was comparable to that found for ΔMRE-c.
As ORF57 appears to preferentially interact with a single-stranded loop structure in the vIL-6 MRE [27] and other targeted RNAs (our unpublished data), and the PAN MRE-II contains an 11-nt loop, we next introduced random point mutations into the MRE-II loop. The result was the creation of an MRE-PM (Fig. 4A and 4B), and we tested its expression in HEK293 cells along with wt PAN, ΔMRE-c and ΔMRE-d.The MRE-PM, with substitution of the 9 nts in the MRE-II loop, was impotent to respond to ORF57, when compared with wt PAN and had a similar expression level observed for both the ΔMRE-c and ΔMRE-d constructs (Fig. 4D). Our results indicate that the 9 nts (UAUGGAUUU) in the MRE-II loop function as an MRE core for ORF57 interaction in enhancing PAN expression.
To rule out any transcriptional effect caused by the CMV promoter in the tested PAN constructs, we replaced the CMV promoter from the CMV-PAN constructs with a native PAN promoter and evaluated ORF57 regulation of PAN expression from its native promoter in HEK293 cells (Fig. 4E, pJM53 for wt PAN and pJM54 for mt PAN [MRE-PM]). KSHV ORF50 (Rta) was used to transactivate PAN expression from its native promoter [30,47] in the presence or absence of ORF57. In the absence of KSHV Rta, the CMV promoter maintained the ability to drive PAN expression, whereas the native promoter did not (Fig. 4E, compare lane 3 to lane 1). Consistent with previous work [30,47], wt PAN, but not its mutant, could be transactivated in a dose-dependent manner from its native promoter by Rta in the absence of ORF57 (Fig. 4E, compare lane 4 to lanes 6, 8, 10, 12, 14, and 16). However, the expression of Rta-transactivated wt PAN, but not its mutant, from the native promoter could be remarkably increased in the presence of ORF57 (Fig. 4E, compare lanes 5 to 7, lanes 9 to 11, and lanes 13 to 15). Our data indicate that the identified 9-nt core in the MRE-II is indeed a functional core in the context of PAN and its native promoter. Moreover, Rta in a high dose (1000 ng) was found to transactivate the expression of wt PAN, but not the mt PAN, to a prominent level (Fig. 4E, compare lane 14 to lane 16) in the absence of ORF57, suggesting instability of the mt PAN lacking the 9-nt core in the MRE-II even when the PAN promoter is highly active. Together, we conclude that, regardless of the promoter used or its transcription efficiency, a functional MRE-core in PAN RNA is essential for stabilization of PAN RNA and for interaction with ORF57.
PAN MRE displays a limited function in the context of heterologous transcripts
To determine whether the PAN MRE can function in the context of a heterologous transcript, we examined PAN expression using the vGPCR construct, an ORF57 insensitive responder (Fig. 1C). We first introduced either the entire MRE or the MRE-II upstream of the vGPCR sequence (Fig. 5A) to mimic its native position in PAN and found, in cotransfected HEK293 cells, that the vGPCR, regardless of whether accompanied with the insertion of the MRE-II (sense or antisense [as]) or the entire MRE, remained to be insensitive to ORF57(Fig. 5B). A minimal increase (~3 fold) of vGPCR RNA expression in the presence of ORF57 was found in all vGPCR expression vectors with or without insertion of the MRE (compare pJM6 to pJM18) and was independent of the MRE-II orientation (compare pJM12 to pJM13).
Motif II (MRE-II) of the PAN 5' MRE is necessary for ORF57 enhancement of PAN expression. (A) Schematic diagrams of the 5' MRE and its mutants. Shown on the top indicates an 84-bp MRE in the 5' PAN which contains three potential motifs (stem-loops) marked as I, II and III, with the sequences underlined. A series of successive 5' to 3' deletions in the 5' MRE, an internal deletion or point mutations in the MRE-II are shown as ΔMRE-a, -c, -d and MRE-PM. Nucleotide substitutions in the MRE-II sequence in an MRE-PM mutant are boxed. (B) Predicted secondary structures of the MRE. On the left is the secondary structure of wt MRE with a folding energy of -19.5 Kcal/mol, consisting of 3 stem-loops I, II, III as described in (A). On the right show the point mutations introduced into the MRE-II loop to create an MRE-PM construct. (C, D) Mapping of a 9-nt core in the MRE-II responsible for ORF57 enhancement of PAN expression. Total RNA of HEK293 cells transfected separately with an indicated PAN construct in (A), together with an ORF57 expression vector (+) or an empty vector (-) was examined for PAN expression by Northen blot with a 32P-labeled T7 probe. (E) The MRE element in the presence of ORF57 promotes PAN accumulation in the context of its native promoter. The upper panel show schematic diagrams of PAN expression driven by a CMV promoter (pJM1) or by a native PAN promoter (PANPr) (pJM53 for wt PAN and pJM54 for mt PAN [MRE-PM]). HEK293 cells transfected with each of these constructs in the presence (+) or absence (-) of ORF57 were analyzed for PAN expression by Northern blot with a 32P-labeled PAN-specific probe oJM7 (Supplementary Material: Table S2). Relative PAN expression levels (fold) were quantified based on the density of each band after normalization to GAPDH for sample loading and were shown at the bottom of each blot, with the PAN level in the absence of ORF57 in (C, D) as 1 for each PAN construct and wt PAN level in the absence of both ORF57 and ORF50 in (E) as 1 for comparison.
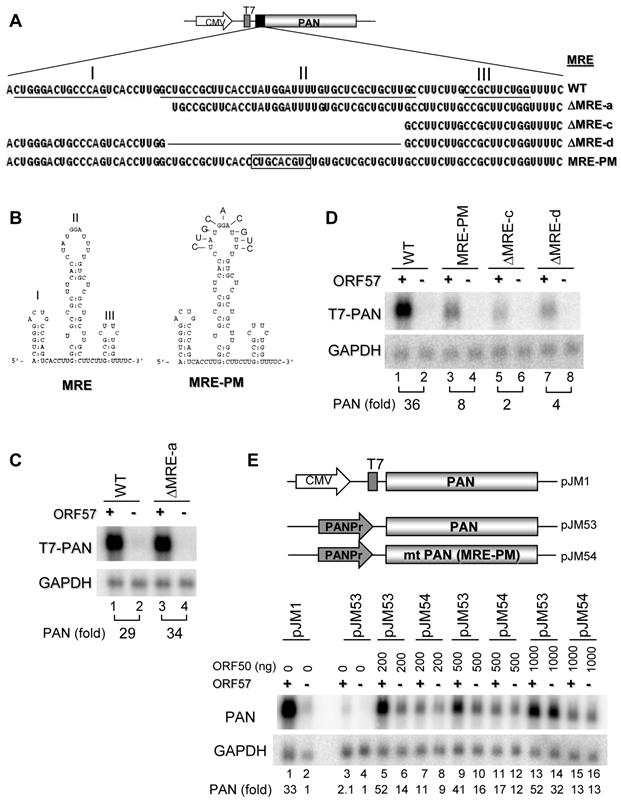
The identified PAN MRE does not function efficiently in the expression of heterologous genes vGPCR and Luciferase. (A) Schematic diagrams of the vGPCR constructs. The MRE-II motif or the entire MRE was inserted upstream of vGPCR (pJM6, Fig. 1C) either in the sense or antisense (As) orientation as indicated by the arrow. (B) Effect of the MRE on vGPCR expression in the presence or absence of ORF57. Total RNA of HEK293 cells transfected with each of the constructs in (A), together with an ORF57 expressing vector (ORF57 +) or an empty vector (-), was examined for vGPCR expression by Northern blot with a 32P-labeled T7 probe. Relative vGPCR expression level (fold) is shown at the bottom of Northern blots after normalization to GAPDH. (C) Schematic Diagrams of the firefly luciferase constructs. The entire MRE and/or ENE elements were inserted upstream (MRE) or downstream (ENE) of the firefly luciferase ORF, respectively, to mimic their native locations in PAN. (D) Effect of the MRE on luciferase expression in the presence or absence of ORF57. HEK293 cells were cotransfected with each firefly luciferase reporter construct, together with a Renilla luciferase pRL-TS construct [42] and a FLAG-tagged ORF57 expressing vector or an empty FLAG vector (FLAG). 24 h after transfection cells were lysated and luciferase activity was measured by a dual luciferase assay. Relative luciferase activity in each sample was calculated by dividing the light unit readings obtained from a tested firefly luciferase reporter construct by the light unit readings obtained from the Renilla luciferase reporter (F/R ratio). Luciferase activities (mean ± 95% confidence limits) from a mixed model analysis of variance are presented along with an adjusted two-tailed p-value.
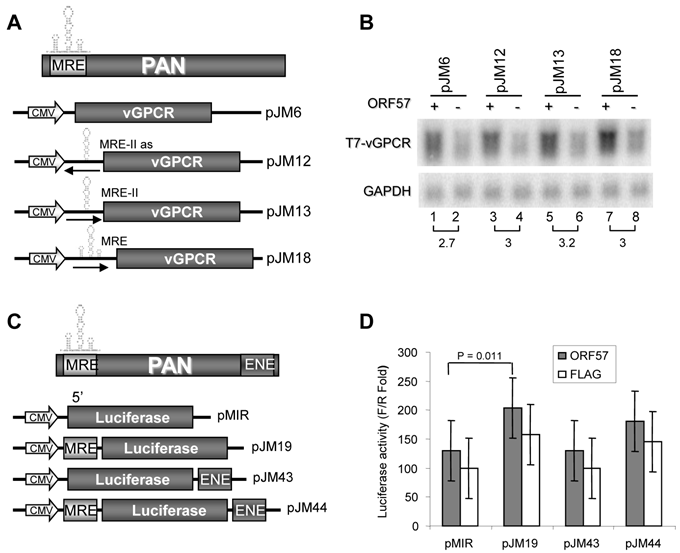
Given that MRE functions in PAN containing an ENE on the 3' end, we then examined the MRE action in the context of the PAN ENE in different luciferase reporters as diagramed in Fig. 5C. The first set (Set 1) of the expression vectors were derived from a pMIR-REPORT-Luciferase vector, with the MRE upstream and the ENE downstream of the firefly luciferase ORF. A Renilla luciferase for dual luciferase assays was expressed from a separate reporter, pRL-TS [42]. The second set (Set 2) of expression vectors not diagramed in Fig. 5C was derived from a psiCHECK2 vector expressing both Renilla luciferase and firefly luciferase from the same vector, with the MRE (sense for pJM47 or antisense for pJM48) upstream and/or the ENE (sense for pJM49 or antisense for pJM50) downstream of the Renilla luciferase ORF. When each of the set 1 reporters was transfected into HEK293 cells, together with a Renilla luciferase reporter pRL-TS and a FLAG-ORF57 or empty FLAG vector (Fig. 5D), the PAN-MRE upstream (pJM19), but not the PAN ENE downstream (pJM43), of the lucifease ORF was found to increase (P=0.011) luciferase activity only in the presence of ORF57. Under the same condition, no synergistic effect was seen for the MRE upstream and the ENE downstream of luciferase on the same vector (pJM44) (Fig. 5D). These data indicate that the PAN MRE could function weakly in a selected heterologous transcript in the presence of ORF57 and other sequence motif(s) in PAN other than the ENE are needed for the MRE to fully function. This interpretation was further verified in Dox-induced TREx BCBL1-Rta cells by transfection of the set 2 expression vectors (data not shown).
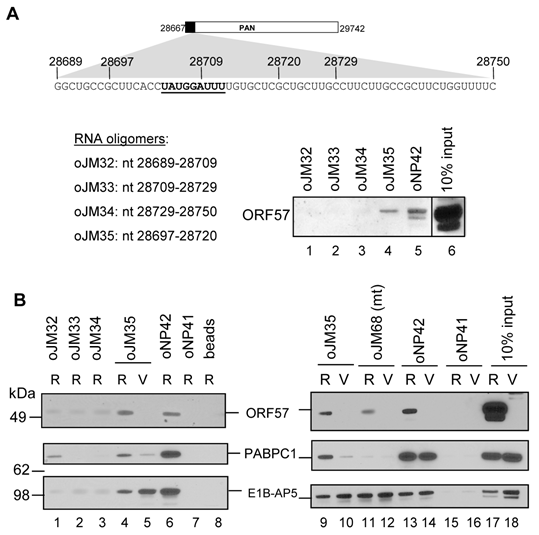
A 9-nt core in the MRE-II loop interacts with ORF57, PABPC1 and E1B-AP5
As shown in Fig. 4, PAN stability requires a functional MRE-II and ORF57. To provide direct biochemical evidence of the 9-nt core RNA interacting with ORF57 protein, we performed a biotinylated-RNA pull-down assay [27,43] using a series of biotinylated RNA oligomers covering the entire PAN MRE-II and MRE-III (oJM32, oJM33, oJM34, oJM35) (Fig. 6A). RNA oligomers from the MRE-I stem-loop were excluded since deletion of this stem-loop from PAN did not affect PAN accumulation in response to ORF57 (ΔMRE-a in Fig. 4A and 4C). When these RNA oligomers were compared in pulling down ORF57 expressed from Dox-induced TREx BCBL1-Rta cells, the oJM35 oligomer which covers the entire 9-nt core of MRE-II loop was found to efficiently bind ORF57 in a level comparable to an vIL-6 RNA oligomer oNP42, a positive ORF57-binding oligomer [27], but all other oligomers showed less binding affinity to ORF57 (Fig. 6A).
Subsequently, oJM35 was used to search for ORF57 protein partners capable of binding to the MRE. Proteins pulled down with oJM35 from total cell extract of Dox-induced TREx BCBL1-Rta cells was identified by Nano LC-MS/MS peptide sequencing technology. We found oJM35 interacting efficiently with cellular PABPC1 and a heterogeneous nuclear ribonucleoprotein E1B-AP5 (E1B 55-kilodalton-associated protein or hnRNPUL1). This could be further verified by RNA pulldown and Western blot analyses (Fig. 6B), showing that only oJM35 containing an intact 9-nt core in the MRE-II could interact efficiently with ORF57 and PABPC1 from TREx BCBL1-Rta cell extract (Fig. 6B, compare lane 4 to lane 5). Other RNA oligomers oJM32, oJM33 and oJM34 do not have a 9-nt core and showed less affinity to these three proteins (Fig. 6B, compare lanes 1-3 to lane 4). However, oJM35 appeared to interact with E1B-AP5 independently of ORF57 because it bound E1B-AP5 in TREx BCBL1-vector cells expressing no ORF57 (Fig. 6B, compare lane 4 to lane 5). A reduced level of E1B-AP5 interaction with oJM35 in the pulldowns with TREx BCBL1-Rta cell extract (R) than with TREx BCBL1-vector cell extract (V) (Fig. 6B, compare lanes 4 to 5 and lanes 9 to 10) suggests a reduced expression of E1B-AP5 in TREx BCBL1-Rta cells (Fig. 6B, compare lane 17 to 18 for the input). More importantly, we found that PABPC1, similar to ORF57, bound wt MRE-core RNA (oJM35), but not its mutant form (oJM68) (Fig. 6B, compare lines 4-5 and 9-10 to lanes 11-12), which is in contrast to E1B-AP5 binding both wt and mt RNA oligomers. The oJM68 has the same sequence as oJM35, but its 9-nt core UAUGGAUUU was changed to CUGCACGUC and this mutation in the context of PAN gene was sufficient to inactivate PAN expression in response to ORF57 (Fig. 4D and 4E). In addition, we found that PABPC1 binding to oJM35 took place more efficiently with the ORF57 expressing cell extract than with the ORF57 lacking one, but its binding to oNP42, a vIL-6 MRE-derived RNA oligomer [27], could be found for both cell total extracts independent of the ORF57 presence (Fig. 6B, compare lanes 4-5 and 9-10 to lanes 13-14), suggesting PABPC1 as an ORF57 partner protein specific in PAN MRE-II interaction.
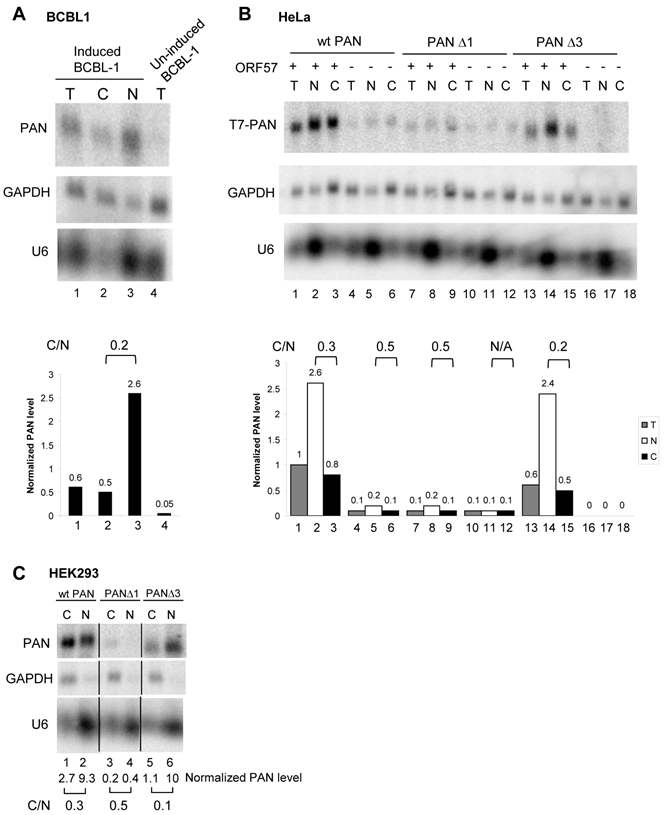
PAN RNA is exportable in the presence of ORF57
PAN was initially discovered as a nuclear non-coding RNA [28,32]. In chemical-induced BCBL1 cells with ORF57 expression, we found that PAN RNA was partially exported, resulting in ~20% of PAN RNA in the cytoplasm (Fig. 7A). This was specially true when PAN RNA was expressed in HeLa cells and HEK293 cells. By careful fractionation of HeLa cells and HEK293 cells with cotransfection of ORF57 and PAN as shown in Fig. 7B and 7C, a small fraction (~30%) of wt PAN were found in the cytoplasmic fraction of both HeLa (compare lane 2 to lane 3 in Fig. 7B) and HEK293 (compare lane 1 to lane 2 in Fig. 7C) cells in the presence of ORF57. The similar results were also obtained from both cell lines by using a PANΔ3 mutant (see Fig. 2A) with deletion of the entire 3'-half region including the ENE (compare lane 14 to lane 15 in Fig. 7B and lane 5 to lane 6 in Fig. 7C). Together, these data suggest a likelihood consequence of nuclear PAN accumulation in the presence of ORF57.
MRE-II RNA interacts in vitro with ORF57 and other cellular proteins. (A) Mapping of an MRE binding site for ORF57. Shown on the top is a PAN MRE sequence lacking the MRE-I region in Fig. 4A, with the 9-nt core of MRE-II loop sequence bolded and underlined and the nt positions of biotinylated RNA oligomers employed in RNA pull-down assays. A cell lysate prepared from TREx BCBL-1-Rta cells [39] induced with Dox for 24 h was used for the RNA pulldown assays with an indicated RNA oligomer. The RNA oligo oNP42 derived from vIL-6 RNA [27] which binds ORF57 was used as a positive control. ORF57 associated with RNA oligos in the pulldowns was immunoblotted using an anti-ORF57 antibody. The cell lysate (10%) before the pulldown was loaded as a Western blot control. (B) Biotinylated RNA affinity pulldown analysis by Western blot using anti-ORF57, PABPC1 and E1B-AP5 antibodies. Total cell extract from TREx BCBL-1-Rta (R) or -vector (V) cells induced by Dox for 24 h was used for the RNA pulldown assays with each biotinylated RNA oligomer. RNA oligos oNP41 and oNP42 derived from vIL-6 RNA [27] were used, respectively, as a negative and positive oligomer control for ORF57 interaction and 10% of the cell lysate from each cell line before the pulldown was loaded for Western blotting control. RNA oligo oJM68 is a copy of oJM35 with the point mutations in the MRE-II loop described in Fig. 4B. Control beads indicate no RNA oligomer on the beads.
A small proportion of PAN are exportable in ORF57-coexpressing cells. Total (T) and fractionated cytoplasmic (C) or nuclear (N) RNA from BCBL1 cells treated with or without valproate (VA, 1 mM) for viral lytic induction (A) or from HeLa cells (B) cotransfected with pJM1 (wt PAN), pJM2 (mt PANΔ1), or pJM4 (mt PANΔ3) in the presence (ORF57+) or absence (an empty vector control, ORF57-) of ORF57, were examined by Northern blot for PAN RNA expression. HEK293 cells cotransfected with pJM1 (wt PAN), pJM2 (mt PANΔ1), or pJM4 (mt PANΔ3) in the presence of ORF57 were also fractionated and examined by Northern blot for PAN RNA expression (C). GAPDH RNA served as a control for sample loading and U6 RNA served as a fractionation efficiency control. The relative levels of PAN in each sample were calculated after normalization to GAPDH.
Discussion
We previously developed a high-throughput and genome-wide CLIP assay as a powerful tool to study ORF57 protein-RNA interactions in living cells [27]. This CLIP assay allowed us to identify PAN RNA as the main target of ORF57. In this report we show that ORF57 binds to a 9-nt core sequence (UAUGGAUUU, KSHV nt 28704-28712) in the loop structure of the PAN MRE-II to promote PAN accumulation by increasing its half-life. Moreover, we found that the PAN MRE-II also interacts with PABPC1 and E1B-AP5. More specifically, the 9-nt core sequence in the PAN MRE-II was identified for the binding of PABPC1, but not for E1B-AP5 [48], suggesting that these two proteins by binding to different regions of the MRE-II may form a stable protein-RNA complex with ORF57 in accumulation of PAN RNA. Recently, we showed that ORF57 stabilizes ORF59 RNA by preventing aberrant ORF59 polyadenylation [26] and promotes the expression of vIL-6 and cellular IL-6 by preventing interactions of vIL-6 and cellular IL-6 RNAs with miR-1293 or miR-608, respectively [27]. In this report, we showed that another function of ORF57 is to prolong RNA half-life by binding to a 5' PAN MRE. Together with folding of all ORF57 CLIP sequences published in our previous report [27], these studies clearly indicate that ORF57 binding is RNA structure- and context-dependent and requires alternative proteins and different response elements to function.
Although our CLIP assays identified three regions of PAN RNA, including a 3' region encompassing the ENE previously reported in other studies [34-36], in association with ORF57 [27], this study demonstrated that only the 5' end of PAN operates as a functional MRE in ORF57-mediated PAN accumulation. Deletion of other putative MREs from the internal region or the 3' region of PAN, including the ENE, had only a minimal effect on ORF57-mediated PAN accumulation. When inserted into the 5' UTR region of an ORF57 non-responding viral transcript vGPCR, however, the 84-nt MRE identified from the PAN 5' end was unable to efficiently promote accumulation of the tested RNAs in the presence of ORF57. This indicates that other regions associated with ORF57, albeit not being important for ORF57 accumulation of PAN, might be needed to act as an auxiliary element to convert an ORF57 non-responder to an ORF57 responder. Moreover, further insertion of a 79-nt ENE to the 3' UTR of a luciferase transcript containing a PAN MRE in its 5' UTR did not provide additional effect to the PAN MRE on luciferase activity. Thus, the biological relevance of ORF57 binding to other PAN regions remains to be understood.
Consistent with our observations, a recent study also reported by deletion mutation analysis that ORF57 binds to a ~300-nt region (ORE) located on the 5' end of PAN RNA that was responsible for PAN accumulation [38]. By using a PAN-Δ79 (ΔENE) mutant, ORF57 was proven to protect the mutant from RNA decay. Whether ORF57 also affects wt PAN half-life was not included in the report [38]. As we stated in Fig. 2 that ORF57 also binds to the ENE region in our CLIP assays and deletion of this region decreased ORF57 effect on PAN accumulation only by 2-fold. In our study, both wt PAN and mt PANΔ1 lacking the MRE were examined and ORF57 was found to greatly prolong the wt PAN half-life from 3.87 h to 12.2 h, but was impotent for the mt PANΔ1 lacking the MRE-II (Fig. 3). Our results also differ from Sahin's report [38] on the roles of the identified MRE in heterologous reporters. We were unable to find the functional 89-nt MRE being greatly active in the 5' UTR region of vGPCR or luciferase RNA in response to ORF57. Although the ~300-nt ORE inserted to the β-globin 3' end was able to increase the β-globin response to ORF57 by 4-fold in Sahin's study [38], we noticed that an approximate 3-fold increase of a reporter gene expression in the presence of ORF57 is common almost for any testing system. An RNA transcript with this level increase in the presence of ORF57 was viewed as an ORF57 non-responsive transcript, such as vGPCR, K5 and luciferase when compared with PAN which showed a ~30-fold increase in the presence of ORF57 in this report. Alternatively, the difference may be due to a larger piece of the ORE insertion at the 3' end of β-globin in Sahin's study that put the testing ORE different from its native location in the PAN.
PABPC1 is a cytoplasmic protein involved in mRNA translation initiation and stability [49-52]. When shuttled to the nucleus, PABPC1 engages in nuclear RNA biogenesis [50,53,54]. In this study, we found that PABPC1 also binds to the 9-nt core of PAN MRE-II. E1B-AP5 or HNRNPUL1 (heterogeneous nuclear ribonucleoprotein U-like 1) has been implicated in the export of cellular mRNAs [48] by interacting with export protein TAP [55]. In addition, E1B-AP5 activates transcription [56], binds p53 [57] and participates in DNA damage signaling pathways [58]. We found that E1B-AP5 interacts with the PAN MRE-II motif outside of the 9-nt core in the MRE-II loop, indicating that E1B-AP5 binds to a different region in the MRE-II from PABPC1. Whether the binding of these two proteins to the PAN MRE-II is also important for ORF57 accumulation of PAN is under active investigation. Nevertheless, our data clearly suggest that PABPC1 and E1B-AP5 may modulate PAN expression in complex with ORF57.
Supplementary Material
Fig.S1: Alignments of the CLIP sequences to PAN in ORF57 CLIP assays. Table S1: The list of all plasmids constructed or used in this study. Table S2: The list of oligos used in this study.
Acknowledgements
This work was supported by the Intramural Research Program of the NCI, Center for Cancer Research, National Institutes of Health. We thank Dr. Michael Kruhlak for his critical reading of this manuscript, Dr. Jae Jung for providing doxycycline-inducible TREx BCBL1-Rta and -vector cell lines, Yan Yuan for KSHV ORF50 expression vector, Myriam Gorospe, Joan Steitz, and other members of the Zheng laboratory for their assistance and critical comments in the course of this study.
This work was presented at the 13th International Workshop on Kaposi sarcoma-associated herpesvirus and related agents, Aug. 29-Sept. 1, 2010, University of California, Los Angeles, California, USA.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865-1869
2. Viejo-Borbolla A, Ottinger M, Schulz TF. Human herpesvirus 8: biology and role in the pathogenesis of Kaposi's sarcoma and other AIDS-related malignancies. Curr HIV /AIDS Rep. 2004;1:5-11
3. Schulz TF. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J Gen Virol. 1998;79:1573-1591
4. Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186-1191
5. Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276-1280
6. Lagunoff M, Bechtel J, Venetsanakos E, Roy AM, Abbey N, Herndier B, McMahon M, Ganem D. De novo infection and serial transmission of Kaposi's sarcoma-associated herpesvirus in cultured endothelial cells. J Virol. 2002;76:2440-2448
7. Ganem D. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol. 2006;1:273-296
8. Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739-1744
9. Montaner S, Sodhi A, Molinolo A, Bugge TH, Sawai ET, He Y, Li Y, Ray PE, Gutkind JS. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi's sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell. 2003;3:23-36
10. Bais C, Van Geelen A, Eroles P, Mutlu A, Chiozzini C, Dias S, Silverstein RL, Rafii S, Mesri EA. Kaposi's sarcoma associated herpesvirus G protein-coupled receptor immortalizes human endothelial cells by activation of the VEGF receptor-2/ KDR. Cancer Cell. 2003;3:131-143
11. Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304-312
12. Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1998;95:10866-10871
13. Majerciak V, Yamanegi K, Nie SH, Zheng ZM. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J Biol Chem. 2006;281:28365-28378
14. Kirshner JR, Lukac DM, Chang J, Ganem D. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J Virol. 2000;74:3586-3597
15. Gupta AK, Ruvolo V, Patterson C, Swaminathan S. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J Virol. 2000;74:1038-1044
16. Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. Kaposi's Sarcoma-Associated Herpesvirus ORF57 Protein Enhances mRNA Accumulation Independently of Effects on Nuclear RNA Export. J Virol. 2007;81:9990-9998
17. Bello LJ, Davison AJ, Glenn MA, Whitehouse A, Rethmeier N, Schulz TF, Barklie CJ. The human herpesvirus-8 ORF 57 gene and its properties. J Gen Virol. 1999;80:3207-3215
18. Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. Kaposi sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes the expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol. 2008;82:2792-2801
19. Majerciak V, Kruhlak M, Dagur PK, McCoy JPJr, Zheng ZM. Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J Biol Chem. 2010;285:11297-11307
20. Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660-668
21. Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell. 2004;13:713-723
22. Lee YJ, Glaunsinger BA. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 2009;7:e1000107
23. Kumar GR, Glaunsinger BA. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol Cell Biol. 2010;30:4996-5008
24. Malik P, Blackbourn DJ, Clements JB. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J Biol Chem. 2004;279:33001-33011
25. Majerciak V, Deng M, Zheng ZM. Requirement of UAP56, URH49, RBM15, and OTT3 in the expression of Kaposi sarcoma-associated herpesvirus ORF57. Virology. 2010;407:206-212
26. Majerciak V, Uranishi H, Kruhlak M, Pilkington GR, Massimelli MJ, Bear J, Pavlakis GN, Felber BK, Zheng ZM. Kaposi's sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J Virol. 2011;85:1528-1540
27. Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. Kaposi's Sarcoma-Associated Herpesvirus ORF57 Promotes Escape of Viral and Human Interleukin-6 from MicroRNA-Mediated Suppression. J Virol. 2011;85:2620-2630
28. Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1996;93:11883-11888
29. Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci U S A. 1996;93:6641-6646
30. Song MJ, Brown HJ, Wu TT, Sun R. Transcription activation of polyadenylated nuclear rna by rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:3129-3140
31. Zheng ZM. Viral oncogenes, noncoding RNAs, and RNA splicing in human tumor viruses. Int J Biol Sci. 2010;6:730-755
32. Zhong W, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J Virol. 1997;71:1207-1212
33. Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J Virol. 2007;81:1062-1071
34. Conrad NK, Steitz JA. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24:1831-1841
35. Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943-953
36. Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc Natl Acad Sci U S A. 2007;104:10412-10417
37. Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244-1247
38. Sahin BB, Patel D, Conrad NK. Kaposi's sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog. 2010;6:e1000799
39. Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol. 2003;77:4205-4220
40. Yamanegi K, Tang S, Zheng ZM. Kaposi's sarcoma-associated herpesvirus K8beta is derived from a spliced intermediate of K8 pre-mRNA and antagonizes K8alpha (K-bZIP) to induce p21 and p53 and blocks K8alpha-CDK2 interaction. J Virol. 2005;79:14207-14221
41. Zheng ZM, Reid ES, Baker CC. Utilization of the bovine papillomavirus type 1 late-stage-specific nucleotide 3605 3' splice site is modulated by a novel exonic bipartite regulator but not by an intronic purine-rich element. J Virol. 2000;74:10612-10622
42. Tang S, Yamanegi K, Zheng ZM. Requirement of a 12-base-pair TATT-containing sequence and viral lytic DNA replication in activation of the Kaposi's sarcoma-associated herpesvirus K8.1 late promoter. J Virol. 2004;78:2609-2614
43. Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C, Baker CC, Zheng ZM. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol. 2009;83:167-180
44. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408
45. Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010;6:806-826
46. Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A. 1996;93:14862-14867
47. Song MJ, Li X, Brown HJ, Sun R. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 2002;76:5000-5013
48. Gabler S, Schutt H, Groitl P, Wolf H, Shenk T, Dobner T. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J Virol. 1998;72:7960-7971
49. Smith RW, Gray NK. Poly(A)-binding protein (PABP): a common viral target. Biochem J. 2010;426:1-12
50. Lemay JF, Lemieux C, St Andre O, Bachand F. Crossing the borders: poly(A)-binding proteins working on both sides of the fence. RNA Biol. 2010;7:291-295
51. Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010;29:4146-4160
52. Walters RW, Bradrick SS, Gromeier M. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA. 2010;16:239-250
53. Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015-13021
54. Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol Cell Biol. 2006;26:3085-3097
55. Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136-158
56. Kzhyshkowska J, Rusch A, Wolf H, Dobner T. Regulation of transcription by the heterogeneous nuclear ribonucleoprotein E1B-AP5 is mediated by complex formation with the novel bromodomain-containing protein BRD7. Biochem J. 2003;371:385-393
57. Barral PM, Rusch A, Turnell AS, Gallimore PH, Byrd PJ, Dobner T, Grand RJ. The interaction of the hnRNP family member E1B-AP5 with p53. FEBS Lett. 2005;579:2752-2758
58. Blackford AN, Bruton RK, Dirlik O, Stewart GS, Taylor AM, Dobner T, Grand RJ, Turnell AS. A role for E1B-AP5 in ATR signaling pathways during adenovirus infection. J Virol. 2008;82:7640-7652
Author contact
![]() Corresponding author: HIV and AIDS Malignancy Branch, National Cancer Institute, National Institutes of Health, 10 Center Dr. Rm. 6N106, Bethesda, MD 20892-1868. Phone: 301-594-1382. Fax: 301-480-8250. E-mail: zhengtnih.gov
Corresponding author: HIV and AIDS Malignancy Branch, National Cancer Institute, National Institutes of Health, 10 Center Dr. Rm. 6N106, Bethesda, MD 20892-1868. Phone: 301-594-1382. Fax: 301-480-8250. E-mail: zhengtnih.gov
Received 2011-10-3
Accepted 2011-10-15
Published 2011-10-16
