ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2012; 8(1):125-138. doi:10.7150/ijbs.8.125 This issue Cite
Research Paper
Molecular Cloning and Characterization of Ecdysone oxidase and 3-dehydroecdysone-3α-reductase Involved in the Ecdysone Inactivation Pathway of Silkworm, Bombyx mori
1. The Key Sericultural Laboratory of Agricultural Ministry, Southwest University, Chongqing, 400715, China.
2. The Institute of Agricultural and Life Sciences, Chongqing University, Chongqing 400044, China.
Abstract
Molting hormone (ecdysteroid) is one of the most important hormones in insects. The synthesis and inactivation of the ecdysteroid regulate the developmental process of insects. A major pathway of ecdysone inactivation is that ecdysone is converted to 3-dehydroecdysone, and then further to 3-epiecdysone in insects. Two enzymes (ecdysone oxidase: EO and 3DE-3α-reductase) participate in this pathway. In this study, based on the previously characterized cDNAs in Spodoptera littoralis, we cloned and characterized EO and 3DE-3α-reductase genes in the silkworm, Bombyx mori. The heterologously expressed proteins of the two genes in yeast showed the ecdysone oxidase and 3DE-3α-reductase activities, respectively. Expression of BmEO was only detected in the midgut at transcriptional and translational levels. We also localized EO within the midgut goblet cell cavities. For Bm3DE-3α-reductase gene, RT-PCR and western blot showed that it was expressed in the midgut and the Malpighian tubules. Moreover, we localized 3DE-3α-reductase within the midgut goblet cell cavities and the cytosol of principal cells of the Malpighian tubules. These two genes have similar expression profiles during different developmental stages. Both genes were highly expressed at the beginning of the 5th instar, and remained a relative low level during the feeding stage, and then were highly expressed at the wandering stage. All these results showed that the profiles of the two genes were well correlated with the ecdysteroid titer. The functional characterization of the enzymes participating in ecdysone inactivation in the silkworm provides hints for the artificial regulation of the silkworm development and biological control of pests.
Keywords: ecdysone, Bombyx mori, ecdysone oxidase, 3DE, 3DE-3α-reductase.
INTRODUCTION
For most insects, molting and metamorphosis are key stages in the developmental process. Many studies have shown that molting hormone (ecdysteroid) plays an important role in this process [1]. In immature stages of insects, the ecdysteroid titer displays the periodic and precise fluctuations which regulate the development of insects [2]. Besides the synthesis of the ecdysteroid, the degradation of the ecdysteroid is also important for the titer regulation. 3-Epimerization of ecdysteroid is one of the major inactivation pathways of the hormone in insects [3-4]. As shown in Supplementary Material: Figure S1, ecdysone is firstly converted to 3-dehydroecdysone (3DE), and then the 3-dehydroecdysone is converted to 3-epiecdysone [3-7]. Both the 3DE and the 3-epiecdysone have less activity than ecdysone [8]. As the intermediate of this pathway, 3DE has been detected in many insect species, especially in the Lepidopteran insects [5]. However, the proportion of 3DE is variable in different species. For instance, the prothoracic glands of Manduca sexta and Spodoptera littoralis have a predominant proportion of 3DE whereas Pieris rapae has a little 3DE [5]. In conclusion, previous studies have proven the 3-epimerization pathway to be important for ecdysone inactivation [8].
The conversion of active ecdysone to 3-epiecdysone, generally regarded as largely inactive, requires two consecutive enzymes (EO and 3DE-3α-reductase). These two enzymes have been found in the gut of M. sexta [9-10], and S. littoralis [11]. Then the genes which encode the two enzymes have been cloned and characterized in several insects. For example, the genes encoding EO and 3DE-3α-reductase in S. littoralis have been cloned and characterized in previous studies [12-13]. Moreover, EO was also identified in Drosophila melanogaster by the same researchers [14]. These previous studies showed that both genes were mainly expressed in the midgut, and the temporal expression profiles of them corresponded to the fluctuation of the ecdysone titer. For another related enzyme, 3DE-3β-reductase has also been found in different insects [5, 7, 15]. This enzyme can also catalyze reduction of 3DE using NAD(P)H, but produce ecdysone in the hemolymph [3, 5, 7, 9, 11]. Thus, the 3DE-3β-reductase enzyme may be involved in the biosynthetic pathway of ecdysteroids.
The silkworm, Bomyx mori, is the only thoroughly domesticated insect in the world to date. As the release of the whole genome sequence and functional genomics data [16-18], the silkworm functional genomics has made a great progress. For example, the gene underlying non-molting glossy (nm-g) which plays a crucial role in ecdysone synthesis was identified in the silkworm [19]. Although the silkworm has only a little amount of 3DE, the 3DE-3β-reductase activity has been found in several studies (Supplementary Material: Figure S1) [5, 20]. Recently, the 3DE-3β-reductase gene in the silkworm was cloned [21]; one gene believed to encode ecdysone oxidase was also cloned [22]. However, previous studies did not show the enzyme activity of the ecdysone oxidase. In addition, the putative silkworm ecdysone oxidase gene was found to be mainly expressed in the hemocytes [22], which was different from the other studies (Midgut is the predominantly expressed tissue for the EO gene from both S. littoralis and D. melanogaster) [13-14]. Therefore, whether the gene is the homologue of the ecdysone oxidase gene in the silkworm or not is still unknown. In this study, we identified the candidate genes that encode ecdysone oxidase and 3DE-3α-reductase in the silkworm by bioinformatics based largely on the characterized S. littoralis genes. Then, we cloned and sequenced the genes. Both proteins encoded by the two genes exhibit high sequence identities with the respective orthologous proteins from S. littoralis. The heterologous expressed proteins of the two genes also showed ecdysone oxidase and 3DE-3α-reductase activities, respectively. The spatial and temporal expression profiles were also determined and the temporal expression profiles were correlated with the ecdysone titer. The functional characterization of the enzymes participating in the 3-Epimerization of ecdysone in the silkworm provides hints for the artificial regulation of silkworm development and the biological control of pests.
MATERIALS AND METHODS
Silkworm strain
The DaZao strain of the silkworm was reared under normal conditions at 25°C.
cDNA cloning and rapid amplification of cDNA ends (RACE)
Three or four of Day 3 of the 5th instar silkworm larvae were collected and grinded in liquid nitrogen to powders and stored in Trizol reagent (Invitrogen, USA). Total RNA was extracted and treated with DNase I (Takara, Japan) to remove the genomic DNA contamination. The RNA was quantified by the ultraviolet spectrophotometer, and then 5 μg RNA was reverse-transcribed to the first strand of cDNA by the M-MLV Reverse Transcriptase Kit (Invitrogen, USA). Based on the bioinformatics analysis, specific primers were designed (Supplementary Material: Table S1) and used to amplify the DNA sequences of the putative BmEO and Bm3DE-3α-reductase genes. In order to obtain the full-length cDNA of the two genes, the GeneRacer kit (Invitrogen, USA) was used according to the manufacturer's instruction. The procedure was shown in previous analysis [23]. All the amplified products were purified with agrose gel purification kit (OMEGA, USA), and then cloned into PMD-19 vector (Takara, Japan). Orientation and presence of the inserted cDNA were confirmed by sequencing of the recombinant plasmid with automated DNA sequencer (Applied Biosystems 3730, Shanghai, China).
Phylogenetic analysis
In order to construct the phylogenetic tree for EO genes, the protein sequence of the candidate gene was used as the query sequence to search for the homologous sequences of other species in NCBI. Previous study showed that the EO protein belongs to the glucose-methanol-choline (GMC) oxidoreductases family [14]. Thus, several proteins which also belong to the GMC oxidoreductases family were downloaded from NCBI. The protein predicted as the ecdysone oxidase in a recent study was also used (Supplementary Material: Table S2, BmGMC2) [22]. All the sequences were aligned by the MUSCLE algorithm [24] and the alignment was corrected manually. The phylogenetic tree was constructed by the neighbor-joining (NJ) method implemented in the MEGA4 program [25]. The evolutionary distance was poisson-corrected. Gaps were pairwisely deleted. 1,000 iterations were used for calculating bootstrap values. 3DE-3α-reductase belongs to the short-chain dehydrogenases/reductases (SDR) superfamily [12]. Thus, some proteins belonging to the SDR superfamily were used for the phylogenetic analysis. In addition, several enzymes from prokaryotic and mammal which participate in the steroid metabolism were also included (Supplementary Material: Table S2).
Expression profile analysis of putative BmEO and Bm3DE-3α-reductase
The DaZao strain of the silkworm was used to survey the expression profiles of the two genes. For spatial expression analysis, 9 main tissues, head, integument, midgut, silk gland, hemocyte, fat body, Malpighian tubule, testis and ovary, were dissected from Day 3 of the fifth instar larvae, and also immediately frozen in liquid nitrogen, respectively. Every tissue sample was collected from three or four larvae. cDNA was synthesized as described above. The PCR was carried out as follows : 1 cycle of 94°C for 4 min and 28 cycles of 94°C for 45 s, 58 °C for 40 s, 72°C for 2 min and 1 cycle of 72°C for 10 min.
For temporal expression analysis, three larvae or pupae were collected at different developmental time points and then frozen immediately in liquid nitrogen. cDNA was synthesized as described above. In order to detect the relatively accurate pattern of temporal expression, the quantitative RT-PCR was performed using Real-Time PCR Detection System (CFX96, Bio-Rad, USA) with a SYBR Premix EX Taq kit (Takara, Japan). The PCR was carried out as follows: 30 s at 95°C, followed by 40 cycles of 5 s at 95°C, 1 min at 60°C. Eukaryotic translation initiation factor 4A (the silkworm microarray probe ID: sw22934), which has similar expression patterns at different developmental stages, was used as an internal control [26].
The primer sequences used in this part were shown in Supplementary Material: Table S1.
Expression of putative BmEO and Bm3DE-3α-reductase in E. coli
The cDNA sequences of putative BmEO and Bm3DE-3α-reductase were amplified by PCR. The forward and reverse primers contain one restriction enzyme site, respectively (BamH I and Not I). Then, the amplified fragments and the prokaryotic expression vector (pET28a: six His-tagged vector) were digested by the same restriction enzyme (BamH I and Not I), followed by ligation for 3h at 16°C. The recombinant plasmid DNAs (pET-BmEO and pET-Bm3DE-3α-reductase) were transformed into the E. coli BL21 (DE3) strain. The positive clone was cultured in 100 ml Luria-Bertani mediums. After 2h preincubation at 37°C, recombinant proteins with His tag were induced by 1mM IPTG for 4h. The induced cells were collected after centrifugation at 6,000×g for 10 min, and the pellets were washed and re-suspended in 20 ml lysis buffer (1× phosphate-buffered saline PBS PH 7.8). Then the cells were incubated on ice for 1h followed by sonication. The lysates were then centrifuged at 10,000×g for 30 min. The pellets were dissolved in 5 ml 1× PBS buffer (PH 7.8) with 8M urea and 10 mM imidazole at 4°C overnight. The solution was centrifuged at 12,000×g for 30 min. In order to purify the recombinant proteins, the supernatant was incubated with the Ni-NTA Superflow beads (Qiagen, Germany) according to the manufacturer's instruction. Proteins were quantified using the BCA protein assay and visualized after SDS-polyacrylamide gel electrophoresis (PAGE) by staining with Coomassie Blue.
Preparation of antiserum
Polyclonal antibodies against BmEO and Bm3DE-3α-reductase were produced by immunizing mice with purified recombinant proteins. Mice were immunized four times over 40 days. Each mouse was immunized by 100 ng recombinant proteins (in 1 ml of phosphate buffered saline) in complete Freund's adjuvant (1:1, v/v) on Day 0 and in incomplete Freund's adjuvant (1:1, v/v) on Day 10, 20, 30 with syringes. Then the antiserum on Day 40 was collected. The mice injected with PBS were used as negative control.
Expression of putative BmEO and Bm3DE-3α-reductase in Yeast
Because the proteins produced in the prokaryotic expression system showed no enzyme activities (data not shown), the eukaryotic expression system was used to detect the function of the two candidate proteins. The cDNA sequences of the putative BmEO and Bm3DE-3α-reductase genes were amplified using PCR, and the forward and reverse primers contain one restriction enzyme site, respectively: SnaB I and Not I. PCR products and yeast expression vector (pPIC9K) were double digested by SnaB I and Not I for 2h at 37°C, followed by ligation for 3h at 16 °C. The ligated plasmids were confirmed by sequencing. The ligated plasmids linearized by Sac I restriction enzyme, and then transformed into Pichia pastoris (X-33) cells by electroporation. The screening of positive clones (BmEO-pPIC9K-X33 and Bm3DE-3α-reductase-pPIC9K-X33) was performed following protocols recommended by Invitrogen. The obtained clone was cultured in buffered glycerol-complex medium (BMGY). Then 50 μl yeast cells were planted into 5 ml BMGY medium and cultured at 28 ℃ until the OD600 reached 2-6. The yeast cells were collected by centrifugatrion at 12,000×g, re-suspended by BMGY to OD600 reaching 1 and cultured continuously at 28 ℃ for 5 d. One per cent methanol was used to induce the expression of the genes at every 24h. The yeast cells containing empty pPIC9K vector were used as the negative control.
Enzyme assay and HPLC
Assays of the recombinant protein activities were performed by the method described in previous studies [13-14]. After induction by 1% methanol for five days, the yeast cells were removed by centrifugation at 12,000×g for 30 min at 4°C. The proteins in the supernatant were precipitated by 80 per cent saturated ammonium sulfate (SAS) overnight at 4°C, and then centrifuged at 10,000×g for 30 min at 4°C. The pellets were dissolved in 5 ml 200 mM Tris buffer (PH 7.5), followed by dialysis with 500 ml 75 mM Tris buffer (PH 7.5) for 24h at 4°C. The buffer used for dialysis was changed every 6h. In order to detect the activity of putative BmEO, 50 μl reaction, containing 20 μl crude recombinant enzyme solution, 50 μg ecdysone, and 30 μl sodium phosphate buffer (PH 7.5), was incubated for 1h at 37℃. Then the reaction was quenched with 50 μl methanol, and proteins were removed by centrifugation. The supernatant was stored at -20℃. Ecdysteroids in the supernatant were analyzed by reversed-phase HPLC using a C18 Nova-Pak cartridge (4.6 × 250 mm; Waters Associates) on a Waters instrument (Waters Associates) linked to a Dual λ Absorbance UV detector set at 254 nm. The mobile phase consisted of acetonitrile/0.1% (v/v) trifluoroacetic acid in water (22:78, v/v), and the elution rate was set at 1 ml/min. The standard sample of 3-DE which was kindly provided as a gift by Dr. R. Lafont (Universite Pierre et Marie Curie, Paris, France) was used to identify the product of the enzyme assay. As the 3DE is the main product of the EO assay, we separated the 3DE by the HPLC from the product. The purified 3DE was used as substrate in the Bm3DE 3α-reductase reaction. Fifty microlitre reaction, containing 20 μl crude recombinant enzyme solution, 10 μl the purified 3DE, 20 μl sodium phosphate buffer (PH 8.0), and 0.5 mM NAPDH, was incubated for 3h at 37℃[12]. After incubation, the reaction was quenched by 50 μl methanol, and proteins were removed by centrifugation. Reversed-phase HPLC was also used to identify the product of the Bm3DE 3α-reductase reaction as shown above.
Mass spectrometry
Mass spectrometry was performed to identify the product of the 3DE 3α-reductase enzyme assay. Mass spectrometry was conducted by a Finnigan TSQ Quantum Ultra AM spectrometer (Thermo Electron Corp., San Jose, CA) equipped with an ESI source in the positive ion mode. The spray voltage was fixed at 3.0 kV and the capillary temperature at 250°C. The collision energy was fixed at 15 eV. Nitrogen was used as the sheath (20 psi) and auxiliary (8 psi) gas.
SDS-PAGE and Immunobloting
The silkworm was reared until the Day 7 of the 5th instar. Midgut, hemocyte, Malpighian tubules, fat body, head and integument were dissected and homogenized in insect physiological saline. The samples were heated in boiling water for 5 min, followed by centrifugation at 10,000 × g for 10 min. The supernatant was collected for further analysis. BCA protein assay was used to quantify the proteins. Twenty microgram proteins were separated in 10% SDS-PAGE and transferred to PVDF membrane. Then, the membranes were blocked by 5% skim milk in TBST (Tris-butffered saline, PH 8.0, 0.1% Tween-20) for 1h at 37°C. To detect the target proteins, the membranes were incubated with the primary antibody at the relevant dilution (anti-BmEO antiserum, 1:5,000; anti-Bm3DE-3α-reductase, 1:7000) for 1h at 37°C, and then secondarily probed with horseradish peroxidase-labeled Rabbit Anti-Mouse Ig at a dilution of 1:15,000 (Sigma, USA). Bands were detected by ECL Plus Western Blotting Detection Reagents (GE Healthcare). The tubulin protein was used as the positive control in this analysis. The tubulin antibody was purchased from Sigma.
Immunohistochemistry
Sections of the Malpighian tubules and middle part of the midgut were dissected from the Day 7 of the 5th instar silkworm. The sections were fixed in 4% paraformaldehyde at room temperature for 3-4 h. They were then embedded in paraffin, and were cut to 5-6 μm thick sections. The sections were blocked by goat serum for 1h at 37°C. The primary antibodies were a dilution of 1:100 for both proteins, and the FITC-labeled Rabbit Anti-Mouse IgG(H+L) was used as a secondary antibody (Sigma, USA) at a dilution of 1:200. FITC displays green fluorescence under a blue light. The nuclei were labeled with 4',6-diamidino-2-phenylindole (DAPI). DAPI displays blue fluorescence under UV light. The sections were examined under an Olympus fluorescence microscope (BX51).
RESULTS
Sequence analysis
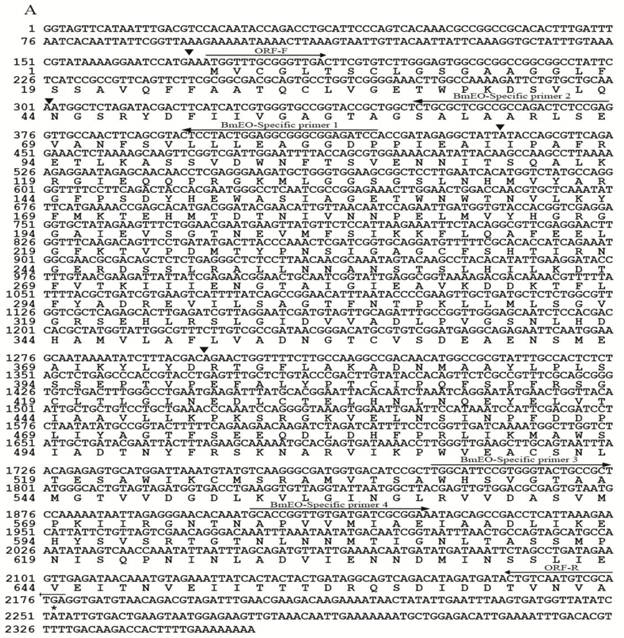
The protein sequence of EO (AY035784) in S. littoralis (SlEO) was downloaded from NCBI. The Blast program was used to search the silkworm genome database (SilkDB: http://silkworm.swu.edu.cn/silkdb/) [27]. One gene showed significant homology to S. littoralis ecdysone oxidase (putative BmEO). The putative BmEO was also found in the expressed sequence tags (ESTs) database (http://silkbase.ab.a.u-tokyo.ac.jp) (EST ID: fdpeP27_F_N19, fdpeP26_F_F03, fdpeP24_F_H08, fdpeP06_F_K03, bmov9m11) [16]. The BmEO candidate gene was located on the 17th chromosome. Based on the nucleotide sequence of the putative BmEO gene, the specific primers were designed. By PCR, cloning and sequencing, the putative BmEO sequence was obtained and confirmed. The putative BmEO cDNA contained an open reading frame of 2, 007 bp. The 5' RACE produced a 171 bp upstream-untranslated region and the 3' RACE produced a 175 bp downstream-untranslated region. Taken together, the complete length of putative BmEO gene was 2, 353 bp and the gene was interrupted by 4 introns (black triangles, Figure 1A). The putative BmEO cDNA encoded a protein of 668 amino acids with a predicted molecular mass of 72, 591.47 Da, and the value of isoelectric point (pI) was 4.97. The deduced amino acid sequence of the putative BmEO showed typical features of the glucose-methanol-choline (GMC) oxidoreductases family. The amino acid identity between putative BmEO and SlEO was 56%. Previous study showed that SlEO had 5 conserved ecdysone-binding residues (Leu96, Met346, Ala445, Leu534 and Trp537) [14]. The putative BmEO sequence had 4 of 5 conserved residues (boldfaces: Leu96, Met346, Ala446, Thr533 and Trp536). However, the silkworm ecdysone oxidase (BmGMC2) defined by previous analysis had only 1 of 5 conserved residues (boldfaces: Leu76, Ala323, Leu410, Ile500 and Phe503) (Supplementary Material: Figure S2A). It should be pointed out that the activity of BmGMC2 using recombinant enzyme has not been proven in their study [22]. Thus, it may not have EO activity. We performed phylogenetic analysis to determine the relationship between the putative BmEO and EOs from other insects (Supplementary Material: Table S1). All the proteins used in phylogenetic analysis belong to the GMC oxidoreductases family. The phylogenetic tree showed that the putative BmEO protein was firstly grouped with SlEO, and then clustered with other silkworm GMC proteins (BGIBMGA005703, BGIBMGA005608, BGIBMGA005609 and BGIBMGA014539). In conclusion, based on the conserved ecdysone-binding sites and the phylogenetic relationships, the putative BmEO in this study is the most likely candidate. Interestingly, the Drosophila EO was in another cluster with some GMC proteins from other insects (Supplementary Material: Figure S3A).
For the candidate gene of the silkworm 3DE-3α-reductase, the protein sequence of 3DE-3α-reductase (AF255341) in S. littoralis (Sl3DE-3α-reductase) was used as the query sequence to blast against the silkworm ESTs database (http://silkbase.ab.a.u-tokyo.ac.jp). Three best contigs were found (EST ID: rbmmt20d13, rbmov9o19, fmgV19n20f). By assembling these EST contigs, we obtained a relative complete gene with the start and stop codon (putative Bm3DE-3α-reductase). Then one fragment was amplified with specific primers by cloning and sequencing. The length of the putative Bm3DE-3α-reductase cDNA was 753 bp. A fragment of 24 bp upstream the 5' cDNA end was identified by 5'RACE, and a 74 bp downstream-untranslated fragment was obtained by 3'RACE. Thus, the complete length of putative Bm3DE-3α-reductase gene was 851 bp (Figure 1B). This gene had no intron. The resulting cDNA sequence encoded a protein of 250 amino acids with a predicted molecular mass of 26, 304.45 Da. The pI of the deduced protein was predicted to be 6.91. By searching the Pfam database, the putative Bm3DE-3α-reductase enzyme belongs to the short-chain dehydrogenases/reductases (SDR) superfamily. Multiple-alignment showed that the putative Bm3DE-3α-reductase had a high level of sequence identity with Sl3DE-3α-reductase (also 56%) (Supplementary Material: Figure S2B).
Nucleotide and deduced amino acid sequences of EO and 3DE-3α-reductase. (A) EO; (B) 3DE-3α-reductase. The cDNA sequence is indicated on the top line, and the deduced amino acid sequence is on the second line. For EO, black triangles represent the intron positions. The positions and directions of primers are indicated as arrows above the nucleotide sequences.
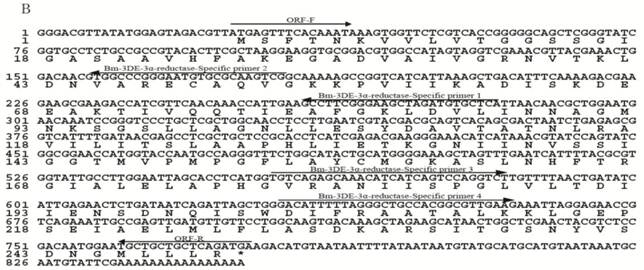
Heterologous expression of the BmEO and Bm3DE-3α-reductase gene in Yeast and Bacteria. (A) The SDS-PAGE analysis of the heterologous expression for the BmEO. (B) The SDS-PAGE analysis of the heterologous expression for the Bm3DE-3α-reductase. (C) The western blotting analysis of the heterologous expression for the BmEO. (D) The western blotting analysis of the heterologous expression for the Bm3DE-3α-reductase. Arrow represents the targeted protein. Lane 1: The supernatant of the pPIC9K-X33 induced by 1% methanol for 5 days. Lane 2: The supernatant of the yeast containing recombinant pPIC9K plasmid without 1% methanol. Lane 3: The supernatant of the yeast containing recombinant pPIC9K plasmid induced by 1% methanol for 5 days. Lane 4: Purified fusion proteins from E. coli with His-tag.
In order to perform the phylogenetic analysis, several proteins belonging to the SDR superfamily were downloaded from SilkDB and NCBI. One ecdysone synthesis related gene (Nm-g/Sro) which belongs to the SDR superfamily was also included in this analysis. In the phylogenetic tree, 3DE-3α-reductases from four insect species together with four silkworm copies formed one clade. This result showed that 3DE-3α-reductase is much conserved in insects. Interestingly, in this clade, the putative Bm3DE-3α-reductase was firstly grouped with another 3 silkworm proteins (BGIBMGA013271, BGIBMGA013270 and BGIBMGA013160) and then clustered with Sl3DE-3α-reductase (Supplementary Material: Figure S3B). Then the 3DE-3α-reductases clade was grouped with three 3α-hydroxysteroid dehydrogenases from prokaryotic. One Nm-g/Sro orthologous clade was conserved in four insect species (Nm-g/Sro clade), indicating its important function in insects. In addition, 3α-hydroxysteroid dehydrogenases (3α-HSDs) from mammals were also used in this analysis, and all these enzymes were clustered together.
The DNA sequences of two genes obtained in this study were deposited in GenBank (Genbank ID, BmEO: JF433972; Bm3DE-3α-reductase: JF433973).
Heterologous expression and enzyme assay
The putative BmEO and Bm3DE-3α-reductase genes were firstly expressed in the E. coli. Both recombinant proteins with His-tag were purified by the Ni-NTA Superflow beads (Qiagen) (Figure 2A, B, the 4th lane). Then the purified fusion proteins were injected into the mice to generate the specific antibodies, respectively. The purified proteins were used in western blotting analyses to determine the specificity of the antibodies. Both the analyses resulted in one specific band corresponding to molecular weights of 72 kDa (putative BmEO) and 26 kDa (putative Bm3DE-3α-reductase), respectively (Figure 2C, D, the 4th lane). Therefore, the antibodies were suitable to do further research.
Yeast cells containing recombinant pPIC9K plasmid and pPIC9K plasmid were induced by 1% methanol, respectively. After 5 days, the proteins in the supernatant were collected. From the SDS-PAGE, compared with the products of pPIC9K-X33 (Figure 2A, B, the 1st lane), there is no significant additional band corresponding to the molecular masses of 72 kDa and 26 kDa in the induced yeast containing recombinant pPIC9K (Figure 2A, B 3rd lane). But one specific band was detected for the products of BmEO-pPIC9K-X33 and Bm3DE-3α-reductase-pPIC9K-X33 in the western blotting analysis, respectively (Figure 2C, D 3rd lane). This result indicated that both the putative BmEO and Bm3DE-3α-reductase genes were expressed in the yeast.
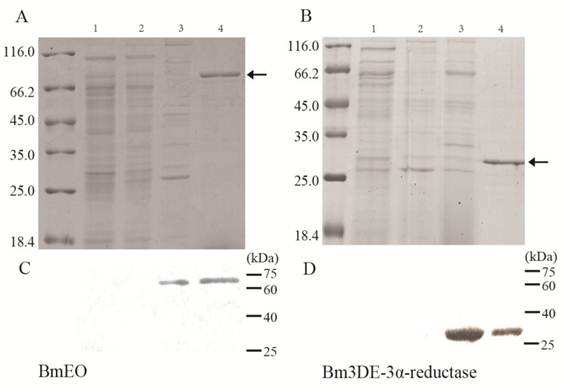
Because the low expression level of the putative BmEO and Bm3DE-3α-reductase genes in yeast, it is very difficult to purify the recombinant proteins. Thus, the crude recombinant proteins in yeast were used to identify their activities. As shown in the reversed-phase HPLC UV chromatograms, there was appreciable conversion of ecdysone into 3-dehydroecdysone from the product of the putative BmEO -pPIC9K-X33 (Figure 3B, C), with no detectable transformation in the case of the products of the control yeast pPIC9K-X33 (Figure 4A, C). Thus, our results clearly showed that the product of the putative BmEO-pPIC9K-X33 was functional in oxidizing ecdysone to 3-dehydroecdysone. The putative silkworm BmEO identified in this study is EO.
For the putative Bm3DE-3α-reductase, we firstly extracted the 3DE from the product of the BmEO-pPIC9K-X33 assay. Then we used the purified 3DE as the substrate to analyze the activity of the Bm3DE-3α-reductase produced in yeast. The results were shown in Figure 4D. Indeed, we can detect two apparent peaks. The second peak is the 3-dehydroecdysone according to elution time of the standard sample (Figure 3C, D). The first additional peak (FAP) is 3-epiecdysone according to several previous studies [12-14]. As shown in Supplementary Material: Figure S1, the molecular formula and the elemental composition of the ecdysone (E) and the 3-epiecdysone is the same, except the epimerization of one hydroxyl group on the C3 position. A previous study showed that the ecdysone and the 3-epiecdysone have similar mass spectra [28]. Thus, we used the mass spectrometry to further identify the product of the putative Bm3DE-3α-reductase enzyme assay, FAP (Figure 4D and Supplementary Material: Figure S3). Firstly, we extracted the first additional peak (FAP in Figure 4D) from the product of the 3DE-3α-reductase enzyme assay. Ecdysone and FAP were eluted at different time points (Supplementary Material: Figure S4A). Then we used mass spectrometry to analyze these two eluted products (E and FAP), respectively. The results showed that they have two main ion peaks at m/z 371 and m/z 445 (Supplementary Material: Figure S4B, C). As the similar mass spectra of ecdysone and 3-epiecdysone, the FAP should be the 3-epiecdysone. Thus, the putative silkworm Bm3DE-3α-reductase has the 3DE-3α-reductase activity.
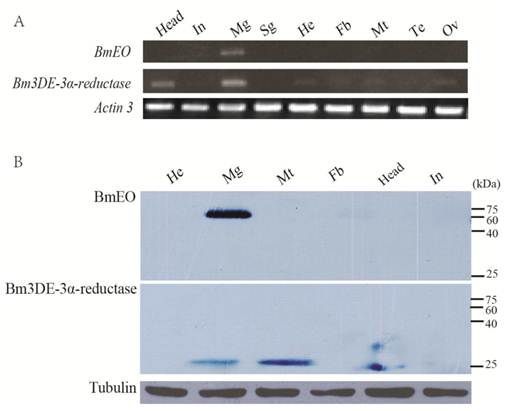
The expression level of BmEO and Bm3DE-3α-reductase in different tissues and developmental stages
The expression profiles of the two genes at the transcriptional and translational levels were examined (Figure 4). Nine main tissues from the Day 3 of the 5th instar silkworm larvae were used to examine the expression profiles of BmEO and Bm3DE-3α-reductase genes. BmEO gene was only expressed in the midgut of the tissues examined (Figure 4A). The expression signal of BmEO gene was not detected in other tissues such as head, integument, silk gland, hemocyte, fat body, Malpighian tubule and gonad (Figure 4A). This result was also confirmed by the western blotting analysis (Figure 4B). Bm3DE-3α-reductase was mainly expressed in the midgut and the head, and weak signals were also detected in the hemocyte, the fat body, the Malpighian tubule and the gonad (Figure 4A). However, the translational expression profile had a little difference from the transcriptional analysis. The Bm3DE-3α-reductase protein was mainly detected in the midgut and the Malpighian tubules, and the weak signal was also detected in the head, which is consistent with the mRNA analysis. But in other tissues, such as the hemocyte and the fat body which had mRNA signals, Bm3DE-3α-reductase protein was not expressed (Figure 4B).
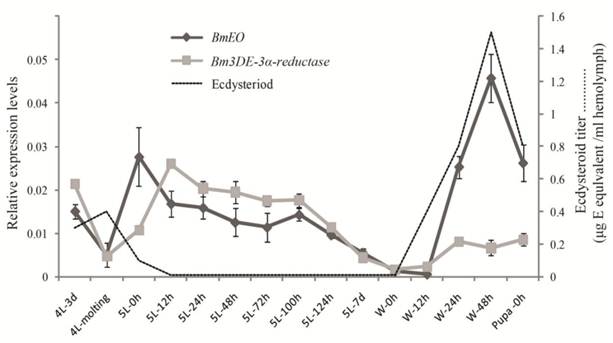
Then, the temporal expression profiles of the two genes were further examined from the 4th instar to the pupal stage by the quantitative RT-PCR (Figure 5). BmEO gene was detected in the late 4th instar, and disappeared during the 4th molting stage. Then BmEO was highly expressed at the early 5th instar (5L-0h), and then the transcriptional level of the gene decreased from the 12 h after the 5th instar to the beginning of the wandering stage (5L 12h-W 12h), with a sharp increase at the late wandering stage (W 24h-W 48h) (Figure 5). Overall, the expression level of BmEO correlates with changes in the hemolymph ecdysteroid titer during silkworm development [29-30]. For the Bm3DE-3α-reductase gene, the temporal expression pattern was similar to that of BmEO. A significant characteristic of this gene is that the expression level of the Bm3DE-3α-reductase gene increased following the BmEO gene in different developmental stages (Figure 5).
Enzymic activity of recombinant EO and 3DE-3α-reductase proteins. (A) the enzyme assay of the supernatant of the pPIC9K-X33 with the ecdysone. (B) the enzyme assay of the supernatant of the BmEO-pPIC9K-X33 with the ecdysone. (C) the standard sample of the 3DE. (D) the enzyme assay of the supernatant of the Bm3DE-3α-reductase-pPIC9K-X33 with the 3DE. 3DE means the 3-dehydroecdysone. FAP means the first additional peak.
Tissue distribution of BmEO and Bm3DE-3α-reductase. (A) The spatial expression profile at the transcriptional level. Total RNA of different tissues from the 3rd Day of the fifth instar larvae were used in the RT-PCR analysis. Silkworm Actin 3 gene was used as the control. (B) The spatial expression profile at the translational level. Western blotting analysis were performed to detect the expression of the protein. Total protein of different tissues from the 3rd Day of the fifth instar larvae were used in this analysis. Tubulin was used as the positive control. In: Integumen; Mg: Midgut; Sg: Silk gland; He: Hemocyte; Fb: Fat body; Mt: Malpighian tubule; Te: Testis; Ov: Ovary.
The temporal expression profiles of BmEO and Bm3DE-3α-reductase. The total RNA of silkworm from 4th instar larvae to the pupal stage were used in the quantitative RT-PCR analysis. 4L or 5L represents the 4th or 5th instar larvae. W represents the wandering stage. The data indicate means and S. E. M. of three samples after independent treatments. The dot line represents the developmental changes in hemolymph ecdysteroid titer based on the data as previously described [30].
Localization of BmEO and Bm3DE-3α-reductase
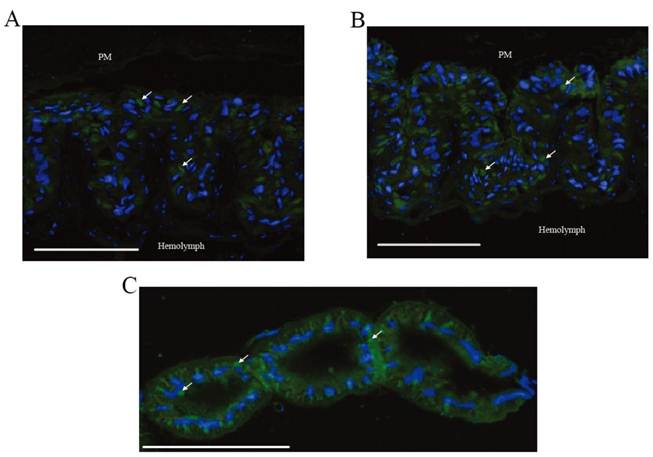
Immunohistochemical analysis was performed to investigate the location of BmEO and Bm3DE-3α-reductase. According to the results of western blots, BmEO was expressed in the midgut, and Bm3DE-3α-reductase was mainly detected in the midgut and the Malpighian tubules, thus only these two tissues were used to show the distribution of the two proteins in immunohistochemical analysis. In the midgut, strong signals for both proteins were detected (Figure 6A, B), and no signal was detected in the negative control (Supplementary Material: Figure S5). One previous study localized two kinds of proteins: soluble alkaline phosphatase (sALP) and membrane-bound alkaline phosphatase (mALP), and showed that the signals of sALP were mainly in the goblet cell cavities whereas the signals of mALP were in the apical microvilli of the midgut cells [31]. Compared with these results, we found that the locations of BmEO and Bm3DE-3α-reductase were very similar to that of sALP. In other words, both proteins were localized in the goblet cell cavities (white arrows). Consistent with the western blotting analysis, strong signals of Bm3DE-3α-reductase were also detected in the Malpighian tubules (Figure 6C). According to the cell type and cellular structure in the Malpighian tubules [32], we speculated that Bm3DE-3α-reductase was expressed in the cytosol of principal cells of the main segment (Figure 6C). However, whether this protein was expressed in the other sections of Malpighian tubules, such as stellate cells, apical, and basolateral membrane of tubule, cannot be identified in our results.
Localization of BmEO and Bm3DE-3α-reductase proteins. (A) Immunohistochemical analysis of the BmEO protein in midgut. (B) Immunohistochemical analysis of the Bm3DE-3α-reductase protein in midgut. (C) Immunohistochemical analysis of the Bm3DE-3α-reductase protein in Malpighian tubules. Arrows represents the localization of the protein; bar, 200 μm. PM represents the peritrophic membrane. Hemolymph represents the hemolymph side of the section. Blue fluorescence represents the nuclei. Green fluorescence represents the positive protein.
DISCUSSION
Ecdysone is one of the key hormones in insects. Periodic and precise fluctuations in the titer of ecdysone regulate development and metamorphosis of insects. 3-Epimerization of ecdysone is one of major inactivation pathways of the hormone [3-4]. As shown in Supplementary Material: Figure S1, ecdysone oxidase converts ecdysone into 3-dehydroecdysone, and then the 3-dehydroecdysone is converted into 3-epiecdysone by 3DE-3α-reductase.
In this study, we identified and cloned the complete cDNA of the putative BmEO and Bm3DE-3α-reductase gene. The BmEO candidate encodes an approximate 72 kDa protein that has 56% identity with SlEO. The protein had almost all 5 conserved ecdysone-binding residues as SlEO (boldfaces: Leu96, Met346, Ala446, Thr533 and Trp536). Moreover, the similarity between the Bm3DE-3α-reductase candidate and Sl-3DE-3α-reductase was also 56%. Both genes were heterologously expressed in the yeast expression system. The enzyme assay indicated that the supernatant from recombinant plasmid-transformed yeasts had ecdysone oxidase and 3DE-3α-reductase enzymatic activities, respectively (Figure 3). Thus, all these results indicated that both candidate genes were the silkworm EO and 3DE-3α-reductase genes, respectively. A recent study cloned another ecdysone oxidase gene (BmGMC2 in this study) [22]. The amino acid sequence of protein encoded by BmGMC2 only had 34% identity with SlEO, and only had 1 conserved ecdysone-binding residue as SlEO (boldfaces: Leu76, Ala323, Leu410, Ile500 and Phe503) (Supplementary Material: Figure S2A). In addition, BmGMC2 gene was mainly expressed in the hemocyte and lowly expressed in the midgut. The expression pattern of this gene is much different from previous researches [13-14]. Thus, whether this gene is also the homologue of the SlEO needs further research.
BmEO protein belongs to the GMC oxireductase family. One study showed that there are 15-23 GMC genes in four insect genomes (Drosophila melanogaster, Anopheles gambiae, Apis mellifera, and Tribolium castaneum), and there is a highly conserved cluster of 10 to 12 GMC genes located within the second intron of a non-GMC gene [33]. Recently, we annotated 43 GMC genes in the silkworm genome (unpublished data). The number of silkworm GMC genes was the largest in all insect genomes sequenced to date. Similar to a previous study, there was also a conserved GMC cluster which is located on the 16th chromosome in the silkworm [33]. As the Drosophila melanogaster EO (DmEO) was located in the GMC cluster, it was speculated that the genes in the cluster may correspond to the developmental process [33]. Interestingly, BmEO is outside of the GMC conserved cluster in the silkworm. The phylogenetic analysis also showed that the DmEO did not cluster with BmEO and SlEO (Supplementary Material: Figure S3A). In addition, Bm3DE-3α-reductase and another three silkworm proteins clustered in a group, and then clustered with Sl3DE-3α-reductase. The sequence identities between these three proteins and Bm3DE-3α-reductase are very high (~93%), and these three genes are tandem located in the same scaffold. These results indicated that the 3DE-3α-reductase gene might have experienced a recent duplication that generated three copies after the split of B. mori and S. littoralis. As shown above, Bm3DE-3α-reductase can convert the 3-dehydroecdysone into the 3-epiecdysone, and it can participate in the ecdysteriod metabolism in the silkworm. Howbeit, the function of another three proteins is still unknown. Whether they may also participate in ecdysteriod metabolism as does Bm3DE-3α-reductase remains to be further studied. The phylogentic tree of 3DE-3α-reductase also showed that insect 3DE-3α-reductases have a close relationship with prokaryotic 3α-hydroxysteroid dehydrogenases (3α-HSDs) which also belong to the SDR superfamily. However, insect 3DE-3β-reductases and mammalian 3α-HSDs belong to the aldo-keto reductase (AKR) superfamily, and all the enzymes clustered in another clade (Supplementary Material: Figure S3B). This suggested that 3DE-3α-reductase and 3DE-3β-reductase may have different originations although both can act on the same substrate (3DE).
In some previous studies, EO activity was predominantly detected in the midgut of some species during the late stage of the last larval instar, such as M. sexta [10], S. littoralis [13] and D. melanogaster [14]. Both the RT-PCR and the western blotting results also showed that the protein and mRNA of BmEO were highly expressed in the midgut, while almost not expressed in other tissues (Figure 4A, B). But in a recent study [22], the expression signal of the putative EO gene they identified was strongly detected in the hemocyte and almost not in the midgut. The Bm3DE-3α-reductase was also highly expressed in the midgut and Malpighian tubules, which is consistent with the observation in S. littoralis. Furthermore, the larval midgut was the only organ showing the 3DE-3α-reductase activity in another Lepidoptera, M. sexta [9-10, 34]. The reason for the various tissue distributions of the enzyme in different species is still unknown. The midgut, fat body and Malpighian tubule are the tissues that are most active in metabolizing ecdysteroids [34]. Thus, the 3-epimerization of ecdysone may play an important role in the reduction of the ecdysone in the midgut and Malpighian tubule.
For the temporal expression, high transcription of BmEO was detected at the beginning of the 5th instar (Figure 5). Bm3DE-3α-reductase also showed a similar expression pattern, while the titer of the ecdysteriod decreased during these periods [29-30]. In addition, expression levels of both genes remain a relative low plateau during the feeding stage (5L 12h-5L 124h) (Figure 5). These results suggested that BmEO and Bm3DE-3α-reductase may function in keeping low ecdysteriod level during the feeding stage. At the beginning of wandering stage, the ecdysteriod titer began to sharply increase until the titer reached its peak on the 48 h of the prepupal stage. During this period, expression levels of BmEO and Bm3DE-3α-reductase were also increasing, especially for the BmEO gene. The highest expression level of the BmEO gene also occurred at the 48h of the prepupal stage (Figure 5). In M. sexta, EO activity increased at a similar developmental stage as shown in the silkworm and reached the peak at the stage with the highest ecdysteroid titer [10]. These results indicate an increased capacity for molting hormone metabolism in the midgut at the time of the molting hormone titer peak (W-48h). Because declining of ecdysteroid titer is as necessary as its rise to insect metamorphosis [35-36], the high expression level of EO gene at W-48h would assure a rapid decline of the high ecdysteroid titer.
The rise in expression level of EO gene is correlated with the ecdysteriod titer, indicating that the EO gene is ecdysteriod responsive. Some previous studies have shown that the transcription level of EO gene may be induced by the ecdysone agonist, RH5849 or RH0345. Several binding motifs for ecdysone induced transcript factor (FTZ-F1 and Broad-complex) in the 5' flanking region of the gene may regulate the transcription of EO gene. In the silkworm, there are also some ecdysone related binding motifs, e.g. POU, Broad-complex and E74A in the 5' flanking region of the silkworm EO gene (Data not shown). Because of the important role in decreasing ecdysone titer, it is necessary to identify the cis-regulatroy elements that direct the EO expression in the future.
Immunohistochemical analyses were performed to localize the proteins (Figure 6 A, B, C). The expression signals of both BmEO and Bm3DE-3α-reductase were detected in the midgut goblet cell cavities. Although primary role of the midgut goblet cell is active transport of potassium from the hemolymph into the gut lumen, several digestive enzymes were also localized within it, such as trehalase that can hydrolyze insect blood sugar, trehalose, into glucose [37]. In addition, 3-epiecdysone is mainly present in the feces when ecdysone is injected into the silkworm [38]. Thus, we speculated that the ecdysone in the hemolymph is firstly transported into the goblet cells, and then metabolized by EO and 3DE-3α-reductase. Finally, the inactive 3-epiecdysone is excreted with the feces from the silkworm body.
Domestic silkworm is a very important economic insect for some developing countries. The silkworm rearing and the silk producing technologies are correlated with the developmental characters of the silkworm. For example, after the pupa turns into a moth, the moth will emerges from the cocoon by releasing proteolytic enzymes to make a hole in the cocoon, and this process may break down the silk fibers [39-40]. Thus, the cocoon is always baked to kill the pupa in the silk industry. However, baking the cocoon is a disadvantage to the quality of the silk. As shown in a previous study, an ecdysteriod peak occurs before pupal differentiation [29]. Therefore, degrading the ecdysone titer at this stage may artificially kill the pupa or delay its maturation. In this study, we cloned and identified the two genes encoding ecdysone oxidase and 3DE-3α-reductase that participate in ecdysone inactivation in the silkworm. In future, it may be possible to overexpress these two genes in the pupal stage to regulate the development of the silkworm by the transgenic technique. In addition, most of the Lepidopteran insects are a pest for humans. Artificial regulation of the ecdysone titer using the two genes may also be of benefit to the biological control of pests.
Supplementary Material
Figures S1-S5 and Tables S1-S2.
Acknowledgements
We would like to thank Prof. Rene Lafont (Universite Pierre et Marie Curie, Paris, France) for generously providing us the standard sample of the 3DE. This research was supported by a grant from National Science Foundation of China (No. 30970409), the Program of Introducing Talents of Discipline to Universities (B07045) and the Doctorial Innovation Fund of Southwest University (Kb2009019).
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Koolman J, Becker JL. Ecdysone. Thieme. 1989
2. Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1:453-465
3. Sakurai S, Warren JT, Gilbert LI. Mediation of ecdysone synthesis in Manduca sexta by a hemolymph enzyme. Arch Insect Biochem. 1989;10:179-197
4. Warren JT, Sakurai S, Rountree DB. et al. Regulation of the ecdysteroid titer of Manduca sexta: reappraisal of the role of the prothoracic glands. Proc Natl Acad Sci U S A. 1988;85:958-962
5. Kiriishi S, Rountree D, Sakurai S. et al. Prothoracic gland synthesis of 3-dehydroecdysone and its hemolymph 3β-reductase mediated conversion to ecdysone in representative insects. Cell Mol Life Sci. 1990;46:716-721
6. Blais C, Lafont R. Biosynthesis of ecdysteroids by prothoracic glands in Pieris brassicae (Lepidoptera insect). Conversion in vitro of radiolabelled precursor of 3-dehydroecdysone. Comptes rendus de l'Academie des sciences. Serie III, Sciences de la vie. 1991;313:359
7. Chen JH, Webb TJ, Powls R. et al. Purification and characterisation of haemolymph 3-dehydroecdysone 3β-reductase in relation to ecdysteroid biosynthesis in the cotton leafworm Spodoptera littoralis. Eur J Biochem. 1996;242:394-401
8. Rees H. Ecdysteroid biosynthesis and inactivation in relation to function. Eur J Entomol. 1995;92:9-9
9. Weirich GF, Thompson MJ, Svoboda JA. Ecdysone oxidase and 3-oxoecdysteroid reductases in Manduca sexta midgut: Kinetic parameters. Arch Insect Biochem. 1989;12:201-218
10. Weirich GF, Feldlaufer MF, Svoboda JA. Ecdysone oxidase and 3-oxoecdysteroid reductases in Manduca sexta: Developmental changes and tissue distribution. Arch Insect Biochem. 1993;23:199-211
11. Milner N, Rees H. Involvement of 3-dehydroecdysone in the 3-epimerization of ecdysone. Biochem J. 1985;231:369-374
12. Takeuchi H, Chen JH, O'Reilly DR. et al. Regulation of ecdysteroid signalling: molecular cloning, characterization and expression of 3-dehydroecdysone 3 alpha-reductase, a novel eukaryotic member of the short-chain dehydrogenases/reductases superfamily from the cotton leafworm, Spodoptera littoralis. Biochem J. 2000;349:239-245
13. Takeuchi H, Chen JH, O'Reilly DR. et al. Regulation of ecdysteroid signaling: cloning and characterization of Ecdysone Oxidase: A novel steroid oxidase from the cotton leafworm, Spodoptera littoralis. J Biol Chem. 2001;276:26819-26828
14. Takeuchi H, Rigden DJ, Ebrahimi B. et al. Regulation of ecdysteroid signalling during Drosophila development: identification, characterization and modelling of ecdysone oxidase, an enzyme involved in control of ligand concentration. Biochem J. 2005;389:637-645
15. Watson R, Spaziani E, Bollenbacher W. Regulation of ecdysone biosynthesis in insects and crustaceans: a comparison. In: (ed.) Koolman J. Ecdysone, from chemistry to mode of action. Thieme: Stuttgart. 1989:188-203
16. Mita K, Kasahara M, Sasaki S. et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27-35
17. Xia Q, Zhou Z, Lu C. et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. 2004;306:1937-1940
18. Xia Q, Cheng D, Duan J. et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007;8:R162
19. Niwa R, Namiki T, Ito K. et al. Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the "Black Box" of the ecdysteroid biosynthesis pathway. Development. 2010;137:1991-1999
20. Nomura Y, Komatsuzaki M, Iwami M. et al. Purification and characterization of hemolymph 3-dehydroecdysteroid 3β-reductase of the silkworm, Bombyx mori. Insect Biochem Mol Biol. 1996;26:249-257
21. Yang H, Liu X, Zhou F. et al. Cloning and expression pattern of 3-dehydroecdysone 3β-reductase (3DE 3β-reductase) from the silkworm, Bombyx mori L. Arch Insect Biochem. 2010;76:55-66
22. Yang HJ, Wang MX, Zhang P. et al. Cloning and characterization of the Bombyx mori ecdysone oxidase. Arch Insect Biochem. 2011;78:17-29
23. Ayumi T, Hiroaki Y, Jun Y. The MC1R gene in the guppy (Poecilia reticulata): Genotypic and phenotypic polymorphisms. BMC Res Note. 2011;4:31
24. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792-1797
25. Tamura K, Dudley J, Nei M. et al. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-1599
26. Wang GH, Xia QY, Cheng DJ. et al. Reference genes identified in the silkworm Bombyx mori during metamorphism based on oligonucleotide microarray and confirmed by qRT PCR. Insect Sci. 2008;15:405-413
27. Duan J, Li R, Cheng D. et al. SilkDB v2. 0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010;38:D453
28. Thompson MJ, Kaplanis JN, Robbins WE. et al. 3-Epi-20-hydroxyecdysone from meconium of the tobacco hornworm. Steroids. 1974;24:359-366
29. Mizoguchi A, Dedos SG, Fugo H. et al. Basic pattern of fluctuation in hemolymph PTTH titers during larval-pupal and pupal-adult development of the silkworm, Bombyx mori. Gen Comp Endo. 2002;127:181-189
30. Kiguchi K, Agui N. Ecdysteroid levels and developmental events during larval moulting in the silkworm, Bombyx mori. J Insect Physiol. 1981;27:805-812
31. Daimon T, Taguchi T, Meng Y. et al. β-Fructofuranosidase Genes of the Silkworm, Bombyx mori. J Biol Chem. 2008;283:15271-15279
32. Beyenbach KW, Skaer H, Dow JAT. The developmental, molecular, and transport biology of Malpighian tubules. Annu Rev Entomol. 2010;55:351-374
33. Iida K, Cox-Foster DL, Yang X. et al. Expansion and evolution of insect GMC oxidoreductases. BMC Evol Biol. 2007;7:75
34. Lafont R, Dauphin-Villemant C, Warren J. et al. Ecdysteroid chemistry and biochemistry. Comp Mol Insect Sci. 2005;3:125-195
35. Apple R, Fristrom JW. 20-Hydroxyecdysone is required for, and negatively regulates, transcription of Drosophila pupal cuticle protein genes. Dev Biol. 1991;146:569-582
36. Guittard E, Blais C, Maria A. et al. CYP18aA1, a key enzyme of Drosophila steroid hormone inactivation, is essential for metamorphosis. Dev Biol. 2010;349:35-45
37. Mitsumasu K, Azuma M, Niimi T. et al. Membrane-penetrating trehalase from silkworm Bombyx mori. Molecular cloning and localization in larval midgut. Insect Mol Biol. 2005;14:501-508
38. Zhang M, Kubo I. Metabolic fate of ecdysteroids in larval Bombyx mori and Heliothis virescens. Insect Biochem Mol Biol. 1993;23:831-843
39. Al-Khateeb T, Nusair Y. Effect of the proteolytic enzyme serrapeptase on swelling, pain and trismus after surgical extraction of mandibular third molars. Int J Oral Max Surg. 2008;37:264-268
40. Mecikoglu M, Saygi B, Yildirim Y. et al. The effect of proteolytic enzyme serratiopeptidase in the treatment of experimental implant-related infection. J Bone Joint Surg. 2006;88:1208-1214
Author contact
![]() Corresponding author: Dr. Ze Zhang, The Key Sericultural Laboratory of Agricultural Ministry, Southwest University, Chongqing 400715, China. Tel: +86-23-68251029; Fax: +86-23-68251128; Email: zezhangedu.cn; http://silkworm.swu.edu.cn/zezhang/
Corresponding author: Dr. Ze Zhang, The Key Sericultural Laboratory of Agricultural Ministry, Southwest University, Chongqing 400715, China. Tel: +86-23-68251029; Fax: +86-23-68251128; Email: zezhangedu.cn; http://silkworm.swu.edu.cn/zezhang/
Received 2011-9-19
Accepted 2011-11-30
Published 2011-12-8
