ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2017; 13(11):1361-1372. doi:10.7150/ijbs.21657 This issue Cite
Research Paper
Co-expression network analysis identified FCER1G in association with progression and prognosis in human clear cell renal cell carcinoma
1. Department of Urology, Zhongnan Hospital of Wuhan University, Wuhan, China
2. Laboratory of Precision Medicine, Zhongnan Hospital of Wuhan University, Wuhan, China
3. Department of Endocrinology, The First Affiliated Hospital of Zhejiang University, Hangzhou, China
4. Department of Radiation and Medical Oncology, Zhongnan Hospital of Wuhan University, Wuhan, China
5. Department of Pathology, Lombardi Comprehensive Cancer Center, Georgetown University Medical School, Washington DC, USA
6. Department of Biological Repositories, Zhongnan Hospital of Wuhan University, Wuhan, China
Abstract
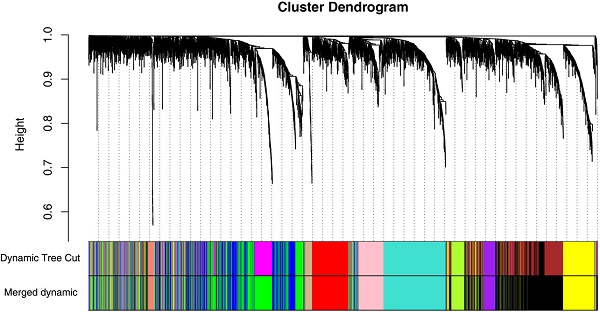
Clear cell renal cell carcinoma (ccRCC) is the most common solid lesion within kidney, and its prognostic is influenced by the progression covering a complex network of gene interactions. In current study, the microarray data GSE66272 containing ccRCC and adjacent normal tissues was analyzed to identify 4042 differentially expressed genes, on which weighted gene co-expression network analysis was performed. Then 12 co-expressed gene modules were identified. The highest association was found between blue module and pathological stage (r = -0.77) by Pearson's correlation analysis. Functional enrichment analysis revealed that biological processes of blue module focused on inflammatory response, immune response, chemotaxis (all p < 1e-10). In the significant module, a total of 38 network hub genes were identified, FCER1G exhibited the highest correlation (r = 0.95) with ccRCC progression. In addition, FCER1G was hub node in the protein-protein interaction network of the genes in blue module as well. Thus, FCER1G was subsequently selected for validation. In the test set GSE53757 and RNA-sequencing data, FCER1G expression was also positively correlated with four stages of ccRCC progression (p < 0.001). Receiver operating characteristic (ROC) curve indicated that FCER1G could distinguish localized (pathological stage I, II) from non-localized (pathological stage III, IV) ccRCC (AUC=0.74, p < 0.001). Besides, FCER1G could be a prognostic gene in clinical practice as well, revealed by survival analysis based on RNA-sequencing data (p < 0.05). In conclusion, using weighted gene co-expression analysis, FCER1G was identified and validated in association with ccRCC progression and prognosis, which might improve the prognosis by influencing immune-related pathways.
Keywords: clear cell renal cell carcinoma (ccRCC), weighted gene co-expression network analysis (WGCNA), FCER1G, prognosis, pathological stage, Receiver operating characteristic (ROC).
