ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2018; 14(7):760-774. doi:10.7150/ijbs.25052 This issue Cite
Research Paper
miR-487b-3p Suppresses the Proliferation and Differentiation of Myoblasts by Targeting IRS1 in Skeletal Muscle Myogenesis
1. Institute of Cellular and Molecular Biology, Jiangsu Normal University, Xuzhou, Jiangsu, 221116, China
2. Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining, Qinghai, 810001, China
Abstract
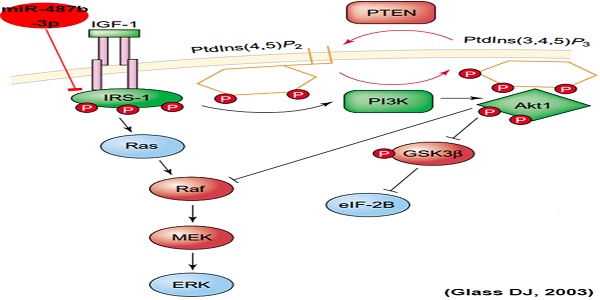
MicroRNAs are endogenous, small non-coding RNAs that can play critical gene-regulatory roles during skeletal muscle development and are highly conserved. miR-487b-3p is expressed in muscle, and the detailed mechanism by which it regulates myoblast proliferation and differentiation has not been explored. Here, we found that miR-487b-3p expression was significantly higher in goat muscle tissues than in other tissues and was higher in fetal goat muscle tissues than in mature goat tissues, suggesting that miR-487b-3p has an important effect on skeletal muscle myogenesis. Functional studies showed that miR-487b-3p overexpression significantly suppressed C2C12 myoblast proliferation and differentiation, which was accompanied by the down-regulation of functional genes related to proliferation (MyoD, Pax7 and PCNA) and differentiation (Myf5, MyoG and Mef2c), whereas the inhibition of miR-487b-3p accelerated C2C12 myoblast proliferation and differentiation and was accompanied by the up-regulation of functional genes. Using Target-Scan and David, we found that miR-487b-3p targeted the 3'-UTR of IRS1, an essential regulator in the PI3K/Akt and MAPK/Erk pathways. We then confirmed the targeting of IRS1 by miR-487b-3p using dual-luciferase assays, RT-qPCR and western blotting. Furthermore, IRS1 silencing markedly inhibited proliferation and differentiation in cultured C2C12 myoblasts, confirming the important role of IRS1 in myogenesis. These results reveal an IRS1-mediated regulatory link between miR-487b-3p and the PI3K/Akt and MAPK/Erk pathways during skeletal muscle myogenesis.
Keywords: miR-487b-3p, IRS1, myoblast proliferation, myoblast differentiation, myogenesis
Introduction
Skeletal muscle is one of three major muscle types, is involved in body metabolism, and comprises approximately half of the total somatic mass [1]. Skeletal muscle myogenesis is a complex process in which mononucleated myoblasts first withdraw from the cell cycle and fuse with each other to form multinucleated myotubes, after which multinucleated myotubes undergo further cell fusion and express contractile proteins to form mature myotubes, which are subsequently transformed into contractile myofibers [2-4]. The process of skeletal muscle myogenesis is regulated by the MRF family, the Mef2 family and other related transcriptional factors. These regulators, including Pax3, Pax7 [5,6], MyoD, Myf5 [7, 8], Myf6 or MRF4 [9], MyoG [10] and Mef2c [11], all play crucial roles in muscle growth and myogenesis. MicroRNAs (miRNAs) are short (20-24 nt) non-coding RNAs that negatively regulate gene expression via post-transcriptional mechanisms such as degradation or translational repression of target mRNAs [12]. Thus far, many miRNAs have been shown to regulate the proliferation and differentiation of myoblasts during muscle myogenesis. Previous studies have reported that several miRNAs, including miR-1 [13,14], miR-133 [15,16] and miR-206 [17,18], are muscle-specific miRNAs because their key regulators are involved in muscle myogenesis in humans, mice and cows. Some non-specific miRNAs in muscle, such as miR-24 [19], miR-26a [20], miR-29 [21], miR-125b [22], miR-181 [23], miR-214 [24] and miR-491 [25], are also essential for the regulation of muscle myogenesis. For example, miR-24 can promote skeletal muscle differentiation by targeting TGF-β1 [19]; miR-181 has been shown to promote myoblast differentiation by down-regulating homeobox protein Hox-A11, a myogenic inhibitor [23]; and the overexpression of miR-491 in C2C12 myoblasts represses myogenic differentiation by targeting myomaker [25].
miR-487b has been identified as a tumor-related miRNA that might play an independent role in glioneuroma [26]. The 14q32 miRNA cluster, which includes miR-487b, is highly involved in neovascularization, and inhibition of miR-487b may be a promising strategy for future therapeutic neovascularization and blood flow recovery [27]. Furthermore, miR-487b also acts as a negative regulator during macrophage activation [28]. miR-487b-5p and miR-487b-3p comprise the miR-487b family and are derived from the miR-487b precursor (based on the miRbase). However, little is known about the function of miR-487b-3p and its regulation in skeletal muscle myogenesis. Insulin and insulin-like growth factor-1 (IGF-1) receptors are tyrosine kinases that can phosphorylate various signaling proteins, including insulin receptor substrate 1 (IRS1). IRS1 plays a key role in insulin-Ras-MAPK signaling, which transmit signals from IGF-1 receptors to the intracellular PI3K/Akt and MAPK/Erk pathways [29]. A recent study showed that IRS1 up-regulation promoted cell proliferation and metastasis in pancreatic cancer cells. Conversely, IRS1 inhibition considerably reduced pancreatic cancer cell proliferation, migration and invasion [30]. Moreover, IRS1 also serves as an enhancer during the IGF-I response in mutant breast cancer cells [31]. These reports demonstrate that IRS1 plays a significant role in the regulation of cell growth and development.
According to our previous high-throughput sequencing results, miR-487b-3p was significantly differentially expressed at two distinct stages of goat muscle development and myogenesis (from the fetal stage to the six-months postnatal stage) [32]. In the present study, we explored the role of miR-487b-3p in regulating C2C12 myoblast proliferation and differentiation. We found that miR-487b-3p serves as a negative regulator of skeletal myogenesis. Furthermore, we confirmed that miR-487b-3p inhibits C2C12 myoblast proliferation and differentiation by directly targeting the 3'-UTR of IRS1 mRNA. Additionally, we demonstrated the function of IRS1 during C2C12 myoblast proliferation and differentiation by using siRNA to silence endogenous IRS1. In total, our studies offer numerous insights into the regulation of miR-487b-3p in skeletal muscle myogenesis and reveal a miRNA-related regulatory link between miR-487b-3p and the PI3K/Akt and MAPK/Erk pathways during myogenesis.
Materials and Methods
Animal tissue sample collection
Xuhuai goats were obtained from a farm (Suining Country, Jiangsu Province, China) at the goat breeding cooperation center (Xuzhou, Jiangsu Province, China). Heart, liver, spleen, lung, kidney, small intestine, longissimus dorsi and leg muscle tissue samples were collected from three Xuhuai goat fetuses (90 days) and three mature goats (2 years). All tissue samples were immediately frozen in liquid nitrogen after harvesting.
Expression profiling and miRNA target prediction
Total RNA was extracted from the tissue samples using TRIzol (Takara; Japan) and reverse transcribed to cDNA using the PrimeScript RT reagent kit (Perfect Real Time, Takara; Japan), which was used to perform the RT-qPCR of all tissues at both stages. The primers for miR-487b-3p are listed in Table 1, and 18S-rRNA was used as an internal normalization control for miRNA measurements.
The TargetScan program (http://www.targetscan.org) was employed to predict the targets of miR-487b-3p. The target gene IRS1 was selected based on TargetScan, KEGG (http://www.genome.jp/kegg) and David Bioinformatics analysis (https://david.ncifcrf.gov/).
Plasmid construction
The stem-loop sequence of miR-487b-3p from miRBase (http://www.mirbase.org) was cloned into the HindIII and XbaI sites of pcDNA3.1 to construct pcDNA3.1(+)-miR-487b-3p. The 3'-UTR of IRS1 from NCBI (https://www.ncbi.nlm.nih.gov/) containing the binding site of miR-487b-3p was amplified from Xuhuai goat genomic DNA and then cloned into the luciferase reporter vector psiCheck-2 (Promega; USA) using the XhoI and NotI restriction sites. The mutant IRS1 3'-UTR luciferase reporter was obtained by changing the miR-487b-3p binding site from GTACGAT to GATGGAT. The primers used in plasmid construction are shown in Table 2.
Primer information for miRNA and mRNA quantitative reverse transcription
| Gene | Primer Name | Primer Sequence (5' to 3') |
|---|---|---|
| miR-487b-3p | Stem-loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCG |
| RT-miR-487b-3p1 | CACTGGATACGACAAGTGGAT | |
| miR-487b-3p Stem-loop -F | AATCGTACAGGGTCATC | |
| miR-487b-3p Stem-loop -R | GTGCAGGGTCCGAGGT | |
| 18S-rRNA | 18S-rRNA-F | GTGGTGTTGAGGAAAGCAGACA |
| 18S-rRNA-R | TGATCACACGTTCCACCTCATC | |
| Myf5 | Myf5-F | TGAAGGCTCCTGTATCCCCT |
| Myf5-R | GAAGGCTGCTACTCTTGGCT | |
| Mef2c | Mef2c-F | AGCACTGACAAAGGTCTGGT |
| Mef2c-R | TCCCTCTGCACAAGTGTCTG | |
| MyoG | MyoG-F | GAGACATGAGTGCCCTGACC |
| MyoG-R | AGGCTTTGGAACCGGATAGC | |
| MyoD | MyoD-F | GCTCTGATGGCATGATGGAT |
| MyoD-R | CACTGTAGTAGGCGGTGTCG | |
| Pax7 | Pax7-F | GCTGCCGGATCTTACCTACC |
| Pax7-R | CCAGCACAGCGGAGTGTTCC | |
| PCNA | PCNA-F | TGAACCTCACCAGCATGTCCAAAAT |
| PCNA-R | TTCAAATACTAGTGCCAACGTGTCCG | |
| IRS1 | IRS1-F | AGAACGAGAAGAAGTGGCGG |
| IRS1-R | GCCTTTGCCCGATTATGCAG | |
| β-actin | β-actin-F | GGCACCACACCTTCTACAATG |
| β-actin-R | GGGGTGTTGAAGGTCTCAAAC |
1 Stem-loop RT-miR-487b-3p was applied for reverse transcription of miR-487b-3p.
Primers used to construct the plasmid
| Gene | Primer Name | Primer Sequence (5' to 3') |
|---|---|---|
| pcDNA3.1(+) -miR-487b-3p | Pre-miR-487b-3p-F | CCCAAGCTTCACTAACCCTCAGCGTCTAA |
| Pre-miR-487b-3p-R | GCTCTAGATAAGTGTCAGAGGCGGGAT | |
| Wild-IRS1 | Wild-IRS1-F | CCGCTCGAGACCTCAGCAAATCCTCCTT |
| Wild-IRS1-R | ATAAGAATGCGGCCGCCCAACGCAACAGTTCCATA | |
| Mutant-IRS1 | Mutant-IRS1-F | CCGCTCGAGTCAGTAGATGGATGGATGCACCCATTTCA |
| Mutant-IRS1-R | ATAAGAATGCGGCCGCCCAACGCAACAGTTCCATA |
Note: HindIII and XbaI restriction sites are italicized; XhoI and NotI restriction sites are underlined.
Cell culture and transfection
HEK293T and murine C2C12 myoblasts were cultured in GM consisting of DMEM (Gibco; USA) supplemented with 10% FBS (Gibco; USA) and 1% penicillin-streptomycin (Invitrogen; USA) at 37°C with 5% CO2. Differentiation of C2C12 myoblasts was induced by DM containing DMEM supplemented with 2% horse serum (Gibco; USA) and 1% penicillin-streptomycin. All cell cultures were performed at least in triplicate.
C2C12 myoblasts were cultured in 6-well plates (Corning; USA). The effects of miR-487b-3p were examined during skeletal myogenesis by transfecting pcDNA3.1(+)-miR-487b-3p (Table 2) and miR-487b-3p mimics (double-stranded RNA oligonucleotides), double-stranded negative control (NC), 2′-O-methylated miR-487b-3p inhibitors (inhibitors) and anti-negative control (anti-NC) purchased from GenePharma (GenePharma; China) (Table 3) into the C2C12 myoblasts using Lipo (Lipofectamine 2000 Invitrogen; USA) following the manufacturer's instructions. Transfection was performed when the cell density reached 70-80% confluence, and the cells were then cultured in Opti-MEM medium (Gibco; USA). After 6 h, the medium was replaced with DM for 7 days. The cells were collected at 2-day intervals.
Sequence information for RNA oligonucleotides
| Name | Sequence Name | Sequence Information (5' to 3') |
|---|---|---|
| miR-487b-3p mimics | AAUCGUACAGGGUCAUCCACUU (sense) | |
| GUGGAUGACCCUGUACGAUUUU (antisense) | ||
| miR-487b-3p | miR-487b-3p NC | UUCUCCGAACGUGUCACGUTT (sense) |
| ACGUGACACGUUCGGAGAATT (antisense) | ||
| miR-487b-3p Inhibitors | AAGUGGAUGACCCUGUACGAUU | |
| miR-487b-3p Inhibitor NC | CAGUACUUUUGUGUAGUACAA |
Cell proliferation assay
First, C2C12 myoblasts were cultured in 6-well plates (Corning; USA) with GM. When the cell density reached 70-80% confluence, pcDNA3.1(+)-miR-487b-3p, miR-487b-3p mimics, NC, miR-487b-3p inhibitors and anti-NC were separately transfected using Lipo following the manufacturer's instructions. After transfection for 6 h, the medium was changed to fresh GM for 24h and 48 h. The cells were collected at 24-h intervals. MyoD, Pax7, and PCNA were detected by RT-qPCR and western blotting to analyze C2C12 myoblast proliferation. Second, C2C12 myoblasts transfected with RNA oligonucleotides (Table 3) were seeded at 1.0×105 cells/well in a 24-well plate (Corning; USA) and cultured in GM for 24 h and 48 h. Cell growth was examined by Cell Counting Kit-8 (CCK-8) reagent (EnoGene; China) at 24-h intervals. Then, the absorbance was measured at 450 nm on a Varioskan LUX Multimode Microplate Reader (Thermo Scientific; USA). Third, a 5-ethynyl-2′-deoxyuridine (EdU) cell proliferation assay (RiboBio; China) was used to measure C2C12 myoblast proliferation according to the manufacturer's protocols. C2C12 myoblasts were seeded in a 24-well plate and transfected with RNA oligonucleotides (Table 3) and pcDNA3.1(+)-miR-487b-3p. Briefly, after being maintained in GM for 24 h, the cells were incubated for 2 h with medium containing 50μM EdU prior to immunostaining. Images were collected using a fluorescence microscope (Olympus; Japan). The ratio of EdU-positive cells was calculated as (EdU-positive cells/Hoechst stained cells) ×100%.
RNA isolation, RT-PCR and RT-q-PCR
Total RNA was extracted from HEK293T and C2C12 myoblasts using TRIzol reagent (Takara; Japan) according to the manufacturer's instructions. For mRNA quantification, 1 μL of total RNA (1000 ng/μL) was reverse transcribed into cDNA using the PrimeScript RT kit (Takara; Japan) and then quantified on an ABI Step One Plus Real-Time PCR system (Applied Biosystems; USA) with SYBR Green II Master Mix Reagent Kit (Takara; Japan) according to the manufacturer's protocols. For miR-487b-3p quantification, a miRNA stem-loop primer and a pair of primers (Table 1) were designed for miRNA reverse transcription and RT-qPCR, respectively. The miR-487b-3p stem-loop was reverse transcribed using the PrimeScript RT reagent kit and quantified by RT-qPCR using SYBR Green II Master Mix Reagent Kit. β-actin and 18S-rRNA were used as internal normalization controls. The cycling conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, and then 60°C for 1 min. Relative gene expression was determined using the 2-ΔΔCt method.
RNAi and western blotting
siRNA against mouse IRS1 and a siRNA control (a non-specific duplex) were purchased from RiboBio (China). Transfection was performed using the Lipo reagent combined with 100 nM siRNA. All procedures were performed according to the manufacturer's protocols.
Total protein was extracted from C2C12 myoblasts from each group using RIPA Lysis Buffer (Solarbio; China) supplemented with 1% PMSF (Solarbio; China) on ice for 25 min. Protein fractions were collected by centrifugation at 13,000 g at 4°C for 5 min and qualified using a BCA protein assay kit (Solarbio; China). For each line, 20 μg of proteins was separated via 8% or 12% SDS-PAGE and transferred to polyvinylidene fluoride membranes. Then, the membranes were blocked with 5% skim milk and subsequently incubated overnight at 4°C with primary antibodies against Myf5 (MW: 28 kDa; Abscience; USA; 1:1000 dilution), Mef2c (MW: 51 kDa; Abscience; USA; 1:1000 dilution), MyoG (MW: 25 kDa; Abscience; USA; 1:1000 dilution), MyoD (MW: 35 kDa; Abscience; USA; 1:1000 dilution), Pax7 (MW: 57 kDa; EnoGene; China; 1:1000 dilution), PCNA (MW: 36 kDa; EnoGene; China; 1:1000 dilution), IRS1 (MW:130 kDa; Abscience; USA; 1:1000 dilution) and β-tubulin (MW: 55 kDa; Abscience; USA; 1:1000 dilution). Then, the membranes were washed with 1×TBST and incubated with secondary antibodies (anti-rabbit IgG or anti-Goat IgG; ZSGB-BIO; China; 1:5000 dilution) conjugated to horseradish peroxidase. Protein bands were visualized using enhanced ECL reagents (Willget; China). β-tubulin was used as the loading control. Protein image acquisition was performed on the FluorChem M system (ProteinSimple; USA), and images were analyzed with Image-Pro Plus software.
Dual-luciferase reporter assay
Wild-type and mutant IRS1 3'-UTR sequences were amplified and then cloned downstream of the Renilla luciferase open reading frame. Approximately 0.2 μg of wild-type or mutant 3'-UTR vector and 100 pmol of miR-487b-3p mimics or NC were co-transfected into HEK293T cells using Lipo following the manufacturer's protocol in 24-well plates; 0.2 μg of wild-type or mutant 3'-UTR vector and 0.8 μg of pcDNA3.1(+)-miR-487b-3p plasmid or pcDNA3.1(+) plasmid were co-transfected into HEK293T cells using Lipo following the manufacturer's protocol in 24-well plates. After transfection for 48 h, relative luciferase activity was quantified using a Dual-Luciferase Reporter Assay (TransGen; China) according to the manufacturer's protocols. Firefly and Renilla luciferase imaging was performed using a Varioskan LUX Multimode Microplate Reader (Thermo-Scientific; USA), and firefly luciferase activity was normalized to that of Renilla luciferase.
Statistical analysis
All data are shown as the mean ± SEM and are based on at least three replicates for each treatment. One-way ANOVA and t-tests were used for statistical analysis in SPSS 22 software and Origin 7.5, respectively. Differences were regarded as significant at P < 0.05. *P< 0.05, and **P< 0.01.
Results
Expression profile of miR-487b-3p in Xuhuai goat
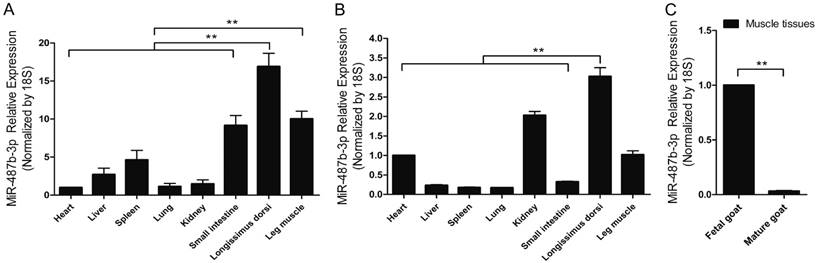
miR-487b-3p is located on Chromosome 21 and the 22 nucleotides comprising goat mature miR-487b-3p are completely conserved across species according to the miRBase database (www.mirbase.org/) and UCSC(http://genome.ucsc.edu/). RT-qPCR analysis of miR-487b-3p levels was performed in various fetal and mature goat tissues to investigate the expression profile of miR-487b-3p in goat tissues and explore the potential function of goat miR-487b-3p. Difference analysis showed that miR-487b-3p expression was high in skeletal muscle tissue and was significantly different from that in most other tissues in fetal goat (Figure 1A) and mature goat (Figure 1B). These results suggested that miR-487b-3p may have potential roles in skeletal muscle development and myogenesis. Moreover, a significant difference in miR-487b-3p expression was observed between muscle tissues from fetal and mature Xuhuai goat (Figure 1C), implying that miR-487b-3p is differentially expressed in muscle cells. Thus, miR-487b-3p was selected as a candidate miRNA for further studies of skeletal muscle myogenesis.
miR-487b-3p negatively regulates myoblast differentiation
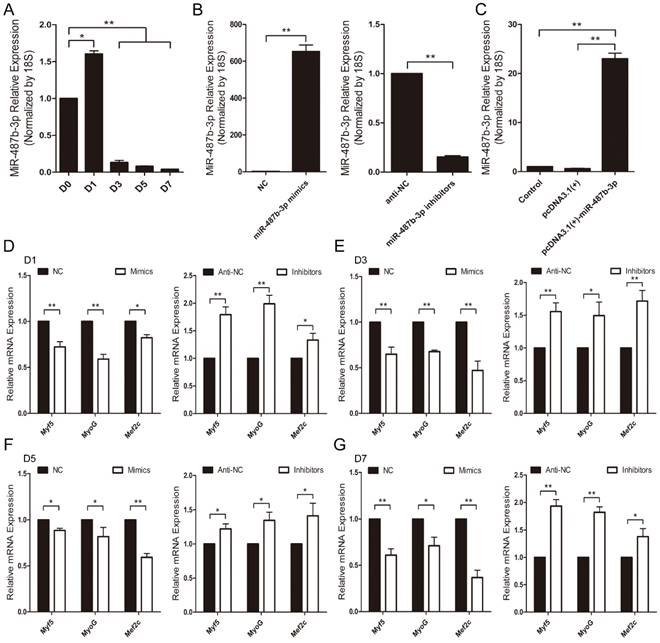
Mouse C2C12 myoblasts were chosen as a myogenesis model to investigate the potential role of miR-487b-3p [33]. First, we examined miR-487b-3p expression levels during C2C12 myoblast differentiation. miR-487b-3p levels increased from D0 to D1 and decreased from D1 to D7 during C2C12 cell differentiation (Figure 2A), which was one period of myogenesis. We detected the effects of a synthetic RNA duplex of miR-487b-3p mimics, inhibitors and the constructed pcDNA3.1(+)-miR-487b-3p plasmid by introducing these reagents into C2C12 myoblasts and then inducing myoblast differentiation. The delivery of these duplexes was highly effective, increasing or reducing cellular miR-487b-3p levels by >10-fold (Figure 2B, C).
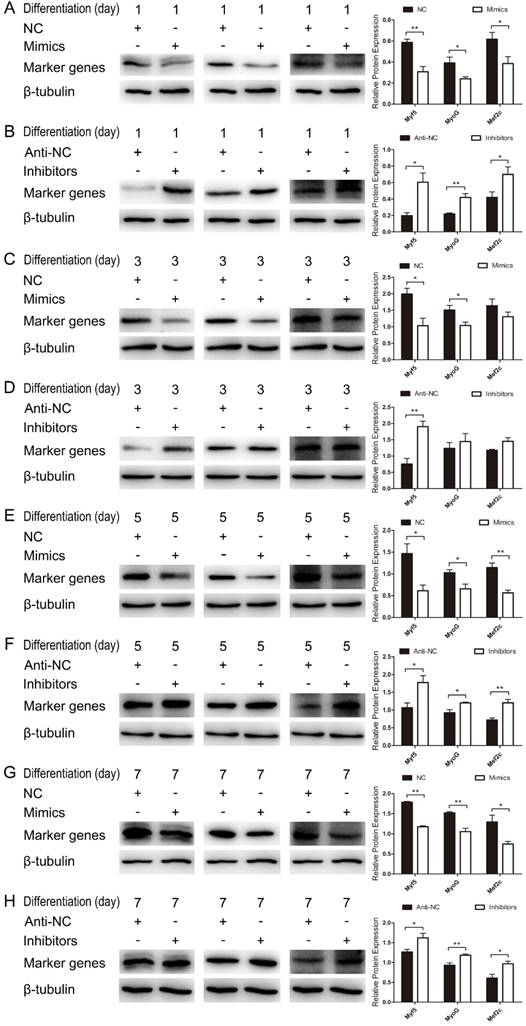
We further examined the function of miR-487b-3p in myogenic differentiation by introducing miR-487b-3p mimics, NC, miR-487b-3p inhibitors or anti-NC into C2C12 myoblasts. C2C12 myoblasts were transfected with miR-487b-3p mimics or inhibitors in GM, and the cells were then induced to differentiate in DM. Cells were collected for RT-qPCR and western blotting analysis against Myf5, MyoG, and Mef2c, which are markers of differentiation, on the 1st, 3rd, 5th and 7th day of differentiation. miR-487b-3p overexpression markedly down-regulated Myf5, MyoG and Mef2c mRNA levels at D1, D3, D5 and D7 in DM. In contrast, miR-487b-3p inhibition enhanced the mRNA expression of myogenic differentiation markers (Myf5, MyoG and Mef2c) at D1, D3, D5 and D7 in DM (Figure 2D-G). Similarly, a significant decrease Myf5, MyoG and Mef2c protein levels was observed on days 1, 3, 5 and 7 of differentiation after miR-487b-3p overexpression, whereas miR-487b-3p inhibition enhanced the protein expression of myogenic differentiation markers, which was accompanied by the enhanced expression of myogenic marker (Myf5, MyoG and Mef2c) mRNA on days 1, 3, 5 and 7 of differentiation (Figure 3). Taken together, these results indicated that miR-487b-3p plays a negative role in the myogenic differentiation erentiation of C2C12 myoblasts.
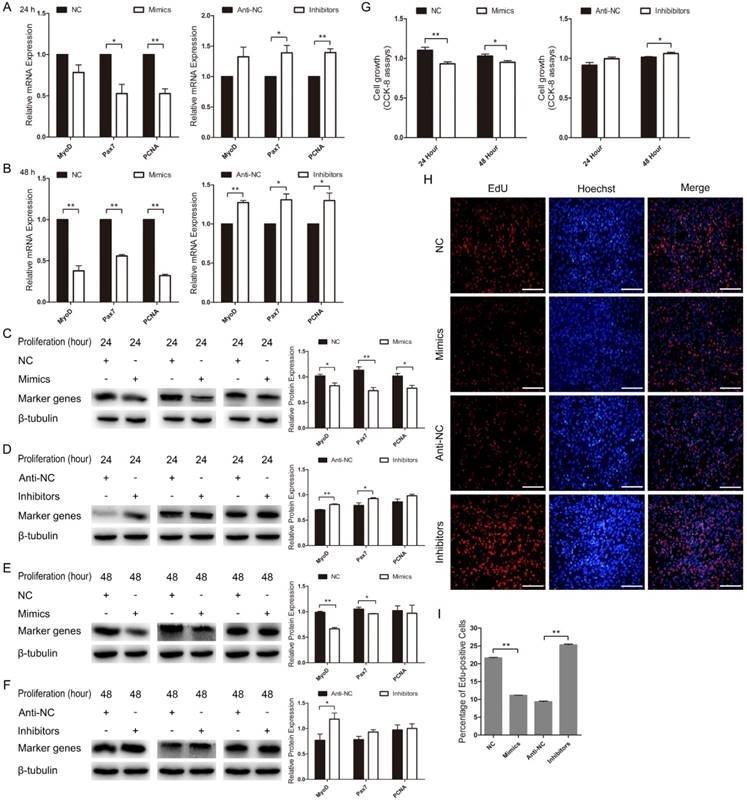
miR-487b-3p suppresses C2C12 myoblast proliferation
Next, we examined the role of miR-487b-3p in C2C12 myoblast proliferation. We found that MyoD mRNA expression decreased in miR-487b-3p mimics-treated cells at 24 h and 48 h but increased with miR-487b-3p inhibitors treatment at 24 h and 48 h in GM. The mRNA levels of Pax7 and PCNA, which are markers of proliferation, significantly decreased in miR-487b-3p mimics-treated cells at 24 h and 48 h but significantly increased with miR-487b-3p inhibitors treatment at 24 h and 48 h in GM (Figure 4A, B). Similarly, MyoD, Pax7 and PCNA protein levels significantly decreased in the miR-487b-3p mimics-treated group but significantly increased in the miR-487b-3p inhibitors-treated group at 24 h (Figure 4C, D) and 48 h in GM (Figure 4E, F). Second, the CCK-8 assay further confirmed that miR-487b-3p mimics inhibited C2C12 myoblast proliferation and that miR-487b-3p inhibitors promoted C2C12 myoblast proliferation at 24 h and 48 h (Figure 4G). Moreover, we also examined the role of miR-487b-3p in C2C12 myoblast proliferation using EdU cell proliferation assays. The number of EdU-positive cells significantly decreased in the miR-487b-3p mimics-treated group compared with that in the miR-487b-3p NC counterparts but was substantially increased in miR-487b-3p inhibitors-treated cells compared with that in the miR-487b-3p anti-NC controls (Figure 4H, I). Together, these results indicated that miR-487b-3p suppresses C2C12 myoblast proliferation.
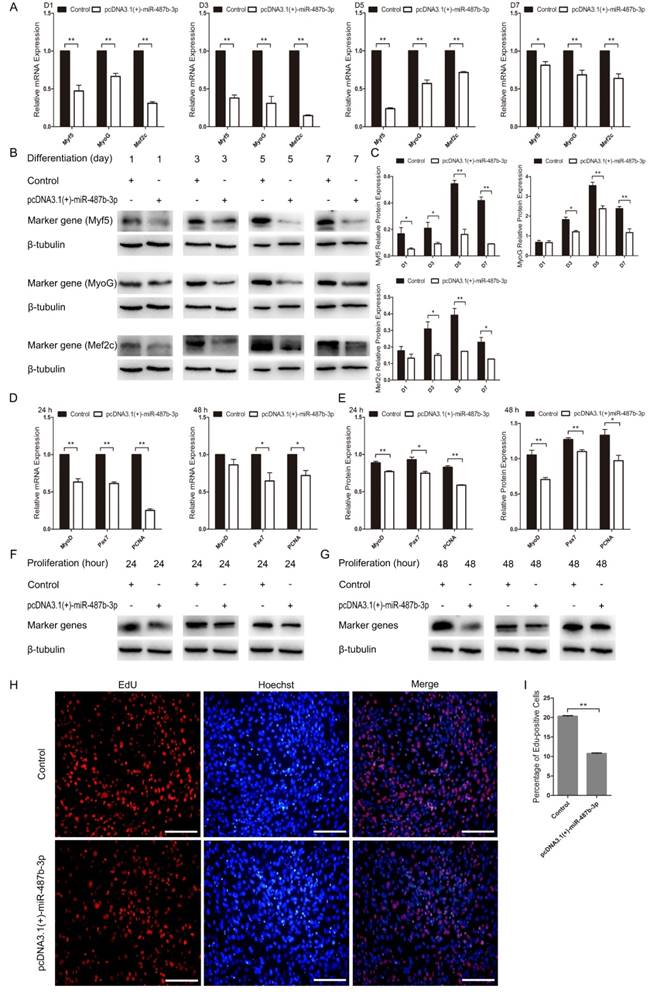
Ectopic miR-487b-3p expression inhibits C2C12 myoblast differentiation and proliferation
We constructed the pcDNA3.1(+)-miR-487b-3p plasmid to detect the role of miR-487b-3p precursor during C2C12 myoblast proliferation and differentiation. pcDNA3.1(+)-miR-487b-3p was transfected into C2C12 myoblasts to increase the expression of miR-487b-3p. miR-487b-3p expression was higher in pcDNA3.1(+)-miR-487b-3p-transfected cells than in the control or pcDNA3.1(+) plasmid groups, as detected by RT-qPCR (Figure 2C). Next, pcDNA3.1(+)-miR-487b-3p was transfected into C2C12 myoblasts in GM, and then the cells were induced to differentiate in DM. Myf5, MyoG and Mef2c mRNA (Figure 5A) and protein (Figure 5B, C) levels were detected to assess the effect of miR-487b-3p on C2C12 myoblasts. pcDNA3.1(+)-miR-487b-3p expression appreciably decreased the expression of Myf5, MyoG and Mef2c at D1, D3, D5 and D7 in DM and inhibited C2C12 myoblast differentiation.
miR-487b-3p expression profile in the developing Xuhuai goat. (A) Tissue distribution of miR-487b-3p detected by RT-qPCR in fetal goat. (B) Tissue distribution of miR-487b-3p detected by RT-qPCR in mature goat. (C) The differences in muscle tissue miR-487b-3p expression between fetal and mature goat. The results are shown as the mean ± SEM of three independent replicates. One-way ANOVA and t-tests were used for statistical analysis. Asterisks indicate significant differences. ** P<0.01.
miR-487b-3p expression and regulation during myogenic differentiation of C2C12 myoblasts. (A) Relative expression of miR-487b-3p at D0, D1, D3, D5, and D7 was examined by RT-qPCR; D0, 1, 3, 5, and 7 indicate DM for 0, 1st, 3rd, 5th, and 7th day, respectively. (B) miR-487b-3p expression after transfection with miR-487b-3p mimics (Mimics), negative control (NC), 2'-O-methylated oligonucleotides against miR-487b-3p (Inhibitors), and single-stranded negative control (anti-NC) determined by RT-qPCR 48 h after transfection. (C) miR-487b-3p expression after transfection with pcDNA3.1(+) plasmid and pcDNA3.1(+)-miR-487b-3p determined by RT-qPCR 48 h after transfection. (D-G) Myf5, MyoG, and Mef2c mRNA expression at D1 (D), D3 (E), D5 (F), and D7 (G) after transfection with Mimics, NC, Inhibitors, and anti-NC determined by RT-qPCR in DM. The results are shown as the mean ± SEM of three independent replicates. One-way ANOVA and t-tests were used for statistical analysis. Asterisks indicate significant differences. * P<0.05, ** P<0.01.
miR-487b-3p inhibits myogenic differentiation in C2C12 myoblasts. (A-G) Protein expression of marker genes (Myf5 (28 kDa; 1:1000 dilution), MyoG (25 kDa; 1:1000 dilution) and Mef2c (51 kDa; 1:1000 dilution)) at D1 (A, B), D3 (C, D), D5 (E, F) and D7 (G, H) was measured after transfection with miR-487b-3p mimics (Mimics), negative control (NC), 2'-O-methylated oligonucleotides against miR-487b-3p (Inhibitors) and single-stranded negative control (Anti-NC) in DM. β-tubulin (55 kDa; 1:1000 dilution) was used as an internal control. D1, 3, 5, and 7 indicate DM for 1, 3, 5, and 7 days, respectively. The results are shown as the mean ± SEM of three independent replicates and t-tests were used for statistical analysis. Asterisks indicate significant differences. * P<0.05, ** P<0.01.
miR-487b-3p suppresses C2C12 myoblast proliferation. (A, B) MyoD, Pax7, and PCNA mRNA expression after transfection with miR-487b-3p mimics (Mimics), negative control (NC), 2'-O-methylated oligonucleotides against miR-487b-3p (Inhibitors), and single-stranded negative control (Anti-NC) in GM at 24 h and 48 h. (C-F) MyoD (35 kDa; 1:1000 dilution), Pax7 (57 kDa; 1:1000 dilution), and PCNA (36 kDa; 1:1000 dilution) protein expression after transfection with miR-487b-3p mimics, NC, Inhibitors and Anti-NC in GM at 24 h (C, D) and 48 h (E, F). β-tubulin (55 kDa; 1:1000 dilution) was used as an internal control. (G) CCK-8 assay of C2C12 myoblasts at 24 h and 48 h after transfection with miR-487b-3p Mimics, NC, miR-487b-3p Inhibitors and Anti-NC. (H) Representative images of EdU assay of C2C12 myoblasts at 24 h after transfection with Mimics, NC, Inhibitors, Anti-NC in GM. Bars, 100 μm. (I) Quantification of EdU-positive cells(n=6). The results are shown as the mean ± SEM of three independent replicates. One-way ANOVA and t-tests were used for statistical analysis. Asterisks indicate significant differences. * P<0.05, ** P<0.01.
pcDNA3.1(+)-miR-487b-3p was transfected into C2C12 myoblasts in GM to explore the role of the miR-487b-3p precursor during C2C12 myoblast proliferation. MyoD, Pax7 and PCNA mRNA and protein expression were examined after transfection with pcDNA3.1(+)-miR-487b-3p at 24 h and 48 h in GM. The pcDNA3.1(+)-miR-487b-3p plasmid significantly down-regulated MyoD, Pax7 and PCNA expression at 24 h (Figure 5D, E) and 48 h (Figure 5F, G). Meanwhile, EdU cell proliferation assay showed that pcDNA3.1(+)-miR-487b-3p decreased the proportion of EdU-positive cells (Figure 5H, I). Taken together, these results suggested that the miR-487b-3p precursor was able to inhibit C2C12 myoblast differentiation and proliferation, consistent with the role of miR-487b-3p mimics in C2C12 myoblast differentiation and proliferation.
Ectopic miR-487b-3p expression inhibits proliferation and differentiation in C2C12 myoblasts. (A) Myf5, MyoG, and Mef2c expression at D1, D3, D5, and D7 after transfection with pcDNA3.1 (+)-miR-487b-3p determined by RT-qPCR in DM. (B, C) Myf5 (28 kDa; 1:1000 dilution), MyoG (25 kDa; 1:1000 dilution) and Mef2c (51 kDa; 1:1000 dilution) protein levels were measured following overexpression of miR-487b-3p with pcDNA3.1(+)-miR-487b-3p in DM at D1, D3, D5 and D7. β-tubulin (55 kDa; 1:1000 dilution) was used as an internal control. (D) MyoD, Pax7, and PCNA mRNA levels after transfection with pcDNA3.1 (+)-miR-487b-3p in GM at 24 h and 48 h. (E) Relative MyoD, Pax7, and PCNA protein expression after transfection with pcDNA3.1 (+)-miR-487b-3p in GM at 24 h and 48 h. (F, G) MyoD (35 kDa; 1:1000 dilution), Pax7 (57 kDa; 1:1000 dilution), and PCNA (36 kDa; 1:1000 dilution) protein expression after transfection with pcDNA3.1 (+)-miR-487b-3p in GM at 24 h and 48 h. β-tubulin (55 kDa; 1:1000 dilution) was used as an internal control. (H) Representative images of EdU assays of C2C12 myoblasts at 24 h after transfection with pcDNA3.1 (+)-miR-487b-3p. Bars, 100 μm. (I) Quantification of EdU-positive cells (n=6). D1, 3, 5, and 7 indicate DM for 1, 3, 5, and 7 days, respectively. The results are shown as the mean ± SEM of three independent replicates. One-way ANOVA and t-tests were used for statistical analysis. Asterisks indicate significant differences. * P<0.05, ** P<0.01.
miR-487b-3p directly targets the 3'-UTR of IRS1
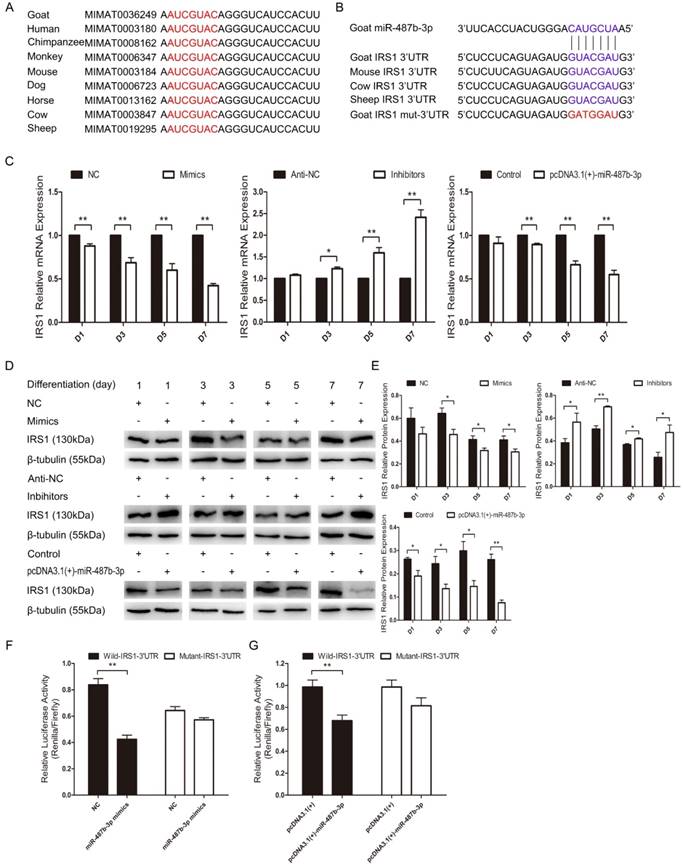
We explored the mechanisms of miR-487b-3p by searching for miR-487b-3p target genes. According to bioinformatics predictions (Target Scan: http://www.targetscan.org/vert_71; David: https://david.ncifcrf.gov/; KEGG Analysis: http://www.genome.jp/kegg/pathway.html), IRS1 was among the target genes of miR-487b-3p. We found that miR-487b-3p seed sequences were conserved and that IRS1 mRNA contained a highly conserved binding site capable of binding to the seed sequence of miR-487b-3p (Figure 6A, B). IRS1 plays a key role in transmitting signals from IGF-1 receptors. Some research has suggested that IRS1-Akt2 signaling plays an important role in myoblast differentiation and glucose metabolism [34,35]. IRS1 is also known to enhance gene transcription by binding to RNA polymerases I and II and can have a positive effect on cell growth and proliferation [36,37].
We investigated whether IRS1 is a direct target gene of miR-487b-3p by introducing miR-487b-3p mimics, NC, miR-487b-3p inhibitors, anti-NC and pcDNA3.1(+)-miR-487b-3p plasmid into C2C12 myoblasts in GM and then inducing cell differentiation in DM. IRS1 mRNA and protein expression was detected to assess the relationship between IRS1 and miR-487b-3p. The results showed that miR-487b-3p up-regulation markedly decreased IRS1 mRNA and protein expression on days 1, 3, 5 and 7 during C2C12 myoblast differentiation. Conversely, miR-487b-3p inhibition significantly accelerated IRS1 mRNA (Figure 6C) and protein expression (Figure 6D, E). These results indicated that miR-487b-3p induction repressed IRS1 mRNA and protein expression in C2C12 myoblasts. Furthermore, we constructed double-luciferase reporters that included a fragment of wild-type or mutant IRS1 3'-UTR. The miR-487b-3p mimics or NC with pcDNA3.1(+) or pcDNA3.1(+)-miR-487b-3p were co-transfected with wild-type or mutant IRS1 3'-UTR double-luciferase reporters into HEK-293T cells, a model cell line that has stable transfection efficiency. The results showed that, compared with NC, miR-487b-3p significantly reduced the luciferase activity of the wild-type IRS1 reporter, whereas no obvious reduction in luciferase activity was observed with the mutant IRS1 reporter (Figure 6F). Similar results were obtained after transfection with pcDNA3.1(+)-miR-487b-3p and pcDNA3.1(+) (Figure 6G). This result confirmed that miR-487b-3p directly targets the 3′-UTR of IRS1. In conclusion, these results suggested that miR-487b-3p regulates IRS1 mRNA and protein expression by directly targeting the mRNA 3'-UTR.
Silencing IRS1 expression represses C2C12 myoblast proliferation and differentiation
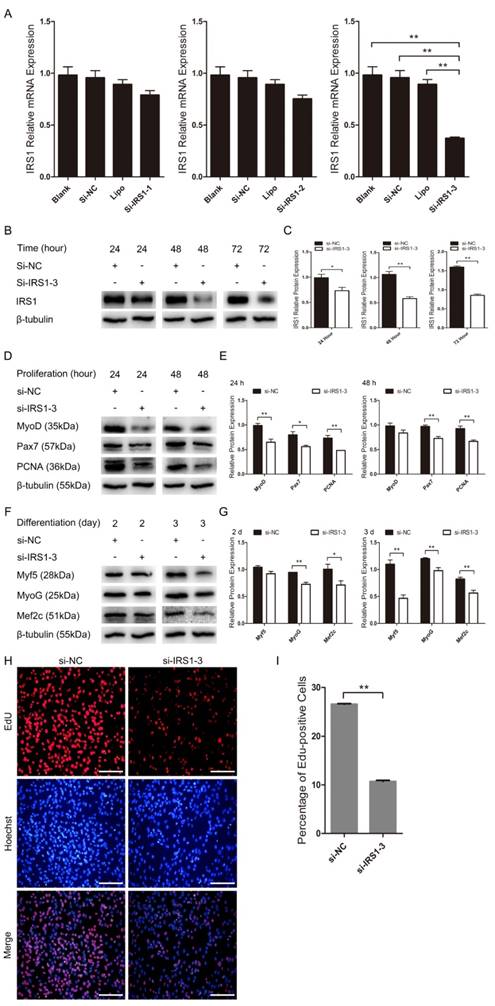
We further examined the role of IRS1 during C2C12 myoblast proliferation and differentiation by silencing endogenous IRS1 in C2C12 myoblasts using synthetic siRNA, the sequences of which were also highly conserved in goat. IRS1 mRNA expression was considerably lower in the si-IRS1-3 group than in the blank, Lipo, si-NC, si-IRS1-1 and si-IRS1-2 groups as detected by RT-qPCR (Figure 7A). Moreover, si-IRS1-3 protein expression was significantly diminished at 24 h, 48 h and 72 h in GM (Figure 7B, C). These results showed that endogenous IRS1 expression was successfully inhibited by si-IRS1-3 in C2C12 myoblasts. Furthermore, IRS1 reduction by transfection with si-IRS1-3 repressed C2C12 myoblast proliferation and differentiation and was accompanied by the down-regulation of proliferation and differentiation marker gene expression. si-IRS1-3 down-regulated MyoD, Pax7 and PCNA protein expression at 24 h and 48 h in GM (Figure 7D, E), which was very similar to the impact of miR-487b-3p overexpression during C2C12 myoblast proliferation. Meanwhile, Myf5, MyoG and Mef2c protein expression was reduced after transfection with si-IRS1-3 on days 2 and 3 of C2C12 myoblasts in DM (Figure 7F, G), consistent with the impact of miR-487b-3p overexpression during C2C12 myoblast differentiation. Moreover, we also examined the effect of si-IRS1-3 on C2C12 myoblast proliferation using EdU cell proliferation assays. Compared with the si-NC control, si-IRS1-3 decreased the proportion of EdU-positive cells (Figure 7H, I). These results indicated that silencing endogenous IRS1 expression repressed C2C12 myoblast proliferation and differentiation.
Discussion
miR-487b-3p expression levels were markedly higher in skeletal muscle than in other examined tissues, which was consistent with our previous high-throughput sequencing results in Xuhuai goat skeletal muscle [32]. These results suggested that miR-487b-3p might be involved in skeletal muscle myogenesis, similar to other muscle-specific miRNAs or muscle non-specific miRNAs [38,39]. Recent work has shown that miR-487b overexpression can suppress the tumorigenesis of colon cancer cells by targeting GRM3 [40], which may be related to the high levels of miR-487b-3p expression in fetal small intestine, as shown in our research (Figure 1A). Moreover, inhibiting miR-487b expression may increase cell invasion, tumorigenicity and proliferation. Consequently, miR-487b expression delays cell growth in cultured lung cancer cells in vitro [41]. However, miR-487b can promote the proliferation, migration, invasion and tube formation of umbilical vein endothelial cells by targeting THSB1 in angiogenesis, suggesting that miR-487b can facilitate recovery after vascular injury. This study also indicated that miR-487b can serve as a potential diagnostic marker for cardiovascular diseases [42]. These studies have shown different functions of miR-487b in cancers or cardiovascular diseases.
miR-487b-3p suppresses IRS1 expression by directly targeting its 3'-UTR. (A) Conservation of miR-487b-3p and its binding site (red) in nine species. (B) The predicted binding site (blue) and mutated site (red) of miR-487b-3p in the 3′-UTR of goat IRS1 and other animals. (C) IRS1 mRNA expression during C2C12 myoblast differentiation after transfection with miR-487b-3p mimics, miR-487b-3p inhibitors or pcDNA3.1(+)-miR-487b-3p by RT-qPCR; total RNA was harvested at D1, D3, D5, and D7 after transfection. (D, E) IRS1 (130 kDa; 1:1000 dilution) protein expression was examined after transfection with miR-487b-3p mimics, miR-487b-3p inhibitors or pcDNA3.1(+)-miR-487b-3p in DM at D1, D3, D5, and D7. β-tubulin (55 kDa; 1:1000 dilution) was used as an internal control. F and G, Dual-luciferase activity assay of wild-type or mutant 3′-UTR of IRS1. (F) miR-487b-3p mimics (Mimics) or NC were co-transfected with wild-type or mutant IRS1 3′-UTR luciferase reporters in HEK293T cells. (G) pcDNA3.1(+)-miR-487b-3p or pcDNA3.1(+) plasmid were co-transfected with wild-type or mutant IRS1 3′-UTR luciferase reporters in HEK293T cells. D1, 3, 5, and 7 indicate DM for 1, 3, 5, and 7 days, respectively. The results are shown as the mean ± SEM of three independent replicates. One-way ANOVA and t-tests were used for statistical analysis. Asterisks indicate significant differences. * P<0.05, ** P<0.01.
IRS1 knockdown inhibits proliferation and differentiation in cultured C2C12 myoblasts. (A) IRS1 expression was measured after transfection with siRNA control (si-NC), Lipo (lipofectamine 2000), si-IRS1-1, si-IRS1-2 and si-IRS1-3 in GM at 24 h. (B, C) IRS1 (130 kDa; 1:1000 dilution) protein expression was examined after transfection with siRNA control (si-NC), si-IRS1-3 in GM at 24 h, 48 h and 72 h. (D, E) MyoD (35 kDa; 1:1000 dilution), Pax7 (57 kDa; 1:1000 dilution), and PCNA (36 kDa; 1:1000 dilution) protein expression was examined after transfection with siRNA control (si-NC), si-IRS1-3 in GM at 24 h (D) and 48 h (E), respectively. (F, G) Myf5 (28 kDa; 1:1000 dilution), MyoG (25 kDa; 1:1000 dilution), and Mef2c (51 kDa; 1:1000 dilution) protein expression was examined after transfection with siRNA control (si-NC), si-IRS1-3 in DM at D2 (F) and D3 (G). β-tubulin (55 kDa; 1:1000 dilution) was used as an internal control. D2, 3 indicate DM for 2 and 3 days. (H) Representative images of EdU assays of C2C12 myoblasts at 24 h after transfection with siRNA control (si-NC), si-IRS1-3. Bars, 100 μm. (I) Quantification of EdU-positive cells (n=6). The results are shown as the mean ± SEM of three independent replicates. One-way ANOVA and t-tests were used for statistical analysis. Asterisks indicate significant differences. * P<0.05, ** P<0.01
miR-487b-3p is a member of the miR-487b family. miR-487b-3p expression was down-regulated during C2C12 myoblast differentiation (Figure 2A), much like miR-221/222 and miR-125b, whose expression has been reported to decrease during myogenic differentiation and to negatively regulate myoblast differentiation [43,22]. These phenomena suggested that miR-487b-3p may also be relevant to skeletal muscle myogenesis. Here, our studies showed that miR-487b-3p overexpression represses the expression of functional genes related to myogenic proliferation (MyoD, Pax7, and PCNA) and differentiation (Myf5, MyoG, and Mef2c), suggesting that miR-487b-3p up-regulation inhibits skeletal muscle proliferation and differentiation; however, miR-487b-3p inhibition accelerated the expression of functional genes related to proliferation (MyoD, Pax7, and PCNA) and differentiation (Myf5, MyoG, and Mef2c) at both the mRNA and protein level, which may show that miR-487b-3p down-regulation can promote skeletal muscle proliferation and differentiation. Meanwhile, EdU assays showed that miR-487b-3p overexpression decreased the proportion of EdU-positive cells, whereas miR-487b-3p inhibition increased the proportion of EdU-positive cells. These results indicate that miR-487b-3p inhibits skeletal muscle proliferation and differentiation. In contrast to miR-487b-3p, miR-214 expression has been shown to increase and to positively regulate C2C12 myoblast proliferation and differentiation [44]. More interestingly, some miRNAs can accelerate myoblast proliferation while negatively regulating myoblast differentiation, or vice versa, such as miR-192 [45] and miR-206 [18]. These facts indicated that different miRNAs play diverse regulatory roles upon skeletal muscle myogenesis.
The PI3K/Akt and MAPK/Erk pathways couple signals from cell-surface receptors to transcription factors, which regulate gene expression. Some studies have reported an effect on PI3K/Akt and MAPK/Erk pathways during myogenesis and myotube fusion [46,47]. Activation or inhibition of PI3K/Akt and MAPK/Erk signaling can stimulate or suppress myogenic cell proliferation, differentiation and survival [48,49]. IRS1, which functions as a crucial signaling effector in the PI3K/Akt and MAPK/Erk pathways [50], was identified as a target gene of miR-487b-3p by bioinformatics analysis, RT-qPCR, western blotting and dual-luciferase reporter gene assay in our study. Previous studies indicated that IRS1 can harmonize skeletal muscle growth and metabolism via the Akt and AMPK pathways and serve as a key factor in inducing myoblast proliferation and myotube hypertrophy by increasing phospho-Akt levels [30,51], suggesting that the miR-487b-3p—IRS1 relationship is a critical regulator of skeletal muscle myogenesis. Here, we used siRNA-mediated IRS1 knockdown experiments to investigate the role of IRS1 in myoblast proliferation and differentiation. We found that knocking down IRS1 suppressed the expression of functional genes related to proliferation (MyoD, Pax7, and PCNA) and differentiation (Myf5, MyoG, and Mef2c) at the mRNA and protein level. EdU assays also showed compared with the si-NC control, IRS1 knockdown decreased the number EdU-positive cells. These results revealed that IRS1 plays a critical role in the regulation of myoblast proliferation and differentiation via the PI3K/Akt and MAPK/Erk pathways.
In conclusion, our results indicated that miR-487b-3p is important for myogenic proliferation and differentiation and can inhibit C2C12 myoblast proliferation, differentiation and myogenesis. Myogenic proliferation and differentiation were delayed by miR-487b-3p overexpression and were promoted by miR-487b-3p inhibition. We also determined that miR-487b-3p can directly target the IRS1 gene during myogenesis and that IRS1 knockdown significantly represses myogenic proliferation and differentiation. These results reveal a miRNA-related regulatory link between miR-487b-3p and the PI3K/Akt and MAPK/Erk pathways during myogenic proliferation and differentiation in which IRS1 plays a vital role, offering crucial insights into the role of miR-487b-3p in skeletal muscle myogenesis. Subsequent studies will replace C2C12 myoblasts with satellite cells from goat skeletal muscles to further characterize the role of miR-487b-3p in skeletal muscle myogenesis (including proliferation and differentiation).
Abbreviations
IRS1: insulin receptor substrate 1; MRFs: myogenic regulatory factors; Pax3: paired box protein pax-3; Pax7: paired box protein pax-7; MyoD: myogenic differentiation 1; Myf5: myogenic factor 5; Myf6: myogenic factor 6; MyoG: myogenin; Mef2c: myocyte-specific enhancer factor 2c; IGF-I: insulin-like growth factor-1; TGF-β1: transforming growth factor beta 1; Hox-A11: homeobox protein A11; HEK293T: human embryonic kidney 293T cells; GM: growth medium; DM: differentiation medium; FBS: fetal bovine serum; DMEM: Dulbecco's modified eagle's medium; Mimics: miR-487b-3p mimics—double-stranded RNA oligonucleotides; NC: double-stranded negative control; Inhibitors: 2′-O-methyl antisense oligonucleotides against miR-487b-3p; Anti-NC: anti-negative control; CCK-8: cell counting kit-8 reagent; EdU: 5-Ethynyl-2′-deoxyuridine; RT-PCR: reverse transcription PCR; RT-qPCR: quantitative real-time PCR; RIPA: radio immunoprecipitation assay; PMSF: phenylmethanesulfonyl fluoride; SDS-PAGE: sodium dodecyl sulfate—polyacrylamide gel electrophoresis; TBST: tris-buffered saline and tween 20; ANOVA: analysis of variance; GRM3: glutamate metabotropic receptor 3; THBS1: thrombospondin 1.
Acknowledgements
This work was supported by grants from the Science and Technology Planning Project of Jiangsu Province (No.SZ-XZ2017016; No.BN2015027), the Science and Technology Planning Project of Xuzhou City (No.KC16NX073; No.KC15N0013), the Natural Science Foundation of Jiangsu Province (No. BK20160217), the Applied Basic Research Program of Qinghai Province (No. 2014-ZJ-710), and Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author Contributions
Jiaoyan Tan, Qi Qi, Lingzhi Yang prepared reagents, Yanhong Wang, Hong Chen collected materials, Chunlei Zhang and Linyong Hu provided technical assistance; Xingtang Fang proof-reading the article. All authors approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Current Opinion in Cell Biology. 2007;19:628-633
2. Yaffe D, Feldman M. The formation of hybrid multinucleated muscle fibers from myoblasts of different genetic origin. Developmental Biology. 1965;11:300-317
3. Glass DJ. Molecular mechanisms modulating muscle mass. Trends in Molecular Medicine. 2003;9:344-350
4. Abmayr SM, Balagopalan L, Galletta BJ, Hong SJ. Cell and molecular biology of myoblast fusion. International Review of Cytology. 2003;225:33-89
5. Shin J, McFarland DC, Velleman SG. Heparan sulfate proteoglycans, Syndecan-4 and Glypican-1, differentially regulate myogenic regulatory transcription factors and paired box 7 expression during turkey satellite cell myogenesis. Poultry Science. 2012;91:201-207
6. Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annual Review of Cell and Developmental Biology. 2007;23:645-673
7. Yablonka-Reuveni Z, Rivera AJ. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Developmental Biology. 1994;164:588-603
8. Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351-1359
9. Moretti I, Ciciliot S, Dyar KA, Abrahsm R, Murqia M, Agatea L. et al. MRF4 negatively regulates adult skeletal muscle growth by repressing MEF2 activity. Nature Communications. 2016;7:12397
10. Brunetti A, Goldfine ID. Role of Myogenin in myoblast differentiation and its regulation by fibroblast growth factor. The Journal of Biological Chemistry. 1990;265:5960-5963
11. McDermott JC, Cardoso MC, Yu YT, Andres V, Leifer D, Krainc D. et al. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Molecular and Cellular Biology. 1993;13:2564-2577
12. Wang XW, Wilfred BR, Xie K, Jennings MH, Hu YH, Stromberg AJ. et al. Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biology. 2010;7:373-380
13. Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B. et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nature Medicine. 2007;13:486-491
14. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genetics. 2006;38:228-233
15. Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S. et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells bytargeting Prdm16. Cell Metabolism. 2013;17:210-224
16. Nariyama M, Mori M, Shimazaki E, Ando H, Ohnuki Y, Abo T. et al. Functions of miR-1 and miR-133a during the postnatal development of masseter and gastrocnemius muscles. Molecular and Cellular Biochemistry. 2015;407:17-27
17. Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Molecular and Cellular Biology. 2011;31(1):203-214
18. Dai Y, Wang YM, Zhang WR, Liu XF, Li X, Ding XB, at al. The role of microRNA-1 and microRNA-206 in the proliferation and differentiation of bovine skeletal muscle satellite cells. In Vitro Cellular & Developmental Biology-Animal. 2016;52:27-34
19. Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G. et al. Transforming growth factor-β-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Research. 2008;36:2690-2699
20. Wong CF, Tellam R. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. The Journal of Biological Chemistry. 2008;283:9836-9643
21. Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J. et al. NF-kB-YY1-miR-29 Regulatory Circuitry in Skeletal Myoge-nesis and Rhabdomyosarcoma. Cancer Cell. 2008;14:369-381
22. Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. The Journal of Cell Biology. 2011;192:69-81
23. Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M. et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mamma-lian myoblast differentiation. Nature Cell Biology. 2006;8:278-284
24. Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent Regulation of the poly-comb protein Ezh2 in skeletal muscle and embryonic stem cells. Molecular Cell. 2009;36:61-74
25. He J, Wang F, Zhang P, Li W, Wang J, Li J. et al. miR-491 inhibits skeletal muscle differentiation through targeting myomaker. Archives of Biochemistry and Biophysics. 2017;625-626:30-38
26. Ames HM, Yuan M, Vizcaíno MA, Yu W, Rodriguez FJ. MicroRNA profiling of low-grade glial and glioneuronal tumors shows an independent role for cluster 14q32.31 member miR-487b. Modern pathology. 2017;30:204-216
27. Welten SM, Bastiaansen AJ, de Jong RC, de Vries MR, Peters EA, Boonstra MC. et al. Inhibition of 14q32 MicroRNAs miR-329, miR-487b, miR-494, and miR-495 increases neovascularization and blood flow recovery after ischemia. Circulation Research. 2014;115:696-708
28. Xiang Y, Eyers F, Herbert C, Tay HL, Foster PS, Yang M. MicroRNA-487b is a negative regulator of macrophage activation by targeting IL-33 production. Journal of Immunology. 2016;196:3421-3428
29. Long YC, Cheng Z, Copps KD, White MF. Insulin Receptor Substrates Irs1 and Irs2 Coordin-ate Skeletal Muscle Growth and Metabolism via the Akt and AMPK Pathways. Molecular and Cellular Biology. 2011;31:430-441
30. Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Proliferation. 2017;50:e12389
31. Li Z, Levine KM, Bahreini A, Wang P, Chu D, Park BH. et al. Upregulation of IRS1 enhances IGF1 response in Y537S and D538G ESR1 mutant breast cancer cells. Endocrinology. 2018;159:285-296
32. Wang Y, Zhang C Fang X, Zhao Y, Chen X, Sun J. et al. Identification and Profiling of microRNAs and their target genes from developing caprine skeletal muscle. PLoS One. 2014;9:e96857
33. Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725-727
34. Karlsson HK, Zierath JR. Insulin signaling and glucosetransport in insulin resistant human skeletal muscle. Cell Biochemistry and Biophysics. 2007;48:103-113
35. Eckstein SS, Weigert C, Lehmann R. Divergent Roles of IRS (Insulin Receptor Substrate) 1 and 2 in Liver and Skeletal Muscle. Current Medicinal Chemistry. 2017;24:1827-1852
36. Reiss K, Del VL, Lassak A, Trojanek J. Nuclear IRS-1 and cancer. Journal of Cellular Physiology. 2012;227:2992-3000
37. Chen J, Wu A, Sun H, Drakas R, Garofalo C, Cascio S. et al. Functional significance of type 1 insulin-like growth factor-mediated nuclear translocation of the insulin receptor substrate-1 and beta-catenin. The Journal of Biological Chemistry. 2005;280:29912-29920
38. Horak M, Novak J, Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Developmental Biology. 2016;410:1-13
39. Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiological Reviews. 2011;91:827-887
40. Yi H, Geng L, Black A, Talmon G, Berim L, Wang J. The miR-487b-3p/GRM3/TGF beta signaling axis is an important regulator of colon cancer tumorigenesis. Oncogene. 2017;36:3477-3489
41. Xi S, Xu H, Shan J, Tao Y, Hong JA, Inchauste S. et al. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. The Journal of Clinical Investigation. 2013;123:1241-1261
42. Feng N, Wang Z, Zhang Z, He X, Wang C, Zhang L. miR-487b promotes human umbilical vein endothelial cell proliferation, migration, invasion and tube formation through regulating THBS1. Neuroscience Letters. 2015;591:1-7
43. Cardinali B, Castellani L, Fasanaro P, Basso A, Alemà S, Martelli F. et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4:e7607
44. Feng Y, Cao JH, Li XY, Zhao SH. Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts. Cell Biochemistry and function. 2011;29:378-383
45. Zhao Q, Kang Y, Wang HY, Guan WJ, Li XC, Jiang L. et al. Expression profling and functional characterization of miR-192 throughout sheep skeletal muscle development. Scientific Reports. 2016;6:30281
46. McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F. et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochimica et Biophysica Acta. 2007;1773:1263-1284
47. Roffe S, Hagai Y, Pines M, Halevy O. Halofuginone inhibits Smad3 phosphorylation via the PI3K/Akt and MAPK/ERK pathways in muscle cells: Effect on myotube fusion. Exprimental Cell Research. 2010;316:1061-1069
48. Kornasio R, Riederer I, Butler-Browne G, Mouly V, Uni Z, Halevy O. β-hydroxy-β-methylbut yrate (HMB) stimulates myogenic cell proliferation, differentiation and survival via the MAPK/ERK and PI3K/Akt pathways. Biochimica et Biophysica Acta. 2009;1793:755-763
49. Madhala-Levy D, Williams VC, Hughes SM, Reshef R, Halevy O. Cooperation between Shh and IGF-I in promoting myogenic proliferation and differentiation via the MAPK/ERK and PI3K/Akt pathways requires Smo activity. Journal of Cellular Physiology. 2012;227:1455-1464
50. Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55:2565-2582
51. Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. Journal of Cell Science. 2013;126:2678-2691
Author contact
![]() Corresponding author: Xingtang Fang, Institute of Cellular and Molecular Biology, Jiangsu Normal University, Xuzhou, Jiangsu, 221116, China. E-mail: xtfangcom
Corresponding author: Xingtang Fang, Institute of Cellular and Molecular Biology, Jiangsu Normal University, Xuzhou, Jiangsu, 221116, China. E-mail: xtfangcom
Received 2018-1-19
Accepted 2018-3-29
Published 2018-5-12
