ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2018; 14(3):237-252. doi:10.7150/ijbs.22868 This issue Cite
Research Paper
Molecular Networks of Postia placenta Involved in Degradation of Lignocellulosic Biomass Revealed from Metadata Analysis of Open Access Gene Expression Data
Department of Biology, Lakehead University, 955 Oliver Road, Thunder Bay, Ontario, P7B 5E1, Canada.
Abstract
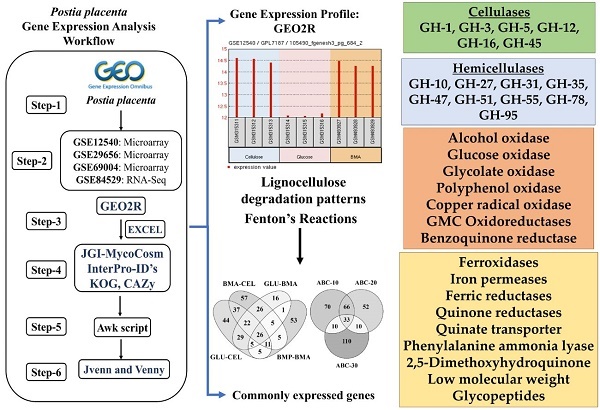
To understand the common gene expression patterns employed by P. placenta during lignocellulose degradation, we have retrieved genome wide transcriptome datasets from NCBI GEO database and analyzed using customized analysis pipeline. We have retrieved the top differentially expressed genes and compared the common significant genes among two different growth conditions. Genes encoding for cellulolytic (GH1, GH3, GH5, GH12, GH16, GH45) and hemicellulolytic (GH10, GH27, GH31, GH35, GH47, GH51, GH55, GH78, GH95) glycoside hydrolase classes were commonly up regulated among all the datasets. Fenton's reaction enzymes (iron homeostasis, reduction, hydrogen peroxide generation) were significantly expressed among all the datasets under lignocellulolytic conditions. Due to the evolutionary loss of genes coding for various lignocellulolytic enzymes (including several cellulases), P. placenta employs hemicellulolytic glycoside hydrolases and Fenton's reactions for the rapid depolymerization of plant cell wall components. Different classes of enzymes involved in aromatic compound degradation, stress responsive and detoxification mechanisms (cytochrome P450 monoxygenases) were found highly expressed in complex plant biomass substrates. We have reported the genome wide expression patterns of genes coding for information, storage and processing (KOG), tentative and predicted molecular networks involved in cellulose, hemicellulose degradation and list of significant protein-ID's commonly expressed among different lignocellulolytic growth conditions.
Keywords: Postia placenta, Brown-rot decay, Gene expression, NCBI-Gene Expression Omnibus (GEO), Lignocellulose, Fenton's reaction
