ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2016; 12(11):1319-1331. doi:10.7150/ijbs.14464 This issue Cite
Research Paper
RNA interference of chitin synthase genes inhibits chitin biosynthesis and affects larval performance in Leptinotarsa decemlineata (Say)
1. Education Ministry Key Laboratory of Integrated Management of Crop Diseases and Pests, College of Plant Protection, Nanjing Agricultural University, Nanjing 210095, China.
2. Department of Plant Protection, Xinjiang Academy of Agricultural Sciences
Urumqi 830091, China.
Abstract
Dietary introduction of bacterially expressed double-stranded RNA (dsRNA) has great potential for management of Leptinotarsa decemlineata. Identification of the most attractive candidate genes for RNA interference (RNAi) is the first step. In the present paper, three complete chitin synthase cDNA sequences (LdChSAa, LdChSAb and LdChSB) were cloned. LdChSAa and LdChSAb, two splicing variants of LdChSA gene, were highly expressed in ectodermally-derived epidermal cells forming epidermis, trachea, foregut and hindgut, whereas LdChSB was mainly transcribed in midgut cells. Feeding bacterially expressed dsChSA (derived from a common fragment of LdChSAa and LdChSAb), dsChSAa, dsChSAb and dsChSB in the second- and fourth-instar larvae specifically knocked down their target mRNAs. RNAi of LdChSAa+LdChSAb and LdChSAa lowered chitin contents in whole body and integument samples, and thinned tracheal taenidia. The resulting larvae failed to ecdyse, pupate, or emerge as adults. Comparably, knockdown of LdChSAb mainly affected pupal-adult molting. The LdChSAb RNAi pupae did not completely shed the old larval exuviae, which caused failure of adult emergence. In contrast, silencing of LdChSB significantly reduced foliage consumption, decreased chitin content in midgut sample, damaged midgut peritrophic matrix, and retarded larval growth. As a result, the development of the LdChSB RNAi hypomorphs was arrested. Our data reveal that these LdChSs are among the effective candidate genes for an RNAi-based control strategy against L. decemlineata.
Keywords: Leptinotarsa decemlineata, chitin synthase, RNA interference, control
Introduction
The occurrence of the Colorado potato beetle Leptinotarsa decemlineata (Say), a notorious defoliator of potato, often causes serious yield loss to potato production in most major potato-growing areas of the world. Chemical control of this beetle has led to development of resistance, resurgence of non-target pests, contamination of food and environment, and destruction of beneficial insects like honeybees, pollinators, parasites and predators [1, 2]. Therefore, it is necessory to explore novel strategies to efficiently control the damage of L. decemlineata.
Among the potential control strategies, RNA interference (RNAi), an RNA-dependent gene silencing process induced by double-stranded RNA (dsRNA), is at the developmental stage [3, 4]. Up to now, three methods are being developed for economical production of dsRNA: expression in plants, chemical synthesis and production in bacteria and other microorganisms. Expression in transgenic plants might work well for commercial crops. In contrast, either a bacterially expressed or a chemically synthesized dsRNA may be a better choice for pest on crop that is directly consumed by humans, such as potato [4]. In L. decemlineata, dietary introduction of bacterially expressed dsRNAs is able to effectively knock down target genes [4], demonstrating its great potential for management of L. decemlineata. The first step towards the development of control strategy is to identify the most attractive candidate genes for RNAi.
Chitin synthase (ChS, EC 2.4.1.16) catalyzes the final biosynthesis step of chitin [5, 6], a linear polysaccharide that is widely distributed in the cuticle of epidermal cells, the linings of trachea, salivary gland, foregut and hindgut, and the peritrophic matrix (PM) in midgut [5]. In general, insect ChSs have been segregated into A and B classes [6-12]. Moreover, in most insect species ChSA contains alternative exons which lead to the production of two splicing mRNA variants, ChSAa and ChSAb [7, 10, 12-17]. Functional analyses using RNAi in different insect species show that ChSs are required for survival, ecdysis, oviposition and egg hatching [7, 8, 17-24]. Furthermore, chitin is absent in plants and vertebrates. All these results indicate that ChS provides an important target for a potential pest control strategy through RNAi [18, 25, 26].
We have identified two ChS genes (LdChSA and LdChSB) in L. decemlineata. Moreover, LdChSA contains two splicing variants, LdChSAa and LdChSAb [26]. In the present paper, we found that RNAi of both LdChSA isoforms, or each of LdChSAa, LdChSAb or LdChSB severely affected larval growth, caused larval lethality, and impaired larval-larval molting, larval-pupal ecdysis and adult emergence. Our results imply that LdChS genes serve as potential targets for a dsRNA-based control method in L. decemlineata.
Materials and methods
Insects
L. decemlineata larvae and adults were kept in an insectary as previously described [27], and were supplied with potato foliage at vegetative growth or young tuber stages in order to assure sufficient nutrition. At this feeding protocol, L. decemlineata larvae progressed through four distinct instars, with the approximate periods of 2, 2, 2 and 4 days, respectively. Upon reaching full size, the fourth-instar larvae stopped feeding, dropped to the ground, burrowed to the soil and entered the prepupae stage. The prepupae spent an approximately 4 days, and then pupated. The pupae developed in about 6 days and the adults emerged. The adults spent an average of 7 days to become sexually mature.
Cloning of the three LdChS cDNAs
A TBLASTN search of the L. decemlineata transcriptome and genome data (https://www.hgsc.bcm.edu/arthropods/colorado-potato-beetle-genome-project) was carried out using the amino acid sequences of Tribolium castaneum ChSs as the queries. This resulted in the identification of three cDNAs that we have named as LdChSAa, LdChSAb and LdChSB. A second set of searches was done using the amino acid sequences of the three L. decemlineata ChSs as queries in an attempt to identify additional genes encoding ChS-related proteins.
The correctness of the putative ChS sequences was substantiated by polymerase chain reaction (PCR) using primers in Table S1. This was followed by 5'- and 3'-RACE to complete these sequences, with SMARTer RACE cDNA amplification kit (Takara Bio., Dalian, China) and SMARTer RACE kit (Takara Bio.). The antisense/sense gene-specific and the nested primers corresponding to the 5'-end and 3'-end of the sequences were listed in Table S1. After obtaining the full-length cDNAs, primer pairs (Table S1) were designed to verify the complete open reading frames. The full-length LdChS cDNAs were submitted to GenBank (LdChSAa, KT964740; LdChSAb, KT964741; LdChSB, KT964742).
Preparation of bacterially expressed dsRNA
The same method as previously described [27] was used to express dsChSA-1, dsChSA-2, dsChSAa, dsChSAb, dsChSB-1, dsChSB-2 and dsegfp derived from a 307 and a 398 bp common fragments in both LdChSAa and LdChSAb, a 89 bp fragment of LdChSAa, a 153 bp fragment of LdChSAb, a 337 bp and a 500 bp fragments of LdChSB, and a 414 bp fragment of enhanced green fluorescent protein (egfp) gene (Figure S1). The seven dsRNAs were individually expressed with specific primers in Table S1, using Escherichia coli HT115 (DE3) competent cells lacking RNase III. Individual colonies were inoculated and grown until cultures reached an OD600 value of 1.0. The colonies were then induced to express dsRNA by the addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.1 mM. The expressed dsRNA was extracted and confirmed by electrophoresis on 1% agarose gel. Bacteria cultures were centrifuged at 5000 ×g for 10 min, and resuspended in an equal original culture volume of 0.05 M phosphate buffered saline (PBS, pH 7.4). These bacterial suspensions (at a dsRNA concentration of about 0.5 μg/ml) were used for bioassay.
RNA interference bioassays and sampling
Two independent bioassays were carried out as previously described [28] using newly-ecdysed second- and fourth-instar larvae, and starved for 4 h before bioassay. Ten second- or fourth-instar larvae per replicate were allowed to feed foliage immersed with one of the following preparations: (1) PBS, (2) dsegfp, (3) dsChSA-1, (4) dsChSA-2, (5) dsChSAa, (6) dsChSAb, (7) dsChSB-1 and (8) dsChSB-2. Each treatment was repeated 21 (for the second-instar larvae) or 15 (for the fourth-instar larvae) times. For the bioassay involving the second-instar larvae, three replicates (30 larvae) were collected to extract total RNA after continuously fed for 3 days. Three replicates were used to extract whole-body chitin and another three replicates were dissected to collect integument, midgut and tracheae after consumption of dsRNA for 3 days and normal foliage for an additional 2 days. For the bioassay involving the fourth-instar larvae, three replicates were collected to extract total RNA after continuously fed for 3 days. The remaining 12 replicates of both bioassays were used to observe pupation and adult emergence as previously described [28]. The consumed foliage areas per replicate (10 larvae) were measured on day 3 after the initiation of bioassay. The growth of all the surviving larvae was examined at 4-h intervals. Instars were identified by head capsule width, the appearance of exuviae, the black color of the pronotum, and the anterior beige and posterior black stripe visible on the pronotum of the third and fourth instars respectively. Prepupae were distinctive from larvae by their disappearance of black pigmentation, their relative inactivity and their curved body shapes. The pupation and the adult emergence were recorded during a 4-week trial period. If necessary, the resulting adult females were individually coupled with normal males. The egg numbers were recorded within a period of 10 days after the emergence. For each bioassay, three biological replicates were carried out.
Chitin analysis
The same method as described [7] was used to test chitin contents from the whole body, integument (removal of the internal organs and muscles) or midgut samples. Briefly, the samples from three replicates (30 larvae) were individually mixed with 0.5 g zirconium beads (0.7 mm diameter, BioSpec Products, Bartlesville, OK) and 0.5 ml of 6% KOH, and were homogenized. The samples were then heated at 80 °C for 90 min, and were centrifuged at 12000 ×g for 20 min and the supernatants were removed. The pellet was suspended in 1 ml PBS, was centrifuged again at 12000 ×g for 20 min and the PBS was discarded. Each pellet was then resuspended in 200 µl of Mcllvaine's buffer (0.1 M citric acid, 0.2 M NaH2PO4, pH 6) and 5 µl of Streptomyces plicatus chitinase-63 (5 mg/ml in PBS) was added to hydrolyze chitin to N-acetylglucosamine (GlcNAc) by incubation for 72 h at 37 °C.
GlcNAc concentrations were individually measured using a modified Morgan-Elson assay [29]. In a 0.2 ml PCR tube, 10 µl of 0.27 M sodium borate and 10 µl of sample supernatant (12000 ×g, 1 min centrifugation) were combined. In a thermocycler, samples were heated to 99.9 °C for about 60 s, mixed gently, and incubated at 99.9 °C for 10 min. Immediately upon cooling to room temperature, 100 µl of diluted dimethylaminobenzaldehyde (DMAB) solution (10% w/v DMAB in 12.5 ml concentrated HCl and 87.5 ml of glacial acetic acid stock, diluted 1:10 with glacial acetic acid) was added, followed by incubation at 37 °C for 20 min. Eighty µl of each sample was transferred to 96-well low-evaporation microtitre dish, and the absorbance at 585 nm was recorded. Standard curves were prepared from stocks of 0.075-2.0 mM of GlcNAc.
Analysis of midgut PM and tracheae for integrity
The larvae having ingested dsRNAs were dissected. Guts and tracheae were observed under a light microscope for retention/loss of structural integrity of the chitin-containing structures. Moreover, the tracheae were incubated with 10 M of NaOH at 95 °C for 2 h to solubilize the cuticular proteins [30], and the treated tracheae were then seen under the light microscope.
Real-time quantitative PCR (qRT-PCR)
For tissue-biased gene expression analysis, RNA samples were extracted from the foregut, midgut, hindgut, Malpighian tubules, epidermis, trachea, fat body, hemocyte and ventral ganglion of the day 1 third-instar larvae. Moreover, samples from the larval survivors of the bioassays were prepared. Each sample contained 5-30 individuals and repeated three times. The RNA was extracted using SV Total RNA Isolation System Kit (Promega). Purified RNA was subjected to DNase I to remove any residual DNA according to the manufacturer's instructions. Quantitative mRNA measurements were performed by qRT-PCR, using four internal reference genes (LdRP4, LdRP18, LdARF1 and LdARF4, the primers listed in Table S1) according to our published results [31]. An RT negative control (without reverse transcriptase) and a non-template negative control were included for each primer set to confirm the absence of genomic DNA and to check for primer-dimer or contamination in the reactions, respectively. Each sample was technically repeated three times. Data were analyzed by the 2-ΔΔCT method, using the geometric mean of the four internal reference genes for normalization. All methods and data were confirmed to follow the MIQE (Minimum Information for publication of Quantitative real time PCR Experiments) guidelines [32].
Data analysis
The data were pooled from three independent biological replicates, given as means ± SE, and analyzed by ANOVAs followed by the Tukey-Kramer test, using SPSS for Windows (SPSS, Chicago, IL, USA). Since there were no significant differences between dsRNAs targeting two different regions of either LdChSA or LdChSB (LdChSA-1/LdChSA-2, or LdChSB-1/LdChSB-2), the data of each LdChS genes were combined.
Results
Tissue-biased expression of LdChSs
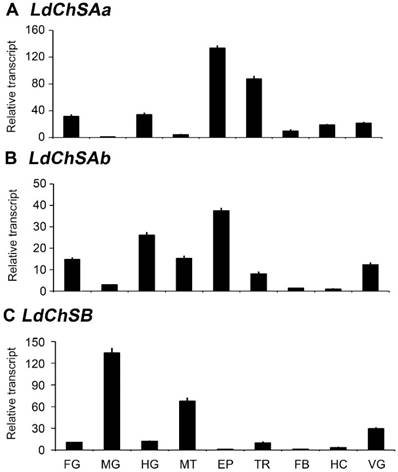
The mRNA levels of LdChSAa, LdChSAb, and LdChSB were detectable in the larval foregut, midgut, hindgut, Malpighian tubules, epidermis, trachea, fat body, hemocyte, and ventral ganglion. LdChSAa was highly expressed in the epidermis, trachea, foregut and hindgut (Figure 1A). LdChSAb was greatly transcribed in the epidermis, foregut, hindgut and Malpighian tubules (Figure 1B). In contrast, LdChSB was expressed at the highest level in the larval midgut (Figure 1C).
Tissue expression profiles of LdChS genes in L. decemlineata. The cDNA templates are derived from the foregut (FG), midgut (MG), hindgut (HG), Malpighian tubules (MT), epidermis (EP), trachea (TR), fat body (FB), hemocyte (HC) and ventral ganglion (VG) of the day 1 third-instar larvae. The lowest expression levels in MG for LdChSAa, in HC for LdChSAb and in FB for LdChSB are set as 1.
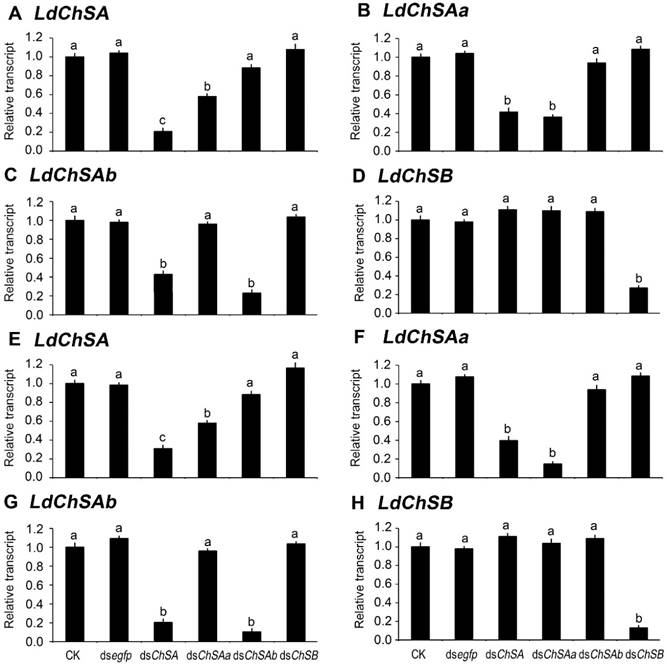
Ingestion of dsChSs at the second and fourth instars silences target genes
We dietarily introduced dsChSA, dsChSAa, dsChSAb and dsChSB into the newly-molted second- and fourth-instar larvae. In order to compare the effeciency of RNAi between LdChSAa and LdChSAb isoforms, three pairs of qRT-PCR primers were designed, the first pair were derived from of the common fragment of LdChSAa and LdChSAb; the second and third pairs were derived from LdChSAa and LdChSAb respectively (Figure S1).
As expected, ingestion of dsChSA knocked down both LdChSAa and LdChSAb (Figure 2). Feeding of dsChSAa silenced LdChSAa (Figure 2B, 2F); the total mRNA level of LdChSAa+LdChSAb (Figure 2A, 2E) was also significantly reduced. In contrast, dsChSAb ingestion only knocked down LdChSAb (Figure 2C, 2G); the total mRNA level of LdChSAa+LdChSAb (Figure 2A, 2E) was not significantly decreased. Consumption of dsChSB downregulated the target LdChSB transcript (Figure 2D, 2H). Conversely, all the four dsRNAs did not knock down non-target LdChS mRNAs (Figure 2).
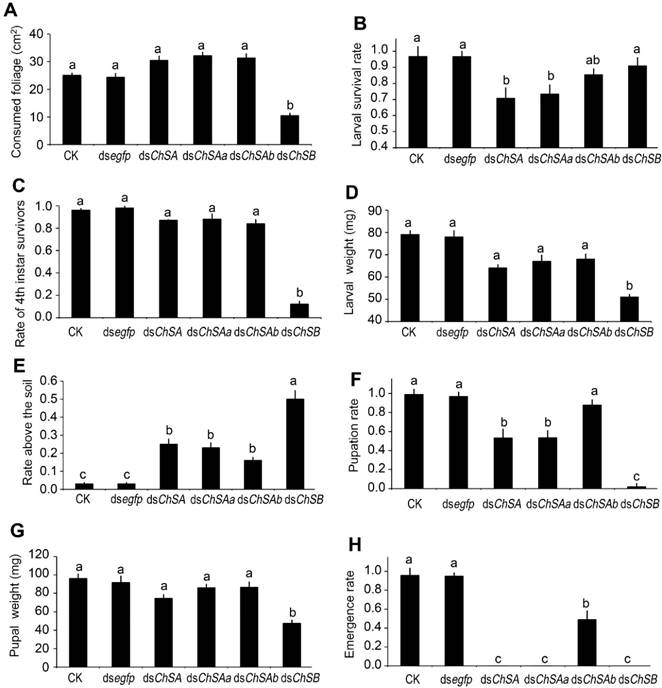
Ingestion of dsChS at the second-instar affects larval performance
After feeding of dsChSB for 3 days, the foliage consumption was inhibited in the treated larvae, in contrast to the beetles that have ingested PBS, dsegfp, dsChSA, dsChSAa and dsChSAb (Figure 3A).
After consumption of treated leaves for 3 days and normal foliage for an additional 2 days, the PBS-, dsegfp- and dsChSAb-fed larvae normally ecdysed to fourth larval instars, with little lethality. In contrast, approximately 30% of the larvae previously exposed to dsChSA and dsChSAa failed to complete the molt and died (Figure 3B). Moreover, most of the larvae previously exposed to dsChSB remained at the third-instar stage (Figure 3C), with lighter fresh larval weight (Figure 3D) and smaller body size (Figure 4B).
The wandering larvae previously fed PBS- and dsegfp-dipped leaves buried into soil. In contrast, approximately 30%-50% of the beetles formerly fed dsChSA, dsChSAa, dsChSAb and dsChSB did not bury themselves into soil normally (Figure 3E), and finally died from dehydration (such as that shown in Figure 4B). The developing periods from the initiation of bioassay to the occurrence of soil-digging behavior were significantly lengthened in the LdChSAa+LdChSAb, LdChSAa, LdChSAb or LdChSB RNAi hypomorphs (Table 1).
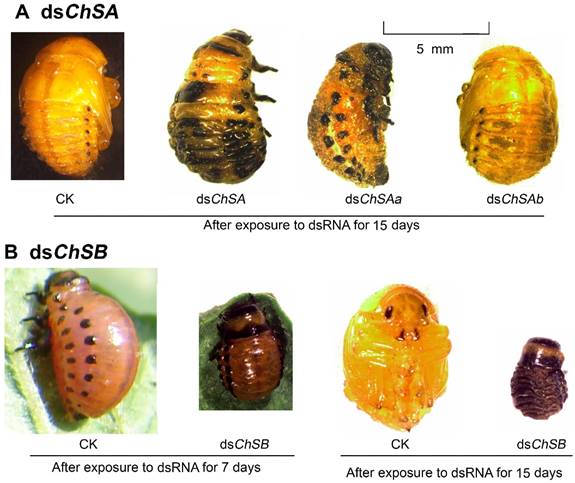
Many LdChSAa+LdChSAb, LdChSAa and LdChSB RNAi larvae did not normally molt to pupae (Figure 3F) and were completely wrapped in larval cuticle (Figure 4A, 4B). After exposure to dsRNA for 15 days, most of the LdChSAa+LdChSAb, LdChSAa and LdChSB RNAi beetles died. No adults emerged from LdChSAa+LdChSAb, LdChSAa and LdChSB RNAi hypomorphs (Figure 3H).
In contrast, approximately 50% of the LdChSAb RNAi pupae emerged as adults (Figure 3H). These adults did not have obvious defective phenotypes and had similar body size, fresh weight and fecundity to control adults (Figure S2).
Silencing of LdChS genes reduces the chitin content
After consumption of dsRNA for 3 days and normal foliage for an additional 2 days, the chitin contents in the whole larval body, integument and midgut samples were determined (Table 2).
The developing period of dsRNA-treated L. decemlineata surviving larvae from the initiation of bioassay to the occurrence of soil-digging behavior.
| Larval instar | 2nd | 3rd | 4th | Total |
|---|---|---|---|---|
| Initiation of the bioassay at the early second instar stage | ||||
| CK | 2.1±0.1 a | 2.1±0.2 a | 4.1±0.2 a | 8.3±0.4 a |
| dsegfp | 2.0±0.2 a | 2.1±0.1 a | 4.0±0.2 a | 8.1±0.5 a |
| dsChSA | 2.2±0.1 a | 2.4±0.2 a | 5.3±0.2 b | 9.9±0.6 b |
| dsChSAa | 2.2±0.1 a | 2.5±0.2 a | 5.2±0.2 b | 9.9±0.6 b |
| dsChSAb | 2.1±0.1 a | 2.1±0.2 a | 4.2±0.2 a | 8.4±0.6 a |
| dsChSB | 2.2±0.1 a | > 12.0 b | ||
| Initiation of the bioassay at the early fourth instar stage | ||||
| CK | 4.1±0.2 a | |||
| dsegfp | 4.2±0.1 a | |||
| dsChSA | 5.7±0.2 b | |||
| dsChSAa | 5.5±0.2 b | |||
| dsChSAb | 4.6±0.2 a | |||
| dsChSB | 5.6±0.2 b | |||
The larval growth is checked at 4-hour intervals. The developmental periods are given as means ± SE, and are subjected one-way ANOVA and followed by the Tukey-Kramer test. Means on the same column followed by the same letters are not significantly different at P <0.05.
Ingestion of LdChS dsRNA by the second (A-D) and fourth (E-H) instar larvae in L. decemlineata. For 3 days, the newly-ecdysed second- and fourth-instar larvae are allowed to feed the foliage immersed with PBS (CK), dsegfp, dsChSA, dsChSAa, dsChSAb or dsChSB. The bars represent mean (± SE). Different letters indicate significant difference at P value < 0.05. The expression levels in blank control (CK) are set as 1.
Effects of knockdown of LdChS genes in L. decemlineata second-instar larvae. The bars represent means (± SE). Different letters indicate significant difference at P value < 0.05.
The chitin concentrations of the whole larval body and integument samples were greatly reduced in the LdChSAa+LdChSAb and LdChSAa RNAi hypomorphs, compared with those in the control specimens. In contrast, the chitin contents of the whole larval body and integument samples in the LdChSAb and LdChSB RNAi hypomorphs were not significantly different from those in the dsegfp-fed controls (Table 2).
Moreover, the chitin content in the midgut sample of the LdChSB RNAi larvae was significantly lower than those in the midguts of control larvae (PBS- and dsegfp-fed specimens), and of the LdChSAa+LdChSAb, LdChSAa and LdChSAb RNAi hypomorphs (Table 2).
Ingestion of chitin synthase dsRNAs affecting chitin content (μg/mg) of L. decemlineata larvae.
| Sample | Whole larvae | Integument | Midgut |
|---|---|---|---|
| CK | 16.9±1.2 a | 91.0±6.7 a | 1.5±0.2 a |
| dsegfp | 17.0±1.4 a | 92.9±7.3 a | 1.6±0.2 a |
| dsChSA | 8.6±1.0 b | 51.8±4.4 b | 1.7±0.1 a |
| dsChSAa | 9.0±0.9 b | 56.3±4.7 b | 1.4±0.2 a |
| dsChSAb | 15.4±1.1 a | 73.2±5.1 ab | 1.5±0.1 a |
| dsChSB | 16.1±1.5 a | 84.5±6.5 a | 0.6±0.1 b |
The samples from three replicates (30 larvae) are individually tested. The chitin contents are given as means ± SE, and are subjected one-way ANOVA and followed by the Tukey-Kramer test. Means on the same column followed by the same letters are not significantly different at P <0.05.
Defective phenotypes of RNAi of LdChS genes in L. decemlineata second-instar larvae. Several defects such as failure of pupation in the LdChSAa+LdChSAb (dsChSA) and LdChSAa (dsChSAa) RNAi larvae (A) and retarded larval growth in the LdChSB RNAi larvae (B) are seen.
Integrity of the tracheae and midgut peritrophic matrix in the RNAi larvae
After ingestion of dsRNA for 3 days and normal foliage for an additional 2 days, the treated larvae were dissected to examine the integrity of the tracheae and the midgut PM (Figure 5 and Figure S3).
Larvae previously fed PBS (Figure 3S, A), dsegfp (Figure 5A), dsChSAb (Figure 3S, C) and dsChSB (Figure 5C) had well developed tracheae, there were distinct taenidia in the tracheae (Figure 5D, 5G, 5F, 5I; Figure 3S, D, G, F, I). The taenidia run around the tracheal tube and formed parallel transverse folds lining the lumen of the tracheae (Figure 5G, 5I; Figure 3S, G, I). In contrast, the dsChSA (Figure 5B)- and dsChSAa (Figure 3S, B)-fed larvae possessed underdeveloped trachea; the taenidia were thinned; no transverse tarnidia stripes were observed (Figure 5E, 5H; Figure 3S, E, H).
Moreover, the larvae previously fed PBS (Figure 3S, J), dsegfp (Figure 5J), dsChSA (Figure 5K), dsChSAa (Figure 3S, K) and dsChSAb (Figure 3S, L) possessed clear gut lumen, which was full of food. In contrast, the dsChSB-fed larvae had indistinct gut lumen, small pieces of food sparsely distributed along the midgut (Figure 5L). After removal of the midgut epithelia cells, intact PM enveloped the food in the larvae previously fed PBS, dsegfp, dsChSA, dsChSAa and dsChSAb (Figure 5M, 5N; Figure 3S, M, N, O). In contrast, only small fragments of the PM were left after removal of the midgut epithelia cells in the larvae formerly fed dsChSB. The food pieces were scattered (Figure 5O).
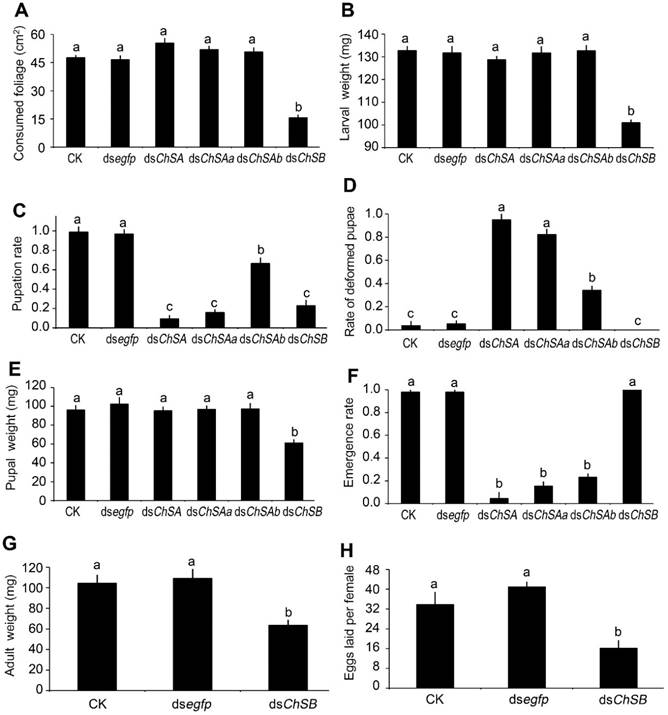
Effects of dsChS ingestion by the fourth-instar larvae
The performance of the larvae that have ingested dsChS at the fourth-instar larvae was also examined.
For the LdChSAa+LdChSAb RNAi larvae, the foliage consumption and larval fresh weight were not affected (Figure 6A, 6B). However, the developing period was lengthened (Table 1). The larval-pupal ecdysis and adult emergence were impaired (Figure 6C, 6D). The resulting moribund beetles were wrapped in larval cuticle (Figure 7A), and finally died (Figure 6F).
For the LdChSAa RNAi hypomorphs, the negative effects and defective phenotypes almost completely mimicked those of the LdChSAa+LdChSAb RNAi larvae (Figure 6A, 6B, 6C, 6D, 6F; Figure 7B; Table 1).
A few LdChSAa+LdChSAb and LdChSAa RNAi pupae successfully emerged as adults. However, the adults died in the soil, with deformed wings (Figure 7D).
LdChSA and LdChSB RNAi on chitin-containing structures in L. decemlineata. The dsegfp (the left column)-, dsChSA (the middle column)-, and dsChSB (the right column)-fed larvae are dissected and observed under a light microscope. The tracheae are visualized using light microscope before (A-F) and after treatment with 10 M of NaOH at 95 °C for 2 hrs (G-I). SP, spiracle; TR, tracheae; L, tracheal lumen; TA, taenidia.

The LdChSAb RNAi larvae consumed a similar amount of foliage and had similar fresh weight (Figure 6A, 6B) to control ones. The number of the pupated LdChSAb RNAi hypomorphs was lower than those in PBS- and dsegfp-fed larvae, but higher than those in dsChSA- and dsChSAa-fed larvae (Figure 6C); The LdChSAb RNAi larvae had less deformed pupae than the LdChSAa+LdChSAb and LdChSAa RNAi larvae (Figure 6D). Most of the LdChSAb RNAi pupae did not completely shed the old larval exuviae, which remained on the tips of the pupal appendages and abdomens (Figure 7C). Consequently, about 70% of the LdChSAb RNAi pupae failed to emerge as adults and finally died (Figure 6F). Since only a small number of females emerged, their fecundities were not measured.
For the LdChSB RNAi larvae, the foliage consumption was inhibited (Figure 6A). The larval growth was retarded, with lighter fresh larval, pupal and adult weights (Figure 6B, 6E, 6G) and smaller pupal and adult sizes (Figure 7E, 7F). Moreover, the developing period was delayed (Table 1). The resulting females laid fewer eggs than controls (Figure 6H).
Effects of silencing of LdChS genes in L. decemlineata fourth-instar larvae. The bars represent means (± SE). Different letters indicate significant difference at P value < 0.05.
Discussion
Two chitin synthase genes (LdChSA and LdChSB) have been identified in L. decemlineata. LdChSA has two splicing variants, LdChSAa and LdChSAb [26]. It is well known that RNAi-mediated knockdown of ChS dramatically affects survival, ecdysis, oviposition and egg hatching in different insect species [7, 8, 17-20]. Therefore, the feasibility of using an RNAi-based pest control strategy targeting LdChS genes was carefully examined in this study.
Many types of L. decemlineata tissues respond robustly to RNAi
RNAi efficiency varied dramatically among insect tissues [3]. In the present paper, we found that LdChSAa and LdChSAb were highly expressed in ectodermally-derived epidermal cells forming epidermis, trachea, foregut and hindgut, whereas LdChSB was mainly transcribed in midgut cells in L. decemlineata. Similar tissue-biased expression profiles have been documented in other insect species [24, 33, 34]. Therefore, we compared the RNAi efficiency in the epidermis and in the midgut by ingestion of dsRNAs respectively targeting LdChSA and LdChSB in L. decemlineata larvae. Our results revealed that ingestion of dsRNAs successfully knocked down LdChSA mainly expressed in the ectodermally-derived epidermal cells, and LdChSB chiefly transcribed in the midgut cells. Moreover, the LdChSA RNAi hypomorphs had lower chitin contents in the integument and whole body samples, and thinner taenidia in tracheae. In contrast, the LdChSB RNAi larvae had a lower chitin content in midgut, ate less foliage, and possessed indistinct gut lumen and incomplete midgut PM. These data demonstrate that ingestion of dsRNA can effectively knock down target gene expressed not only in larval gut, but also in epidermis and trachea.
Previous results revealed that feeding of dsRNA effectively knocked down target genes in alimentary canal, fat body, muscle, neuronmuscular junction, brain and corpora allata in L. decemlineata [27, 28, 35-44]. Therefore, many types of L. decemlineata tissues respond robustly to RNAi. This suggest that dsRNA has great potential for management of L. decemlineata.
Targeting LdChS with dsRNA is a potential control strategy
It has been well documented that the growth and development of insects and fungi are dependent on precisely tuned expression of ChS genes. Conversely, this process is absent in vertebrates and plants [18, 25, 26]. It is accordingly considered that ChS presents an attractive target for combating insect pests and fungi-born diseases.
Injection of dsChSs at an amount from 200 to 3000 ng per individual caused larval lethality, impaired ecdysis, decreased adult fecundity and/or affected egg hatching in Orthopteran Locusta migratoria [17, 22], Hemipteran Rhodnius prolixus [23] and Nilaparvata lugens [45], Coleopteran T. castaneum [7, 8, 21], Lepidopteran Spodoptera exigua [19], Ostrinia furnacalis [14] and Bombyx mori [46, 47], and Dipteran Drosophila melanogaster [48, 49] and Bactrocera dorsalis [24]. These data demonstrate that ChSs are the potential candidate genes for an RNAi-based pest control strategy.
In the present paper, we designed two dsChSAs from the common fragments of LdChSAa and LdChSAb to knock down two splicing variants, and designed two dsChSBs to silence LdChSB at the second and fourth instar stages. We measured that the immersed potato foliage contained approximately 5 ng dsRNA per cm2. Accordingly, a third- and a fourth-instar larva respectively ingested 90 and 150 ng dsRNA after three days' dsRNA exposure based on the consumed foliage area in the present paper, a dose comparable with that injected into a T. castaneum larva [7, 8, 21]. At this dsRNA dose, feeding of dsChSA or dsChSB killed all treated second-instar beetles. Moreover, ingestion of dsChSB significantly reduced foliage consumption, in contrast to feeding of dsChSA. This antifeedant effect may reduce foliage damage when application of dsChSB to potato field. Thus, it can be expected that dsChSB is more effective to protect potato from damage by L. decemlineata young larvae.
Moreover, our data revealed that all the dsChSA-fed fourth-instar larvae do not successfully emerge as adults, whereas the fourth-instar LdChSB RNAi larvae ate less, grew slowly, and obtained lighter fresh larval, pupal and adult weights. The resulting females laid fewer eggs. Therefore, it is expected that foliar spraying of dsChSA is more effective against L. decemlineata old larvae than application of dsChSB.
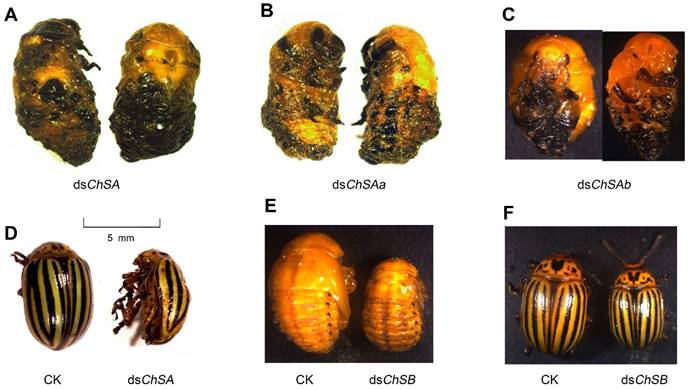
Defects of RNAi of LdChS genes in L. decemlineata fourth-instar larvae. Several defective phenotypes are seen. These include failure of ecdysis in the LdChSAa+LdChSAb (dsChSA) and LdChSAa (dsChSAa) RNAi larvae (A, B), failure of shedding the old larval exuviae in the LdChSAb (dsChSAb) RNAi larvae (C), deformed LdChSAa+LdChSAb and LdChSAa RNAi adults with wrinkled wings (D), and smaller LdChSB RNAi pupae and adults (E, F).
RNAi of each of the two LdChSA splicing variants has different negative effects
In the present paper, we found that knockdown of LdChSAa significantly decreased the transcript level of LdChSAa+LdChSAb, whereas silencing of LdChSAb only slightly affected the mRNA level of LdChSAa+LdChSAb. Providing that dsChSAa and dsChSAb only silenced their respective target mRNAs, these results suggest that LdChSAa is the mainly expressed variant in L. decemlineata larvae. In agreement with the suggestion, the expression levels of LmChSAa were much higher than those of LmChSAb during most developmental stages in L. migratoria [17].
Moreover, RNAi-aided silencing of LdChSAa at the second or fourth instar stages almost completely mimicked the negative effects and defective phenotypes of the LdChSAa+LdChSAb RNAi larvae. In contrast, knockdown of LdChSAb at the second instar stage caused lower larval and pupal mortalities than silencing of LdChSAa. Approximately 50% of the LdChSAb RNAi pupae emerged as adults. These adults showed little defects and had a similar body size, fresh weight and fecundity to control adults. Similarly, silencing of LdChSAb at the fourth instar stage had fewer side effects on larval and pupal survival than knockdown of LdChSAa. However, the resulting pupae did not completely shed the old larval exuviae, which remained on the tips of the pupal appendages and abdomens. About 70% of the LdChSAb RNAi pupae did not emerged as adults and finally died. Therefore, foliar spraying of dsChSAa should be more effective against L. decemlineata larvae than application of dsChSAb.
In other insect species, silencing of the two ChSA variants also caused different defective phenotypes. In T. castaneum, dsTcChSAa disrupted larval-larval, larval-pupal, and pupal-adult ecdysis, while dsTcChSAb only influenced pupal-adult molting [2, 7]. Knockdown of BdChSAa+BdChSAb or only BdChSAa in B. dorsalis had a similar result in that the larvae were trapped in old cuticles and died without completely tanning, whereas injection of dsBdChSAb into B. dorsalis third (final) instar larvae caused no visible abnormal morphological changes, and no influence on pupation [24]. In L. migratoria, dsLmChSAa resulted in three different lethal phenotypes, i.e., translucent new cuticle, trouble shedding old cuticle, and stunted development, whereas dsLmChSAb only caused crumpled cuticle [17]. In O. furnacalis, silencing of OfChSA-2a led to incomplete molting, while knockdown of OfChSA-2b only influenced the formation of head cuticle in the third-instar larvae [14].
Developing stage-dependent RNAi efficiency in L. decemlineata larvae
RNAi efficiency is inconstant between life stages [3]. In Apis mellifera, for example, when a 504 bp of vitellogenin-dsRNA was injected at the preblastoderm stage, 15% workers had strongly reduced levels of vitellogenin mRNA. In contrast, 96% individuals showed the mutant phenotype when dsRNA was introduced in newly emerged bees [50].
Similarly, RNAi efficiency is variable among life stages in L. decemlineata. For a housekeeping gene S-adenosyl-L-homocysteine hydrolase (LdSAHase), dsRNA caused larval lethality, inhibited growth and impaired pupation in an instar-dependent manner: the young larvae are more susceptible to dsRNA than the old ones [51]. In the present paper, our data revealed that knockdown of LdChSB caused more serious defects and more deaths in the young larvae. RNAi of LdChSAa+LdChSAb and LdChSAa resulted in similar negative effects on larval development in young (second-instar) and old (fourth-instar) larvae. Conversely, silencing of LdChSAb produced more serious influences in old larvae than those in young ones. In order to improve RNAi efficiency, the appropriate timing for dsRNA treatment is the young larval instars for a housekeeping gene. As for a non-housekeeping gene, a suitable stage for application of a dsRNA is when the target gene is actually expressing and the protein is playing its role.
In addition, our results revealed that the activities of core RNAi-machinery proteins affect RNAi efficiency in L. decemlineata. Exposed to dsegfp for 6 h significantly elevated the mRNA levels of four core RNAi genes LdDcr2a, LdDcr2b, LdAgo2a and LdAgo2b, in the first-, second-, third- and fourth-instar larvae and increased RNAi efficiency to LdSAHase. When the exposure periods were extended, however, the expression levels of LdDcr2a, LdDcr2b, LdAgo2a and LdAgo2b were gradually reduced. As a result, RNAi efficiency to LdSAHase was decreased [51]. This result indicates that an appropriate application timing must be ascertained before a non-housekeeping gene-derived dsRNA is applied to potato to control L. decemlineata.
Supplementary Material
Supplementary figures and tables.
Abbreviations
ChS: Chitin synthase; PM: Peritrophic membrane; qRT-PCR: Quantitative real-time PCR; dsRNA: double-strand RNA; RNAi: RNA interference; First-strand complementary DNA: cDNA; LdChSAa+LdChSAb RNAi: RNA interference both LdChSAa and LdChSAb
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31272047 and 31360442), and the Fundamental Research Funds for the Central Universities (KYTZ201403).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Alyokhin A. Colorado potato beetle management on potatoes: Current challenges and future prospects. Fruit, Vegetable and Cereal Science and Biotechnology. 2009;3:10-9
2. Alyokhin A, Baker M, Mota-Sanchez D, Dively G, Grafius E. Colorado potato beetle resistance to insecticides. American Journal of Potato Research. 2008;85:395-413
3. Scott JG, Michel K, Bartholomay LC, Siegfried BD, Hunter WB, Smagghe G. et al. Towards the elements of successful insect RNAi. Journal of Insect Physiology. 2013;59:1212-21
4. Palli SR. RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Current Opinion in Insect Science. 2014;6:1-8
5. Moussian B. The apical plasma membrane of chitin-synthesizing epithelia. Insect Science. 2013;20:139-46
6. Merzendorfer H. The cellular basis of chitin synthesis in fungi and insects: common principles and differences. European Journal of Cell Biology. 2011;90:759-69
7. Arakane Y, Muthukrishnan S, Kramer KJ, Specht CA, Tomoyasu Y, Lorenzen MD. et al. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Molecular Biology. 2005;14:453-63
8. Arakane Y, Specht CA, Kramer KJ, Muthukrishnan S, Beeman RW. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochemistry and Molecular Biology. 2008;38:959-62
9. Kato N, Mueller CR, Fuchs JF, Wessely V, Lan Q, Christensen BM. Regulatory mechanisms of chitin biosynthesis and roles of chitin in peritrophic matrix formation in the midgut of adult Aedes aegypti. Insect Biochemistry and Molecular Biology. 2006;36:1-9
10. Hogenkamp DG, Arakane Y, Zimoch L, Merzendorfer H, Kramer KJ, Beeman RW. et al. Chitin synthase genes in Manduca sexta: characterization of a gut-specific transcript and differential tissue expression of alternately spliced mRNAs during development. Insect Biochemistry and Molecular Biology. 2005;35:529-40
11. Chen X, Yang X, Senthil Kumar N, Tang B, Sun X, Qiu X. et al. The class A chitin synthase gene of Spodoptera exigua: molecular cloning and expression patterns. Insect Biochemistry and Molecular Biology. 2007;37:409-17
12. Zimoch L, Hogenkamp DG, Kramer KJ, Muthukrishnan S, Merzendorfer H. Regulation of chitin synthesis in the larval midgut of Manduca sexta. Insect Biochemistry and Molecular Biology. 2005;35:515-27
13. Arakane Y, Hogenkamp DG, Zhu YC, Kramer KJ, Specht CA, Beeman RW. et al. Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochemistry and Molecular Biology. 2004;34:291-304
14. Qu M, Yang Q. A novel alternative splicing site of class A chitin synthase from the insect Ostrinia furnacalis-Gene organization, expression pattern and physiological significance. Insect Biochemistry and Molecular Biology. 2011;41:923-31
15. Qu M, Yang Q. Physiological significance of alternatively spliced exon combinations of the single-copy gene class A chitin synthase in the insect Ostrinia furnacalis (Lepidoptera). Insect Molecular Biology. 2012;21:395-404
16. Rezende GL, Martins AJ, Gentile C, Farnesi LC, Pelajo-Machado M, Peixoto AA. et al. Embryonic desiccation resistance in Aedes aegypti: presumptive role of the chitinized serosal cuticle. BMC Developmental Biology. 2008;8:82
17. Zhang J, Liu X, Zhang J, Li D, Sun Y, Guo Y. et al. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust. Insect Biochemistry and Molecular Biology. 2010;40:824-33
18. Merzendorfer H. Insect chitin synthases: a review. Journal of Comparative Physiology B. 2006;176:1-15
19. Tian H, Peng H, Yao Q, Chen H, Xie Q, Tang B. et al. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One. 2009;4:e6225
20. Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Molecular Biology. 2010;19:683-93
21. Kelkenberg M, Odman-Naresh J, Muthukrishnan S, Merzendorfer H. Chitin is a necessary component to maintain the barrier function of the peritrophic matrix in the insect midgut. Insect Biochemistry and Molecular Biology. 2015;56:21-8
22. Liu X, Zhang H, Li S, Zhu KY, Ma E, Zhang J. Characterization of a midgut-specific chitin synthase gene (LmCHS2) responsible for biosynthesis of chitin of peritrophic matrix in Locusta migratoria. Insect Biochemistry and Molecular Biology. 2012;42:902-10
23. Mansur JF, Alvarenga ESL, Figueira-Mansur J, Franco TA, Ramos IB, Masuda H. et al. Effects of chitin synthase double-stranded RNA on molting and oogenesis in the Chagas disease vector Rhodnius prolixus. Insect Biochemistry and Molecular Biology. 2014;51:110-21
24. Yang W-J, Xu K-K, Cong L, Wang J-J. Identification, mRNA expression, and functional analysis of Chitin Synthase 1 gene and its two alternative splicing variants in Oriental fruit fly, Bactrocera dorsalis. International Journal of Biological Sciences. 2013;9:331-42
25. Kola VSR, Renuka P, Madhav MS, Mangrauthia SK. Key enzymes and proteins of crop insects as candidate for RNAi based gene silencing. Frontiers in Physiology. 2015;6:119
26. Shi J-F, Mu L-L, Guo W-C, Li G-Q. Identification and hormone induction of putative chitin synthase genes and splice variants in Leptinotarsa decemlineata (Say). Archives of Insect Biochemistry and Physiology. 2016;92:242-258
27. Zhou L-T, Jia S, Wan P-J, Kong Y, Guo W-C, Ahmat T. et al. RNA interference of a putative S-adenosyl-L-homocysteine hydrolase gene affects larval performance in Leptinotarsa decemlineata (Say). Journal of Insect Physiology. 2013;59:1049-56
28. Liu X-P, Fu K-Y, Lü F-G, Meng Q-W, Guo W-C, Li G-Q. Involvement of FTZ-F1 in the regulation of pupation in Leptinotarsa decemlineata (Say). Insect Biochemistry and Molecular Biology. 2014;55:51-60
29. Reissig JL, Strominger JL, Leloir LF. A modified colorimetric method for the estimation of N-acetylamino sugars. Journal of Biological Chemistry. 1955;217:959-66
30. Shi J-F, Fu J, Mu L-L, Guo W-C, Li G-Q. The Leptinotarsa uridine diphosphate N-acetylglucosamine pyrophosphorylase genes LdUAP1 and LdUAP2 are specialized for synthesis of chitin in larval epidermal cuticle and midgut peritrophic matrix. Insect Biochemistry and Molecular Biology. 2016;68:1-12
31. Shi X-Q, Guo W-C, Wan P-J, Zhou L-T, Ren X-L, Tursun A. et al. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Research Notes. 2013;6:93
32. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009;55:611-22
33. Tetreau G, Cao X, Chen Y-R, Muthukrishnan S, Jiang H, Blissard GW. et al. Overview of chitin metabolism enzymes in Manduca sexta: Identification, domain organization, phylogenetic analysis and gene expression. Insect Biochemistry and Molecular Biology. 2015;62:114-26
34. Shirk PD, Perera OP, Shelby KS, Furlong RB, LoVullo ED, Popham HJR. Unique synteny and alternate splicing of the chitin synthases in closely related heliothine moths. Gene. 2015 http://dx.doi.org/10.1016/j.gene.2015.08.001
35. Wan P-J, Lü D, Guo W-C, Ahmat T, Yang L, Mu L-L. et al. Molecular cloning and characterization of a putative proline dehydrogenase gene in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Science. 2014;21:147-58
36. Wan P-J, Guo W-Y, Yang Y, Lü F-G, Lu W-P, Li G-Q. RNAi suppression of the ryanodine receptor gene results in decreased susceptibility to chlorantraniliprole in Colorado potato beetle Leptinotarsa decemlineata. Journal of Insect Physiology. 2014;63:48-55
37. Zhu F, Xu J, Palli R, Ferguson J, Palli SR. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Management Science. 2011;67:175-82
38. Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O. et al. Control of coleopteran insect pests through RNA interference. Nature Biotechnology. 2007;25:1322-6
39. Ochoa-Campuzano C, Martinez-Ramirez AC, Contreras E, Rausell C, Real MD. Prohibitin, an essential protein for Colorado potato beetle larval viability, is relevant to Bacillus thuringiensis Cry3Aa toxicity. Pesticide Biochemistry and Physiology. 2013;107:299-308
40. Meng Q-W, Liu X-P, Lü F-G, Fu K-Y, Guo W-C, Li G-Q. Involvement of a putative allatostatin in regulation of juvenile hormone titer and the larval development in Leptinotarsa decemlineata (Say). Gene. 2015;554:105-13
41. Fu K-Y, Guo W-C, Lü F-G, Liu X-P, Li G-Q. Response of the vacuolar ATPase subunit E to RNA interference and four chemical pesticides in Leptinotarsa decemlineata. Pesticide Biochemistry and Physiology. 2014;114:16-23
42. Wan P-J, Fu K-Y, Lü F-G, Guo W-C, Li G-Q. A putative Δ1-pyrroline-5-carboxylate synthetase involved in the biosynthesis of proline and arginine in Leptinotarsa decemlineata. Journal of Insect Physiology. 2014;71:105-13
43. Kong Y, Liu X-P, Wan P-J, Shi X-Q, Guo W-C, Li G-Q. The P450 enzyme Shade mediates the hydroxylation of ecdysone to 20-hydroxyecdysone in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Molecular Biology. 2014;23:632-43
44. Fu K-Y, Li Q, Zhou L-T, Meng Q-W, Lü F-G, Guo W-C. et al. Knockdown of juvenile hormone acid methyl transferase severely affects the performance of Leptinotarsa decemlineata (Say) larvae and adults. Pest Management Science. 2015 DOI 10.1002/ps.4103
45. Wang Y, Fan H-W, Huang H-J, Xue J, Wu W-J, Bao Y-Y. et al. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae). Insect Biochemistry and Molecular Biology. 2012;42:637-46
46. Zhuo W, Fang Y, Kong L, Li X, Sima Y, Xu S. Chitin synthase A: a novel epidermal development regulation gene in the larvae of Bombyx mori. Molecular Biology Reports. 2014;41:4177-86
47. Zhuo W, Chu F, Kong L, Tao H, Sima Y, Xu S. Chitin synthase B: a midgut-specific gene induced by insect hormones and involved in food intake in Bombyx mori larvae. Archives of insect biochemistry and physiology. 2014;85:36-47
48. Ostrowski S, Dierick HA, Bejsovec A. Genetic control of cuticle formation during embryonic development of Drosophila melanogaster. Genetics. 2002;161:171-82
49. Devine WP, Lubarsky B, Shaw K, Luschnig S, Messina L, Krasnow MA. Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proceedings of the National Academy Sciences of the United States of America. 2005;102:17014-9
50. Amdam GV, Simões ZL, Guidugli KR, Norberg K, Omholt SW. Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnology. 2003;3:1
51. Guo W-C, Fu K-Y, Yang S, Li X-X, Li G-Q. Instar-dependent systemic RNA interference response in Leptinotarsa decemlineata larvae. Pesticide Biochemistry and Physiology. 2015;123:64-73
Author contact
![]() Corresponding author: Guo-Qing Li, Email: ligqedu.cn. Tel/Fax: +86-25-84395248.
Corresponding author: Guo-Qing Li, Email: ligqedu.cn. Tel/Fax: +86-25-84395248.
Received 2015-11-18
Accepted 2016-9-10
Published 2016-10-25
