ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2019; 15(6):1225-1239. doi:10.7150/ijbs.30100 This issue Cite
Research Paper
Aplnra/b Sequentially Regulate Organ Left-Right Patterning via Distinct Mechanisms
1. College of Animal Science in Rongchang Campus, Southwest University, Key Laboratary of Freshwater Fish Reproduction and Development, Ministry of Education, Key Laboratory of Aquatics Science of Chongqing, Chongqing 402460, China.
2. Development and Regeneration Key Laboratory of Sichuan Province, Department of Anatomy and Histology and Embryology, Chengdu Medical College, Chengdu 610500, China.
3. UoE Centre for Inflammation Research, Queen's Medical Research Institute, University of Edinburgh, Edinburgh Bioquarter, 47 Little France Crescent, Edinburgh, EH16 4TJ, UK.
4. Ministry of Education Key Laboratory of Child Development and Disorders; Key Laboratory of Pediatrics in Chongqing, CSTC2009CA5002; Chongqing International Science and Technology Cooperation Center for Child Development and Disorders, Children's Hospital of Chongqing Medical University, 400014, Chongqing, China.
5. School of Public Health, Chengdu Medical College , Chengdu 610500, China
6. Department of Nephrology, Institute of Nephrology of Chongqing and Kidney Center of PLA, Xinqiao Hospital, Third Military Medical University, Chongqing, PR China.
7. Renal Division, Brigham and Women's Hospital, Department of Medicine, Harvard Medical School, Boston, Massachusetts. USA.
*These authors contributed equally to this work.
Abstract
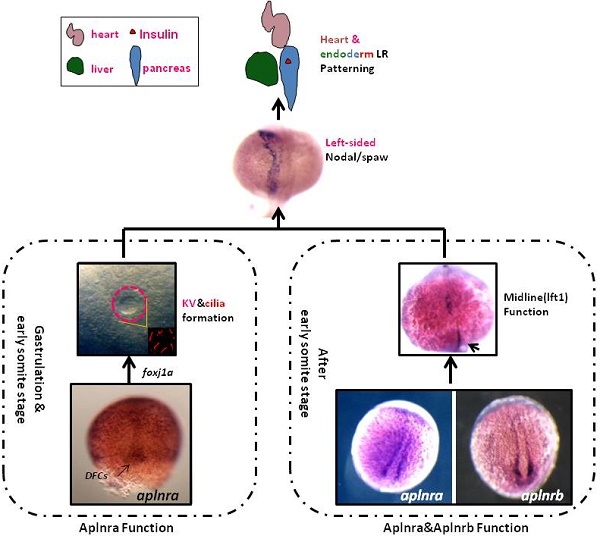
The G protein-coupled receptor APJ/Aplnr has been widely reported to be involved in heart and vascular development and disease, but whether it contributes to organ left-right patterning is largely unknown. Here, we show that in zebrafish, aplnra/b coordinates organ LR patterning in an apela/apln ligand-dependent manner using distinct mechanisms at different stages. During gastrulation and early somitogenesis, aplnra/b loss of function results in heart and liver LR asymmetry defects, accompanied by disturbed KV/cilia morphogenesis and disrupted left-sided Nodal/spaw expression in the LPM. In this process, only aplnra loss of function results in KV/cilia morphogenesis defect. In addition, only apela works as the early endogenous ligand to regulate KV morphogenesis, which then contributes to left-sided Nodal/spaw expression and subsequent organ LR patterning. The aplnra-apela cascade regulates KV morphogenesis by enhancing the expression of foxj1a, but not fgf8 or dnh9, during KV development. At the late somite stage, both aplnra and aplnrb contribute to the expression of lft1 in the trunk midline but do not regulate KV formation, and this role is possibly mediated by both endogenous ligands, apela and apln. In conclusion, our study is the first to identify a role for aplnra/b and their endogenous ligands apela/apln in LR patterning, and it clarifies the distinct roles of aplnra-apela and aplnra/b-apela/apln in orchestrating organ LR patterning.
Keywords: aplnra/b, apela/apln, left right patterning, spaw, midline
Introduction
G protein-coupled receptor (GPCR) APJ, being close to the angiotensin II (Ang II) receptor, was identified as an orphan GPCR[1]. It remained an orphan receptor until a 36-amino acid peptide apelin was discovered [2]. In cardiac development and disease, the function of apelin-APJ has been studied in many cases [3-6]. In apelin and APJ knockout (KO) mice, the sarcomeres of cardiomyocytes are impaired in isolated ventricular myocytes [3]. Apelin/APJ also shows a role in the sustainability and amplification of the cardiac response to stress [5] and in essential hypertension (EHT) [6]. Apelin-APJ plays a role in the Cripto signaling pathway in mammalian cardiac myogenesis by extracellular signal-regulated kinase/ p70S6 kinase [7]. In vascular diseases, the upregulation of apelin in the atherosclerosis of human coronary artery suggested apelin-APJ signaling contributes to coronary vasospasm [8], while conflicting evidence in KO studies has shown antagonistic and inducing roles of apelin-APJ signaling in atherosclerotic formation [9, 10]. All these data indicate that the role of apelin/APJ and its mechanism of action in mammals need to be clarified.
The zebrafish homologs of APJ, aplnra/b and their endogenous ligands apela/apln were identified recently [11-14]. Aplnra/b is involved in regulating gastrulation cell movement [14, 15] and heart [13, 16-18] and vasculature development [19-21], which motivated the study for aplnra/b and its ligands apela/apln in disease and embryonic development. In the zebrafish grinch (grn) mutant, aplnrb loss of function leads to a reduced myocardial progenitor cells (MPCs) via cell-autonomous way [13, 18]. Aplnra/b directly modulates Nodal/TGFβ signaling to determine heart progenitor cells in a another cell-non-autonomous fashion during gastrulation [16], as well as regulating progenitor movement through a G-protein signaling-independent manner in later stages [17]. Detailed analyses showed that overexpression of apln, though not loss of function, phenocopied the heart development defect of the grinch (grn)/or apela mutant [12, 13, 18]. These findings suggested that apln, apela and their receptor aplnra/b might play different roles in heart development, as well as the possibility that another receptor may exist. The ligand Apela but not the receptor Aplnr is expressed in human embryonic stem cells [12], suggesting the distinct roles of apln, apela and receptor aplnra/b in heart or other organ development. All these reports have shown the association between aplnr and the underlying complicated mechanisms during heart development. However, the role of aplnr in cardiovascular development is still not clear.
Left-right (LR) patterning is a fundamental process in early development, and most of its mechanisms are conserved in the animal kingdom [22,23]. In zebrafish, left-sided Nodal/Spaw in the lateral plate mesoderm (LPM) [24] is initiated and amplified by the Node flow in Kupffer's vesicle (KV), which sequentially regulates organ LR patterning [25-29]. In addition to the central role of KV/cilia (or Node/cilia in mouse) in initiating asymmetric Nodal/Spaw, pegasus, nek8 and atp1a1a.1 regulate left-sided Nodal/Spaw expression pattern in a KV/cilia-independent manner [30-32], suggesting that the procedure of initiating and maintaining asymmetric Nodal/Spaw is intricate.
In zebrafish, aplnra/b are zygotic genes and are expressed after blastula stage [11, 18]. At the early gastrulation stage, aplnra loss of function leads to gastrulation movement defect [15] and heart progenitor decreasing [12, 13, 18]. More recently, apela was discovered to work as the ligand for aplnra/b to guide vascular precursors migrate to the midline [33]. During gastrulation, aplnra but not aplnrb is expressed in the cells near dorsal forerunner cells (DFCs) and in DFCs (Fig. S1), the progenitors of KV. At the somite stage, aplnra/b were expressed in the cells near the midline (Fig. S1 and [33]). Since the roles of DFCs/KV and the midline in LR asymmetry patterning are widely reported [24, 26, 28, 29], we hypothesized that aplnra/b might play a crucial role in LR asymmetry patterning. Here, we found that aplnra/b were involved in organ LR patterning via the ligands apela/apln at different developmental stages.
Results
The complementary roles of aplnra/b in organ LR asymmetry patterning
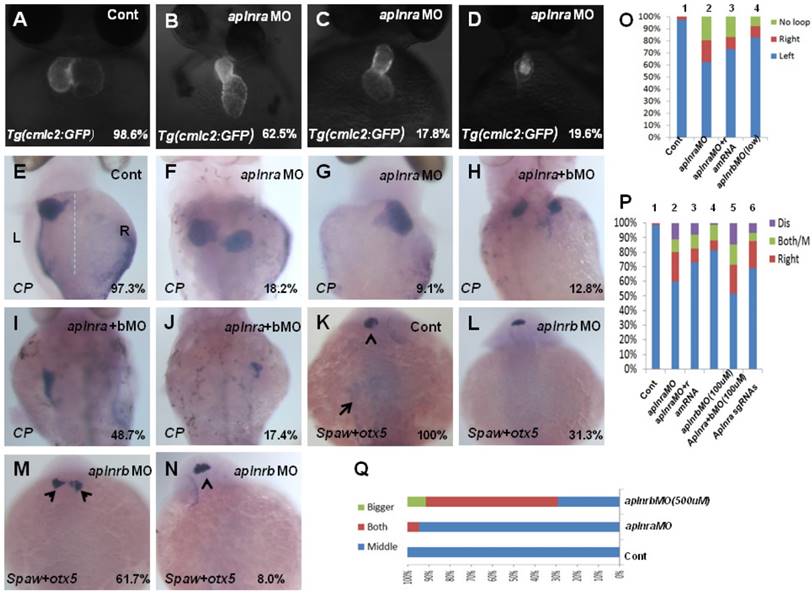
The aplnra/b expression pattern and the critical role in gastrulation cell movement [11, 14] led us to hypothesize aplnra/b play vital roles in LR patterning. To examine this hypothesis, we synthesized the antisense morpholino oligos for zebrafish aplnra/b (MOATG, Gene tools) to block the translation of aplnra/b [13, 18]. Since aplnrb loss of function leads to heart progenitors disappearing or decreasing greatly [13, 18] (Fig. S2 C), which inhibits the analysis of heart LR patterning in aplnrb morphants, we first examined the heart LR patterning in aplnra morphants to evaluate the role of aplnra in LR patterning. Compared with control morphants, the hearts were smaller (Fig. S2 D-E and Fig. 1. B-D) and defective in looping in aplnra morphants (Fig. 1 B-D, O), but no distinct neural epithelium laterality was observed (Fig. 1 Q). At 72 hours post fertilization (hpf), the embryos displayed abnormal morphology, and the liver laterality was also disturbed in aplnra morphants (Fig. 1 F-G, P), displaying right-sided (Fig. 1 G, P) and both-sided liver (Fig. 1 F, P). Since aplnra and aplnrb are involved in regulating heart progenitor development in a redundant way [13, 16], we explored whether aplnrb also contributed to organ LR patterning. The experiments indicated that although aplnrbMO injection (500 µM) resulted in the heart disappearing, deformed embryos ([13, 18] and Fig. S2 C) and head asymmetry (Fig. 1 M, Q), which prevented an analysis of heart laterality, titrating the concentration of aplnrbMO to 100 µM gave rise to 18.2% and 17.5% of embryos displaying heart and liver laterality defects, respectively (Fig. 1 O, P), without clearly deformed embryos (Fig. S2B). Furthermore, coinjecting aplnraMO (400 µM) and aplnrbMO (100 µM) resulted in 40.2% of embryos displaying a liver laterality defect (Fig. 1 H-J, P). This ratio was higher than that in aplnra morphants (Fig. 1 P, 27.3%). These data indicated the redundancy of aplnra/b in organ LR patterning. To confirm the specific role of aplnra in organ LR patterning, we tried to rescue the organ LR patterning defect by coinjecting aplnra MO and aplnra mRNA together into the embryos. After titrating the concentration of aplnra mRNA, we found that aplnra mRNA injection (20 ng/µl) partially restored the LR defect in aplnra morphants (Fig. 1 O, P).
The CRISPR/Cas9 system has been broadly used in gene editing in zebrafish [34-36]. The high editing efficiency gave us the chance to analyze the role of the aplnra gene in Founder(Go) zebrafish embryos [35, 37]. To further confirm the specific role of aplnra in LR patterning, we designed and synthesized 4 guide RNAs for aplnra in vitro and then coinjected them with Cas9 protein into the cytoplasm at the one-cell stage to edit the genome of aplnra gene. The results demonstrated that, when the aplnra gene was edited (Fig. S3), the heart and liver LR defect phenotype in these embryos was also observed (Fig. 1 P and Fig. S3), which further confirmed the role of aplnra in organ LR asymmetry patterning.
All the data above suggest the specific role of aplnra in LR patterning, aplnra and aplnrb regulate organ LR asymmetry patterning in a redundant way.
Organ left-right lateral defect in embryos treated with different ways. (A-D, O) The pattern of heart displayed in controls and aplnra/b morphants. Compared with controls (A, 98.6%, n=72), embryos injected with aplnra MO displayed normal-loop (B, 62.5%, n=112; p<0.01), reversed-loop (C, 17.8%, n=112; p<0.01) and no loop (D, 19.6%, n=112; p<0.01). (O, column 1 to 4) The cartogram of heart LR defect for embryos treated with different MOs or mRNA. Only more than 18.2% of aplnrb morphants displayed heart LR defect (O, column 4, n=115; p<0.05). Aplnra mRNA injection (O, column 3, 26.8%, n=71; p<0.05) partially rescued the heart LR defect in aplnra morphants (O, column 2, 37.5%, n=112). (E-J, P). While 97.3% of control embryos showed left-sided expression of cp (E, n=112), 18.2.0% (F, n=176, p<0.01), 9.1% (G, n=176, p<0.05) and 10.8% (P, n=176, p<0.05) of aplnra morphants showed both sided and right-sided expression of cp, or disappear. (H-J, P). Compared with that in aplnra morphants, aplnra mRNA partially rescued liver LR defect (24%, n=121, p<0.05). In aplnra/b double morphants, the expression of cp was greatly downregulated (100%, n=195, p<0.001), and more embryos displayed liver LR defect (H-J, P, 51.3%, n=195, p<0.05) than that in aplnra morphants (P, 38.1%, n=176). In embryos injected with aplnra sgRNAs and Cas9 protein, 15.8%; and 6.4% of them displayed right sided and both sided/middle liver (P, n=233, p<0.05). (K-N, Q). otx5 was expressed in the middle telencephalon. All the embryos injected with Cont MO displayed the expression of otx5 in the middle telencephalon (K, 100%, n=32, and Q, column 1).While 61.7% of embryos injected with aplnrb MO showed both-sided otx5 in telencephalon (M, Q, n=34), only 5.5% of aplnra morphants displayed both-sided otx5 (Q, 37). For observation, heart, ventral view; cp, spaw and otx5, dorsal view.
Asymmetric Nodal/spaw in the LPM is disturbed when aplnra/b is downregulated
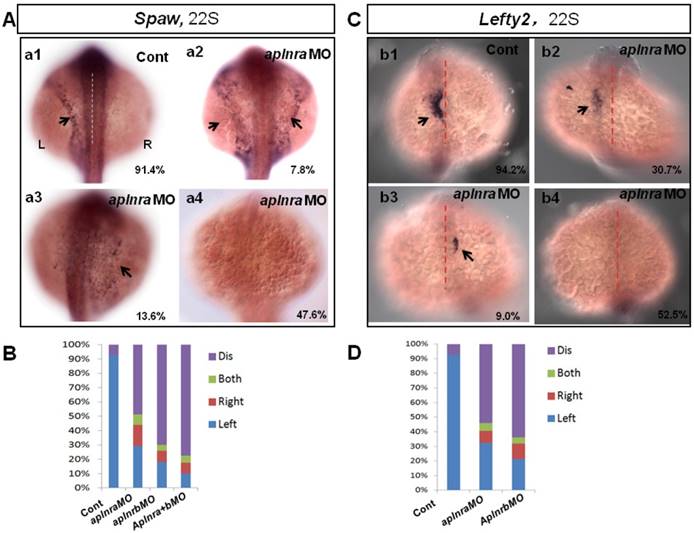
The crucial role of Nodal/spaw in LR patterning was shown in previous studies [21-23]. Downregulation of Nodal/spaw leads to organ laterality defect [38]. To reveal how aplnra/b regulates organ laterality, first we examined the left-sided Nodal/spaw in embryos injected with aplnraMO. The left-sided Nodal/spaw was disturbed in aplnra morphants, displaying both-sided, right-sided and disappeared spaw expression patterns (Fig. 2 A). Since aplnra and aplnrb regulate liver laterality in a redundant way (Fig. 1 P), we continued to evaluate whether left-sided Nodal/spaw was affected in embryos injected with aplnrbMO or aplnraMO+ aplnrbMO. In most embryos injected with aplnrbMO or aplnra+bMO, the left-sided Nodal/spaw disappeared (Fig. 2 B, and Fig. S4). Furthermore, we sequentially checked lefty2, the downstream gene of spaw in aplnra morphants, and found that the left-sided lefty2 expression pattern in the heart field was substantially downregulated (Fig. 2 C, D), which was consistent with that of spaw in aplnra morphants. The substantial downregulation of lefty2 in the heart field coincided with the role of aplnra in heart progenitor development, i.e., aplnra loss of function substantially downregulates heart progenitor development [12, 16]. The data above suggest that the left-sided Nodal/spaw or lefty2 were not only disturbed but also downregulated in aplnra morphants (Fig. 2; Fig. S4). To determine when the downregulation of Nodal signaling was initiated, we examined the expression of lefty1 and other Nodal-related genes at the gastrulation stage. The experiments indicated that the Nodal-related gene sox32, ndr2 and mxtx2 were downregulated at 70% epiboly (Fig. S4). These data demonstrate that, from the late gastrulation stage, Nodal-related genes started to be downregulated. At the somite stage, in addition to the downregulation of Nodal signaling, the left-sided Nodal/spaw and the downstream gene lefty2 were also randomized. These data suggest aplnra/b signaling contributes to organ LR patterning via the Nodal/spaw signaling pathway.
Expression of left-sided Nodal signaling in LPM. (A. a1-a4, B) Expression of left-sided spaw in embryos. In control embryos, near all the embryos expressed left-sided spaw (A. a1, 91.4%, n=93). While in aplnra and aplnrb morphants, left-sided spaw expression pattern was changed (A. a2-a4, B), 13.6% (p>0.05), 7.8% and 47.6% (p<0.001) of aplnra morphants expressed right-sided, both-sided spaw or spaw disappeared (B, middle-left column, n=103). 10.2 % (p<0.05) and 6.1% (p>0.05) of aplnrb morphants showed right-sided and both sided spaw (B, middle-right column, n=98), majority of aplnrb morphants showed no staining for spaw (B, middle-right column, n=98, p<0.001). (C. c1-c4, D) The expression of left-sided lft2 in the heart progenitor field. In the control embryos, lft2 was expressed in the left side of heart progenitor field (C.c1, 94.2%, n=52). Lft2 was down-regulated in aplnra morphants (C. c2-c4), the left-sided expression pattern was also perturbed (C. c2-c4, D), with 9.0%, 6.3% and 52.5% of embryos showing right-sided, both-sided lft2 and lft2 disappeared, respectively (C. c2-c4; D, middle column, n=74). The lft2 expression pattern in aplnrb morphants (D, right column, n=93) was similar to that in aplnra morphants (D, middle column, n=74), but more embryos had no staining for lft2 in aplnrb morphants (D, right column, n=93) than in aplnra morphants (D, middle column, n=74). All the embryos were observed from dorsal view. L, Left; R, Right.
Aplnra but not aplnrb dominantly regulates KV formation and ciliogenesis
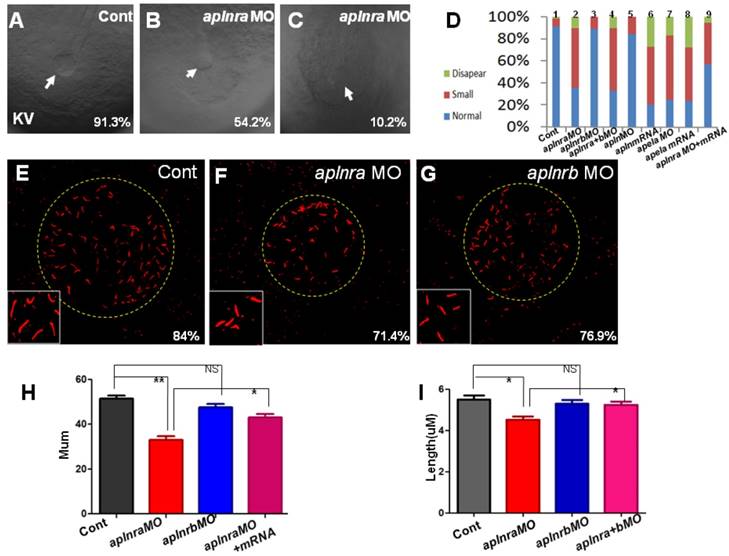
Previous studies and our current research indicated that aplnra but not aplnrb is expressed in the DFCs at the gastrulation stage ([11, 18] and Fig. S1). In zebrafish, DFCs will form the KV at the early somite stage, and defective KV morphogenesis or defective ciliogenesis (or functional defect) will give rise to disturbed left-sided Nodal/spaw and subsequent organ LR defect [24, 26, 27]. To determine whether aplnra/b regulated KV morphogenesis or ciliogenesis, we analyzed the KV/cilia development in aplnra or aplnrb morphants. Compared with control morphants, at the 10-12 somite stage, the KV was smaller in a majority of aplnra morphants (Fig. 3 A-D) while not in aplnrb morphants (Fig. 3 D). Further, we examined the cilia development and found that aplnra morphants displayed a slightly shorter cilia than control morphants (Fig. 3 F, I). The cilia numbers were also decreased in aplnra morphants (Fig. 3 F, H). In aplnrb morphants, no distinct difference in cilia length or cilia number was observed when compared with control morphants (Fig. 3 G, H, I). To assess the specific role of aplnra in KV formation and ciliogenesis, the aplnra mRNA was used to rescue the KV morphogenesis and ciliogenesis defects in aplnra morphants. The results showed that the KV morphogenesis and ciliogenesis defects in aplnra morphants were partially rescued by coinjecting with aplnra mRNA (Fig. 3 D, H, I). Since aplnra expression was also found in DFCs, we studied whether aplnra loss of function in DFCs would lead to KV morphogenesis defect. Indeed, injecting aplnra MO at 256-512 cell to specifically downregulate the function of aplnra resulted in a KV morphogenesis defect (Fig. S5). In addition, DFC-specific downregulation of aplnra resulted in randomized spaw and the sequential organ LR defect (Fig. S5). These data show that aplnra but not aplnrb dominantly regulates KV formation and ciliogenesis during the early somite stage.
KV morphogenesis and Ciliogenesis in treated embryos. (A-D) In control, 91.3% of embryos showed normal KV (A, n=181), but 35.6% (p<0.001), 54.2% (p<0.001) and 10.2% (p<0.001) of embryos injected with aplnra MO showed normal KV, smaller KV and no KV, respectively (B, C and D. column2, n=175), this kind of KV phenotype also can be found in aplnra+b morphants (D. column 4, n=213, p<0.001) or apela morphants (D. column 7, n=208, p<0.001), embryos injected with apela mRNA (D. column 8, n=153, p<0.001) or embryos injected with apln mRNA (D. column 6, n=135, p<0.001) . In aplnrb or apln morphants, no distinct phenotype about KV was discovered (D. column 3(n=147) and column 5, n=193). Injection of aplnra mRNA partially restored the KV phenotype in aplnra morphants (D. column 9, n=116, p<0.05). (E-I) In the KV, compared with that in control morphants (E and H, n=25), the cilia number is decreased in aplnra morphants (F and H, n=28, p<0.01), difference was also found about the length of cilia (F, I, n=12, p<0.05). In aplnrb morphants, only mild difference about cilia number and cilia length was observed (G, H, I, n=9, p>0.05). Co-injection of aplnra mRNA with aplnra MO in the embryos resulted that the number (G, H, n=26, p<0.05) or length of cilia (G, I, n=14, p<0.05) was closed to that in control. *, p<0.05; **, p<0.01; NS, not significant.
Foxj1a is downregulated in aplnra morphants
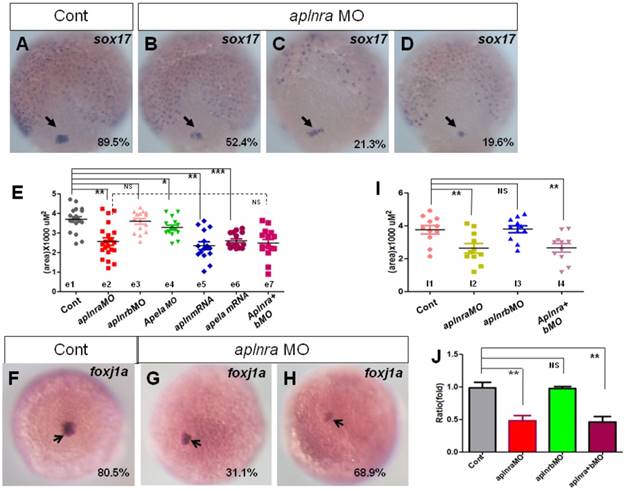
In zebrafish and mouse, the Node/KV plays an important role in initiating asymmetric Nodal/spaw in LPM [3, 29, 39-41]. At the gastrulation stage, the DFCs move together to the tailbud and then form the KV at the early somite stage [42, 43]. During this process, decreasing the DFC number or disturbing the critical signaling pathways, such as FGF or Wnt signaling, leads to deformed KV morphogenesis or a ciliogenesis defect [28, 44]. To determine how KV morphogenesis and ciliogenesis were disturbed in aplnra morphants, we checked whether the primordial cells of KV in aplnra morphants were intact at the late gastrulation stage. In zebrafish, sox17 is generally used as the marker of DFCs. First we examined the expression of sox17 under aplnra loss of function. sox17 showed three kinds of expression pattern in aplnra morphants, 1) similar to that of control (Fig. 4 B), 2) scattered expression (Fig. 4 C) and 3) decreased expression (Fig. 4 D). By measuring the area of sox17 expression, we confirmed that the area of sox17 expression was slightly smaller than that in controls (Fig. 4 E e2, red group). However, the downregulation of sox17 in DFCs was not found in aplnrb morphants (Fig. 4 E e3, pink group).
Fgf8, dnah9 and foxj1a are involved in KV formation and ciliogenesis [20, 28, 33, 39, 45]. dnah9 also lies downstream of FGFR signaling to regulate ciliogenesis [39]. To study how aplnra regulates the development of KV and cilia, we examined the expression of fgf8 and its downstream genes erm, dnah9 and foxj1a. At the 2-somite stage, in situ and Q-PCR experiments showed that there was no distinct difference in fgf8 (Fig. S6 A, C) or dnah9 expression (Fig. S6 D, E) between aplnra morphants and control embryos. Interestingly erm, the downstream gene of fgf8, was upregulated in aplnra morphants (Fig. S6 B, C). Finally, we found that foxj1a was greatly downregulated (Fig. 4 F-H), which was also confirmed by analyzing the area of foxj1a expression (Fig. 4 I) and Q-PCR (Fig. 4 J).To examine whether foxj1a mediates the role of aplnra in KV/cilia development, we performed rescue experiments. The results indicated that transient expression of foxj1a by injecting foxj1a mRNA partially rescued the KV phenotype and the heart LR patterning defect in aplnra morphants. In aplnra morphants, 54.2% of embryos displayed smaller KV (Fig. S 7 B), while 78.6% of embryos coinjected with aplnraMO and foxj1a mRNA displayed slightly larger KV than that in embryos injected with aplnraMO (Fig. S7 C). For heart LR patterning, only 22.1% of embryos coinjected with aplnraMO and foxj1a mRNA displayed the LR patterning defect (Fig. S 7 E, F). This ratio was lower than that in aplnra morphants (Fig. 1 O, column 2, 37.5%). All these data demonstrate that foxj1a but not fgf8 was downregulated in aplnra morphants and that foxj1a possibly at least partially mediated the role of aplnra to regulate KV morphogenesis and the subsequent organ LR patterning.
Our early data in this study showed that injection of aplnrbMO enhanced the organ LR patterning defect in aplnra morphants (Fig. 1 P, column 5). Although aplnrb loss of function did not lead to distinct defective KV and ciliogenesis (Fig. 3), we could not exclude the possibility that aplnrb was partially involved in helping aplnra to regulate the expression of cilia-related genes. Here, we examined this possibility, and the result indicated that no distinct difference in the expression of cilia-related genes was observed between aplnrb morphants and control morphants (Fig. 4 E, I; Fig. S6). We also did not find a significant difference in the expression of cilia-related genes between aplnra morphants and aplnra+b morphants (Fig. 4 E, I and Fig. S6). This result further confirmed that aplnrb contributes to organ LR patterning in a KV/cilia-independent way.
Apela and apln regulate organ LR asymmetry
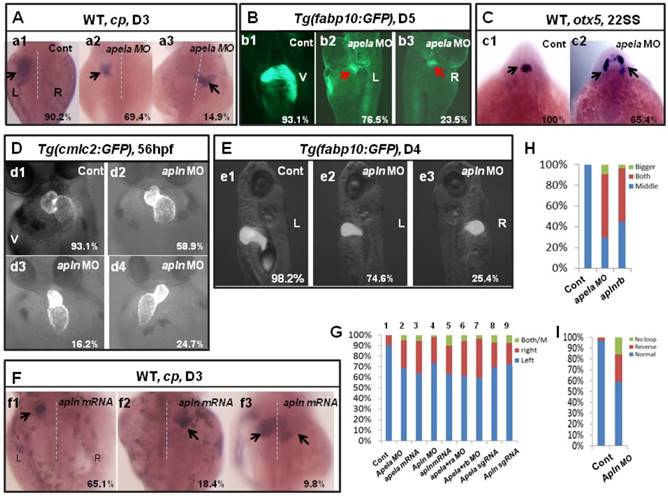
The first identified endogenous ligand of aplnra/b was apelin(apln) [18], the late ligand. More recently, apela was identified to work as the early endogenous ligand of aplnra/b in heart development [12]. To evaluate whether apela/apln work as the ligands of aplnra/b during LR patterning, we examined the expression patterns of apela/apln from gastrulation to somitogenesis. The results demonstrated that, starting from the bud stage, apln was expressed in the midline (Fig. S8 Aa3-a8) and heart progenitors (Fig. S8 Aa7, left arrow; a8, down arrow), while it was not expressed in the KV epithelium (Fig. S8 Aa6, the down black arrow). Apela was expressed ubiquitously at 75% epiboly (Fig. S8 Bb1), in the midline and presomite mesoderm (PSM) at the bud stage (Fig. S8 Bb2-3), in the midline and KV epithelium at the 4-somite stage (Fig. S8 B4-b5, white arrow), and in the midline and heart progenitors at the 15-somite stage (Fig. S8 Bb6-b8, arrow). These expression data demonstrate that the expression patterns of apln/apela are different at early developmental stages. If apln/apela work as the ligands of aplnra/b during LR pattering, they might play different roles during this process. To evaluate this hypothesis, we continued to analyze the organ laterality in apela morphants and apln morphants. In apela morphants, the majority of embryos had no heart or a very small heart (Fig. S9), consistent with a previous report [12]. This phenotype blocked the analysis of heart LR in apela morphants. At day 3, in situ staining for cp demonstrated that liver laterality was perturbed in apela morphants (Fig. 5 Aa2-a3), similar to the phenotype in aplnra morphants (Fig. 1 G,P). In addition, the liver LR patterning defect was also observed in Tg(fabp10:GFP) transgenic embryos injected with apela MO (Fig. 5 Bb2-b3). To confirm the general role of apela in LR patterning, otx5 was used to examine the head laterality(46, 47). We found otx5 was expressed in both sides of the head in major of apela morphants (Fig. 5 C), phenocopying aplnrb morphants (Fig. 1 M, Q). In apln morphants, the heart progenitors were not decreased, but the heart (Fig. 5 D, H and Fig. S10 A) and liver (Fig. 5 E,G and Fig. S10 B) LR patterning were disturbed. To further confirm the specific roles of apela/apln in organ LR patterning, we also used the CRISPR/Cas9 method to edit the apln/apela genes and evaluated the roles of apela/apln in LR patterning. The results indicated that, when apela or apln genomic DNA was edited (Fig. S11), the liver LR defect was also observed (Fig. 5 G, columns 8 and 9 and Fig. S11). These data suggest that, as the endogenous ligands, apela/apln were involved in regulating organ LR patterning. In a previous report, as the ligand of aplnra/b, apela or apln gain of function led to decreased heart progenitor cell number [18], Here, apela or apln gain of function also resulted in a liver LR patterning defect (Fig. 5 F, G; and Fig. S10), implying apela/apln and the receptors aplnra/b are in the same cascade in LR patterning.
Given that apela/apln and aplnra/b work in the same cascade in LR patterning, simultaneously downregulating aplnra/b and their ligands apela/apln will result in a stronger LR defect phenotype than that in aplnra morphants or apela/apln morphants. Indeed, the liver LR asymmetry defect in apela morphants was enhanced by coinjecting with aplnra MO or aplnrb MO (Fig. 5 G column 6 and column 7), further confirming that apela/apln and the receptors aplnra/b were in the same cascade in LR patterning. Since apela was expressed from the early stage and apln was expressed in the midline from the late gastrulation stage [12, 18, 21] (and Fig. S8 Aa3-a8), it seems that the ligands apela and apln sequentially couple with aplnra/b to regulate LR patterning at different stages.
Expression of sox17 and foxj1a. (A-E) Expression of sox17 in DFCs at 90% epiboly. In control embryos, 89.5% of embryos showed normal expression (A, n=88). In aplnra morphants, three kinds of expression pattern were discovered: mild decreased (B, 52.4%, n=89, p<0.001), scattered (C, 21.3%, n=89, p<0.001) and decreased (D, 19.6%, n=89, p<0.01). The area of sox17 expression was measured and in control embryos, the average level was 3.68x103 uM2 (E. e1 group, n=19), the average area in aplnra morphants was 2.55x103 uM2 (E. e2 group, n=22, p<0.05). Sox17 expression areas were also measured for aplnrb morphants, aplnra+b morphants, apela morphants, embryos injected with apln mRNA and embryos injected with apln mRNA, respectively (E, e3-e7 group). The average areas were 3.63x103 uM2 (E, e3 group, aplnrb morphants, n=15, p>0.05), 2.53x103 uM2 (E. e7 group, aplnra+b morphants, n=15), 3.28x103 uM2 (E, e4 group, apela morphants, n=15, p<0.05), 2.46x103 uM2 (E, e5 group, apln mRNA, n=15, p<0.01) and 2.64x103 uM2 (E, e6 group, apela mRNA, n=15, p<0.001), these data indicated sox17 expression was down reglated in all these treated embryos, excepted for aplnrb morphants. (F-J) Analysis of foxj1a expression. Foxj1a was expressed in DFCs at 90% epiboly in control (F) and treated embryos (G, H). Compared with control embryos (F, 80.5%, n=82), foxj1a was greatly down regulated in aplnra morphants (H-I, 68.9%, n=87, p<0.001) as well in aplnra+b morphants (I, 65.7%, n=76, p<0.001), but not in aplnrb morphants (I, 18.9%, n=74, p>0.05). Foxj1a expression area in control embryos, aplnra morphants, aplnrb morphants and aplnra+b morphants were average 3.75 x103 uM2. 2.56 x103 uM2, 3.57 x103 uM2 and 2.39 x103 uM2 respectively (I). Compared with that in control embryos, q-PCR experiment showed the quantity of foxj1a expression in aplnra morphants and aplnra+b morphants were down-regulated with 0.48 folds and 0.46 folds respectively, while the expression of foxj1a in aplnrb morphants was not affected (J). *, p<0.05; **, p<0.01; ***, p<0.001; NS, not significant.
Apela loss of function depresses left-sided Nodal/spaw in the LPM in a KV-dependent way
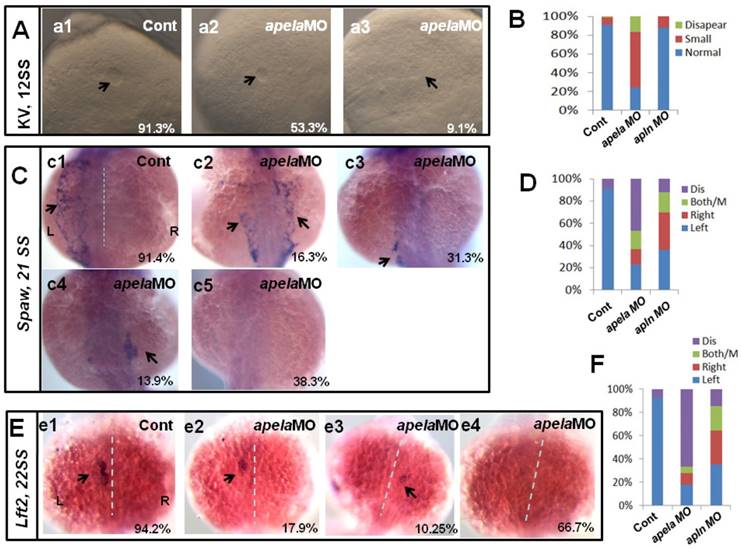
Our current data showed that apela/apln worked as the ligands of aplnra/b to regulate organ LR patterning, but how they regulated LR patterning was unknown. The different expression patterns between apela and apln gave rise to the possibility that the mechanisms by which apela/apln regulate LR patterning are different. To evaluate this possibility, we examined the KV development and the expression pattern of Nodal/spaw. In apela morphants, the KV morphogenesis was affected, most of embryos displayed smaller KV (Fig. 6 Aa1-a3, B), similar to the phenotype in aplnra morphants (Fig. 3 B-D). In contrast, the defective KV formation was not found in apln morphants (Fig. 6 B). This result implied the possibility that apela but not apln works as the ligand of aplnra to regulate KV development and the downstream left-sided spaw in the LPM. Indeed, further experiments showed that left-sided spaw (Fig. 6 C.c2-c5, D) and lft2 (Fig. 6 E.e2-e4, F) were substantially downregulated, and the left-sided expression pattern of both of them was disturbed in apela morphants (Fig. 6 C,E), similar to that in aplnra morphants (Fig. 2). In apln morphants, although the left-sided expression patterns of spaw and lft2 were also disturbed (Fig. 6 D, F), neither of them was substantially downregulated.
Since apela was found to be expressed in KV epithelium, we analyzed whether apela loss of function in DFCs resulted in LR patterning defects. As shown in Fig. S5, the KV development and left-sided spaw expression were all disturbed, but spaw was not downregulated in embryos with apela loss of function in DFCs (Fig. S5 Aa4 and Dd4). Heart and liver LR patterning defect were also observed in these embryos (Fig. S5 Bb4 and Cc4). These data further demonstrate that apela is involved in LR patterning via a KV/cilia-dependent cascade.
Organ LR asymmetry defect in embryos with aplnr ligands loss or gain of function. (A, G) Majority of control embryos expressed left-sided liver marker cp in day 3 (A. a1, G. column 1, 90.2%, n=51). Liver LR asymmetry was disturbed in apela MO injected embryos (A. a2-a3, G. column 2, 30.6%, n=121, p<0.01) and apela mRNA injected embryos (G. column 3, 34.9%, n=103, p<0.01). In transgenic line Tg(fabp10:GFP), the liver LR defect was also observed, displayed left-sided and right-sided liver in 76.5% and 23.5% of apela morphants, respectively (B. b2 and b3, n=51), but the GFP in liver region started to express in day 5 (B. b2 and b3). (C) Head marker otx5 was expressed in the middle telencephalon (C. c1, H. column 1, 100%, n=32), but many of apela morphants (C. c2, H. column 2, 65.4%, n=26) and aplnrb morphants (H. column 3 61.7%, n=34) showed both-sided otx5 in telencephalon. (D, I) Apln loss of function resulted heart LR asymmetry defect (D. d2-d4, I), displayed reversed loop (D. d4, I, 24.7%, n=117, p<0.05), normal loop (D. d2, I, 58.9%, n=117, p<0.01) and linear heart (D. d3, I, 16.2%, n=117, p<0.05). (E. e1-e3, G) In apln morphants, liver development was not delayed (E), but 25.4% liver LR asymmetry defect was found (E. e2-e3, G. column 4, n=185, p<0.001). Apela mRNA or apln mRNA gain of function also resulted in liver LR asymmetry defect, 34.9% (p<0.01) and 37.2% (p<0.01) of embryos displayed right-sided or both-sided expression of cp (G. column 5, n=97). When compared with the liver LR defect in apela morphants (G. column 2, 30.6%, n=121), high ratio of embryos co-injected with apela MO and aplnra MO (G. column 6, 37.1%, n=70), or embryos co-injected with apela MO and aplnrb MO (G. column 7, 40.0%, n=91) showed liver LR asymmetry defect. In embryos injected with apela/apln sgRNAs with Cas9 protein, the livers also showed left right asymmetry defect (G. column 8 and 9). L, left; R, right; V, ventral view; D3, day 3; D5, day 5.
KV morphogenesis and left-sided Nodal signaling in apela morphants. (A-B) In control, 91.2% of embryos showed normal KV (A, n=136), but 37.6%, 53.3% and 9.1% of embryos injected with apela MO showed normal KV, mild smaller KV and no KV, respectively (B, C and D. column2, n=178). (C. c1-c5, D) Majority of control morphants expressed left-sided spaw in the LPM (C. c1, D left column, 91.4%, n=35). In apela MO morphants, 31.3%, 13.9% and 16.3% of embryos showed left-sided spaw, right-sided spaw and both-sided spaw (C. c2-c4, D middle column, n=30), while the spaw expression in most of apela morphants couldn't be found (C. c5, D middle column, 38.9%, n=86). (E-F) The Nodal downstream gene lft2 was checked. Compared with that in control embryos (E. e1, F right column, 94.2%, n=52), lft2 was down regulated and the left-sided expression pattern was disturbed in apela morphants (E. e2-e4, F middle column, n=39), displaying left-sided (17.9%), right-sided (10.25%), both-sided (5.12%) expression and the expression disappeared (66.7%). The disturbed KV morphogenesis and the perturbed spaw or lft2 expression were also observed in apln morphants (D, D, F, right column).
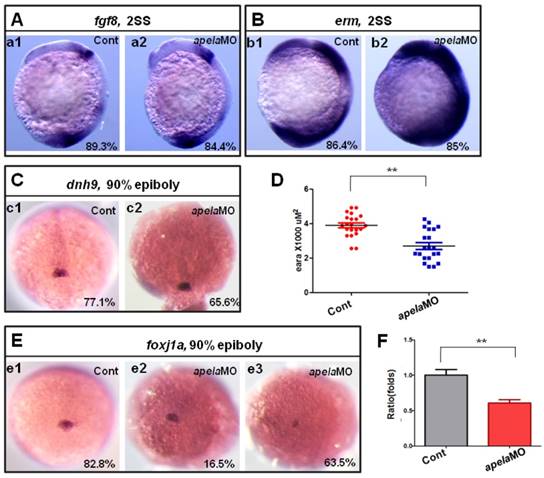
Since aplnra regulated the expression of foxj1a and erm, if the ligand apela but not apln truly couples with aplnra to regulate KV formation and ciliogenesis, the expression of foxj1a and erm in apela morphants should be similar to that in aplnra morphants. Indeed, the expression of fgf8 (Fig. 7 A) and dnah9 (Fig. 7 C) was not affected in apela morphants, while erm (Fig. 7 B) and foxj1a (Fig. 7 D-F) were increased and decreased, respectively, similar to aplnra morphants (Fig. 4 G-I). These data suggest that apela but not apln works as the ligand of aplnra to regulate LR patterning in a KV-dependent way.
Lft1 is downregulated in midline in both apela and apln morphants
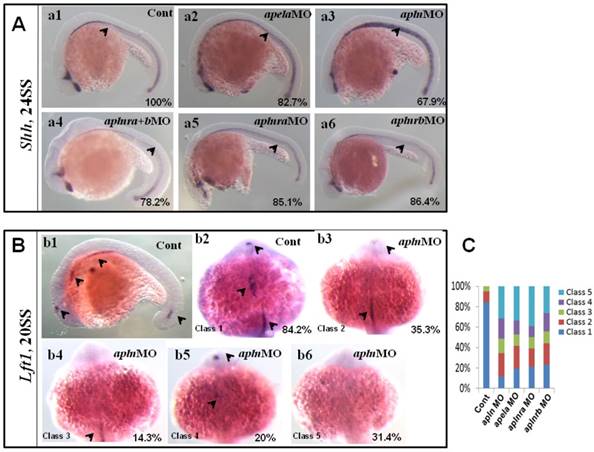
Our data above showed that apln was involved in regulating organ LR patterning and left-sided spaw and lft2, but this process was independent of KV morphogenesis and foxj1a expression. How does apln regulate organ LR patterning? Since midline defects lead to randomized expression of spaw and lft2 in a KV/cilia-independent manner [48], and apln is expressed in the midline from the late gastrulation stage to the somite stage [18, 21] (Fig. S8 Aa3-a8), so we hypothesized that apln might be involved in regulating midline formation or function. To evaluate this possibility, we examined whether apln loss of function affected midline formation. In apln morphants, no deformed midline was observed in the living embryos (data not shown), and no decreased expression of shh was discovered (Fig. 8 Aa3). We also did not find any distinct difference in the expression of shh among the apela morphants, aplnra morphants, aplnrb morphants and aplnra+b double morphants (Fig. 8 Aa2-a6, shown by black arrow head). These data show that apln (including the early ligand apela) and their receptors are not involved in regulating the expression of shh, one of the critical genes in the midline to maintain the left-sided Nodal/spaw in LPM.
Since intact lft1 in the midline is vital to ensure left-sided spaw expression in the LPM and the later organ LR patterning, we examined whether the expression of lft1 was affected in apln morphants. At 20 SS, lft1 was expressed in 4 domains in wild-type embryos, including the left telencephalon, left heart field, trunk midline and tail midline (Fig. 8 Bb1, shown by the black arrow head). In apln morphants, lft1 in the trunk midline disappeared or decreased greatly in the majority of embryos (Fig. 8 Bb4-b6, class 3 to class 5; C, the second column). In the control morphants, the expression of lft1 in the midline was intact (Fig. 8 Bb1, C, column 1). This result indicated the that lft1 in the trunk midline was depressed in apln morphants, implying that lft1 in the midline mediates the effect of apln to regulate LR patterning.
Since apela is expressed in the midline and guides angioblasts to move to the midline [33], and its receptor aplnra/b is expressed close to that of apln/apela, we supposed that apela and the receptors aplnra/b were also involved in regulating the expression of lft1 in the trunk midline. Indeed, further experiments showed that the defective expression pattern of lft1 was also found in apela morphants, aplnra morphants and aplnrb morphants (C, column 3 to 5). Nearly half of embryos showed greatly decreased lft1 in the trunk midline in all the morphants. This result provided additional evidence that apln (and apela) works as the ligand of aplnra/b to regulate organ LR patterning at the somite stage. In summary, at the somite stage, apln/apela-aplnra/b are involved in the regulation of lft1 in the trunk midline to maintain the midline function and the subsequent organ LR patterning.
Discussion
The complementary role of aplnra and aplnrb in organ LR patterning
In vertebrates, aplnr (aplnra/b in zebrafish) has multiple roles in organ development and diseases [5, 21, 39, 40, 49]. In mouse, aplnr regulates cardiac contractility [50], heart looping and vascular maturation [21]. In zebrafish, functions in regulating gastrulation cell movement [14], heart [16-18], angiogenesis [33] and lymphatic system development [19, 20] are also reported. While in regulating heart development, zebrafish aplnra and aplnrb play different roles via multiple mechanisms: In aplnrb mutants, most heart progenitors are absent [18], but in aplnra mutants or morphants, the heart progenitors are only decreased [12]. Similarly, the vasculogenesis defect in aplnra mutants is not as strong as that in aplnrb mutants, but knockdown of aplnra enhances the vasculogenesis defect in aplnrb mutants [33]. It is possible that the different roles of aplnra/b in heart development and angiogenesis come from the different expression patterns of aplnra/b. In our current study, the data demonstrate the critical roles of aplnra/b in organ LR patterning (Fig. 1). During this process, aplnra and aplnrb have redundant roles during liver LR patterning (Fig. 1 H-J, P). Interestingly, the head laterality in aplnrb morphants was more severe than that in aplnra morphants (Fig. 1 K-Q), phenocopied that in squint or MZoep mutants [46]. This result was consistent with a more recent report that Nodal/TGFβ signaling is greatly downregulated in aplnrb mutants [16]. On the role of aplnrb in heart LR patterning, although aplnrb loss of function results in heart absence or heart size decrease in most of embryos (Fig. S2 Cc2-c4), we still believe the role of aplnrb in organ LR patterning is general for all organs, since the left-sided spaw was substantially downregulated or disturbed in aplnrb morphants (Fig. 2 B, Fig. S4 A, B).
By carefully analyzing the ratio of the heart and liver LR patterning defects in embryos injected with the same MOs, we found that the rates of the heart and liver LR defects were different (Fig. 1 and Fig. 5). Our early studies also found this kind of phenotype [38, 51]. Here, we wanted to know, in the embryos injected with aplnra MO, whether the liver LR patterning was also defective in the embryos with a reversed heart loop. We sorted out the Tg(cmlc2:GFP) embryos with a reversed heart loop, incubated them to 5 days post fertilization, and then examined the liver LR patterning using in situ experiments (Fig. S12). The results demonstrated that, in the embryos with a reversed heart loop, 29.4% of embryos displayed right-sided liver (Fig. S12 Aa1 and a3), while in the embryos with a normal heart loop, 18.7% of embryos displayed right-sided liver (Fig. S12 Bb1 and b3). Clearly, this result indicates that the heart and liver LR defects do not always occur at the same time in the aplnra morphants. It also implies that the detailed mechanisms underlying heart and liver LR patterning are at least partially different.
One other interesting phenotype was that aplnrb morphants displayed liver and head laterality defects, but no KV morphogenesis defect. For this result, two kinds of data can explain how the organ LR patterning was affected in aplnrb morphants. First, although aplnrb loss of function did not lead to defective KV development, downregulated Nodal signaling at the gastrulation stage and the somitogenesis stage contributed to the organ LR patterning defect. This explanation is supported by our current data (Fig. 2 B, D and Fig. S4 A,B) and previous reports [16]. Second, at the somite stage, the downregulation of lft1 in the midline also contributed to the organ LR patterning defect in aplnrb morphants (Fig. 8 B, C).
KV/cilia related signaling pathway analysis in apela morphants. (A-C) Compared with the control embryos (A. a1, B. b1 and C. c1), the expression of fgf8 and the downstream gene dnh9 were not changed in apela morphants (A. a2, 84.4%, n=32; C. c2, 65.6%, n=32), but the expression of erm was up regulated in apela morphants (B. b2, 85%, n=20). (E-D) Foxj1a was expressed in DFCs. Compared with that in control embryos (E. e1, 82.8%, n=35), foxj1a was down regulated in apela morphants (E. e3, 63.5%, n=104). The average area of foxj1a expression in control embryos and apela morphants were 3.87 x103 uM2 (n=21) and 2.75 x103 uM2, respectively (D, n=21, p<0.01). Q-PCR experiment indicated that foxj1a expression was down regulated with 0.61 folds compared with that in control embryos (F). *, p<0.05; **, p<0.01.
Aplnra regulates LR patterning in KV/cilia-dependent and -independent ways
The mechanism by which organ LR laterality is established is complicated and needs more study [50, 52]. In zebrafish, two events are critical for LR asymmetric signaling initiating and maintenance: normal KV morphogenesis/ciliogenesis and an intact midline [47, 48, 53-56]. Our results first identify the dominant role of aplnra in LR patterning (Fig. 1 A-H). In this mechanism, the left-sided spaw and lft2 were perturbed (Fig. 2 A-D), which mediated the disturbed organ LR patterning in aplnra morphants. Further data indicated that foxj1a in DFCs was downregulated in aplnra morphants (Fig. 4 F-J), possibly mediating the role of aplnra in regulating KV morphogenesis and ciliogenesis (Fig. 3), the downstream left-sided spaw, lft2 and the subsequent organ LR patterning (Fig. 2 A-D). Since aplnra is expressed ubiquitously in the embryos from the dome stage to the late gastrulation stage [12, 14], and in the early somite stage it is also expressed near the midline [33], these data argue against the idea that aplnra is involved in regulation of LR pattering in only a KV morphogenesis/ ciliogenesis-dependent manner. Indeed, besides the perturbed left-sided spaw expression pattern in aplnra morphants (Fig. 2 A), spaw was downregulated or disappeared in most aplnra morphants (Fig. 2 B) meaning that aplnra loss of function not only randomized the left-sided spaw in LPM but also depressed the expression of spaw in LPM. This result was consistent with the earlier report that Nodal and Nodal-related genes were downregulated in aplnra mutants [16] (Fig. S4 C-F). In addition, our previous data showed that spaw loss of function resulted in randomized organ LR patterning [38], further supporting the hypothesis that aplnra contributes to left-sided spaw expression and the sequential organ LR patterning via not only a KV/cilia cascade but also a KV/cilia-independent signaling pathway.
The expression of aplnra in the midline at the somite stage encouraged us to investigate the role of aplnra in midline development. There was no distinct morphogenesis defect in living aplnra morphants, and the expression of shh in aplnra morphants was intact (Fig. 8 Aa5), while the expression of lft1 in trunk midline was substantially depressed (Fig. 8 B, C). These data suggest that aplnra, besides contributing to KV and cilia development at an early stage, also regulates midline function by regulating the expression of lft1 at the somite stage. In aplnra morphants or mutant embryos, lft1 was unaffected at the early gastrulation stage (50% epiboly) [37]. To assess when lft1 was downregulated in embryos with aplnra loss of function, we examined the expression of lft1 from the shield stage in aplnra morphants. The data indicated that lft1 was slightly downregulated from the shield stage, suggesting downregulation of lft1 in the midline at the somite stage comes from two separate time points: gastrulation and somitogenesis stage. In brief, aplnra contributes to organ LR patterning in KV/cilia-dependent and -independent ways.
Apela but not apln works as the ligand to regulate KV formation and ciliogenesis
Cardiovascular development defects have been reported in APJ KO mice, but not in Apelin KO mice [3, 21, 41]. Heart contractility defects and disturbed looping were also found in APJ KO mice, but these functional and morphogenic defects do not exist in Apelin KO mice, even though the Apelin KO mice obtained a heart contractility defect during aging and severe heart failure was observed in response to pressure overload [5]. These data from mice suggest the possibility that Aplnr regulates heart development in an apelin-independent manner. This possibility was also confirmed by data from zebrafish. In zebrafish, two endogenous ligands apela and apln were discovered [14, 18]. During heart development, aplnra/b contribute to heart progenitors specification and migration [13, 16, 18], while only loss of function for the ligand apela and not apln phenocopies aplnrb loss of function [16, 18]. Here, our data further show the specific role of apela but not apln in KV morphogenesis and ciliogenesis during LR patterning. During the gastrulation and early somite stages, apela loss of function resulted in KV development defect and disturbed ciliogenesis (Fig. 3 D, 4 E, 6 A, B), which contributes to, at least partly, the downstream left-sided gene spaw expression defect (Fig. 6 C,D) and organ LR defect (Fig. 5 A-C). In this process, it is possible that the downregulated foxj1a dominantly contributes to the decreased KV progenitors and the smaller KV in apela morphants (Fig. 7 D-F). On the other hand, apln was mildly expressed from the late gastrulation stage, and downregulation of apln did not lead to abnormal KV morphogenesis (Fig. 3 D), heart or endoderm organ progenitor specification or outgrowth (Fig. 5 D, E and data not shown [18]). Of course, we cannot exclude the possibility that some unknown aplnr ligand is also involved in KV morphogenesis.
Expression of midline related genes in different treated embryos. (A. a1-a6) Shh expression at 24SS. Compared with that in control embryos (A. a1, 100%, n=31), no clearly decreased or increased expression of shh in midline was found in apela morphants (A. a2, 82.7%, n=29), aplnra+b double morphants (A. a4, 78.2%, n=23), aplnra morphants (A. a5, 85.1%, n=27) and aplnrb morphants (A. a6, 86.4%, n=22). In apln morphants, the expression of shh was slightly increased (A. a3, 67.9%, n=28). (B. b1-b6, C) Expression of lft1 at 20SS. In wild type embryos lft1 was expressed in 4 domains, including left telencephalon, left heart field, trunk midline and tail midline (B. b1, black arrow head, n=45), and in dorsal view, lft1 was found to be expressed in left telencephalon, left heart field, trunk midline (B. b2, black arrow head). In apln morphants, lft1 expression was found to be decreased with different phenotypes (B. b3-b6, n=85). Among all the alpn morphants, lft1 in midline was disappeared or decreased greatly in more than half of embryos (B. b4-b6, class 3 to class 5; C, the second column, 64.7%, n=85, p<0.01). The expression pattern of lft1 in apln morphants was also found in apela morphants (C, column 3, 58.2%, n=79, p<0.01), aplnra morphants (C, column 4, 60%, n=75, p<0.01) and aplnrb morphants (C, column 5, 56.2%, n=89, p<0.01), near or more than half of embryos showed greatly decreased lft1 in the trunk midline.
Conclusion
Zebrafish G protein-coupled receptors aplnra /aplnrb were involved in organ LR patterning via an apela/apln ligand-dependent pathway. At the gastrulation and early somite stages, aplnra but not aplnrb was specifically involved in regulating KV/Cilia morphogenesis, coupled by the early ligand apela but not ligand apln. Thereafter, aplnra continued to regulate midline function by specifically regulating lft1 expression in the trunk midline, and this role also depended on the role of the ligands apela/apln. Although aplnrb was not involved in KV/cilia morphogenesis during the gastrulation stage, at the somite stage, aplnrb regulated lft1 expression in the trunk midline, left-sided spaw and lft2 expression in the LPM, and subsequent organ laterality in an apela/ apln-dependent manner. Mechanistically, at the gastrulation and early somite stages, foxj1a potentially lies downstream of the Apela-Aplnra cascade to regulate KV morphogenesis and ciliogenesis. In summary, being coupled with the endogenous ligands apela and apln, aplnr/b sequentially orchestrates organ LR patterning in a KV/cilia-dependent and -independent manner at different stages.
Experimental procedures
Ethics statement
All the experimental protocols were approved by Chengdu Medical College (Sichuan, China) and QMRC, University of Edinburgh. Zebrafish were maintained in accordance with the Guidelines of Experimental Animal Welfare from Ministry of Science and Technology of People's Republic of China (2006) and the Guidelines of Experimental Animal Welfare from Home office in UK.
Zebrafish
Zebrafish (Danio rerio) of the AB genetic background (wild type), Tg(fabp10:GFP) (57) and Tg(cmlc2:GFP) (58) lines were raised and maintained at 28.5 °C, and staged by hours post-fertilization (hpf) by using morphological criteria (59).
Morpholino and mRNA injections
The standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) and the following antisense morpholinos (MOs) were synthesized (Gene Tools) and applied to knock down aplnra (aplnra MO, 5′-TGTATTCCGACGTTGGCTCCATTTG-3′, 400 uM) (13); aplnrb (aplnrb MO, 5′- CAGAGAAGTTGTTTGTCATGTGCTC -3′, 500uM, or 100uM for low concentration) (13, 18), apela (apela MO, 5′- TGGAAGAATCTCATGGTGATGCTCA-3′, 200uM) and apln (apln MO, 5′- AACAGCCGTCACGCTCCCGACTTAC-3′, 300 uM) (13), the plasmids used for synthesizing apela mRNA and apln mRNA were gifts from Ian C Scott lab. aplnra mRNA, aplnrb mRNA, apela mRNA and apln mRNA were synthesized in vitro using mMESSAGE mMACHINE Kit (AM1340, Ambion). The concentration for mRNA injection was as the following: aplnra mRNA (for rescue experiment), 20ng/ul; apln mRNA, 50ng/ul; apela mRNA, 50ng/ul; foxj1a mRNA, 10ng/ul. All the MOs or mRNAs were injected at 1-4 cell stage to downregulate the expression of target genes or to rescue/overexpress the target genes in whole embryos. For specific knockdown for the target genes (aplnra or apela) in DFCs, the MO was injected into the yolk at 256-512 cell stage.
Whole Mount In Situ Hybridization (WISH)
Whole Mount In Situ Hybridization (WISH) was performed according to the established methods (38) using the established antisense probe otx5, apela, aplnrb, aplnra, apln, spaw, sox17, lft1, lft2, shh, fgf8 and erm (12, 13, 25, 60). To prepare the antisense probe of cp, foxj1a,vox, sox32,vmhc and dnh9, we applied the cDNA (prepared from day 3 embryos and 10 hpf embryos) to amplify the template by PCR, then synthesized the antisense probe according to the Kit manual. The embryos for in situ experiments were dechorionated and fixed in 4% PFA overnight at 4 °C, then washed with PBT (PBS-Tween, 0.1%), dehydrated with MeOH(100%) and stored in MeOH at - 20 °C at least for 24 hours.
Antibody staining
The embryos were fixed in 4% PFA at 4 °C for overnight and then washed with PBST (5 minutes/ times, 3 times) and 100% methanol 3 times, stored at -20 °C for at least overnight. Embryos were incubated with antibodies against alpha-tubulin (1:300; Sigma, 6793), then incubated at 4 °C overnight with Alexa fluorescent-conjugated secondary antibodies (1:1000; Invitrogen) which was diluted in the blocking solution. After washed with the PBST, embryos were preceded for mounting and imaging.
Quantitative PCR (Q-PCR)
The cDNA template synthesis was performed in vitro using RT kit (Fermantas). Quantitative PCRs were performed for fgf8, erm and foxj1a. The following primers were used: fgf8 (5-GAGGCTATAAACATGAGACTCATAC-3, 5-CGAACTCGACTCCCAAATGTGTC-3); erm (5-GTGAGAAGCAAGCGACATGGATG-3, 5-GAGTCTCTGCTCTTGTCCACATG-3), foxj1a (5-CTATCGAGGAAGGACAGGATTTG-3, 5- GTCAGTGGCATGCCTATAGACGC -3). mxtx2 (5-CAGCACAATGGCAGTCGTGCAC-3; 5-CTTTCTCCAGCACGTCGATCTG-3); ndnr2(5-CTTGCTACTGCCAGCTGCTG-3; 5-CAGTGCTGTGTTACGCTCAGCAG-3), sox32 (5-CAGCATGTATCTCGACCGGATG-3; 5-CGTCTTACTCGAGTTTCCACG-3) and ism1 (5-GTGAAGAGGATGGTGCGTCTGG-3; 5-GAATATGAGCCACCCGAGTCTG-3). Transcription of beta-actin (5-CATGGATGAGGAAATCGCTGCC-3, 5-GCTCAGGATACCTCTCTTGCTC-3) was used for normalization.
SgRNAs preparing and Cas9 protein co-injection
Four sgRNAs for aplnra, apln and apela were designed according to previous report (61). The individual sgRNA templates for each gene were prepared and then pooled to synthesize the sgRNAs in vitro (NEB #E2040S). The prepared sgRNAs were mixed with Cas9 protein (NEB, #M0646T) (The final concentration for sgRNAs and Cas9 protein were 4uM and 600ng/ul, respectively), and co-injected into the cytoplasm at the one cell stage. For one pool of embryos injected, part of the injected embryos were raised to 27 hpf for analyzing whether the heart cells were decreased greatly, and part of the sorted embryos were used to check genome editing efficiency using PCR. While the remaining embryos injected were raised to stages needed for LR asymmetric phenotype analysis. The sgRNAs targeted for each genes were as: aplnra sgRNA1: ACGGGAATGAGAGAGTAAGA, aplnra sgRNA2: TTTCCAAATGGAGCCAACGT, aplnra sgRNA3: CGCCGTTCCCGGACAAACCG, aplnra sgRNA4: TGTGGCGCGCGAAATCCAAA; apln sgRNA1: CTGGGGAGAGGAGGGAAA, apln sgRNA2: GACTGGCAGGGAAACGGA, apln sgRNA3: CGCTGGTGATTGTGCTGG, apln sgRNA4: AGCGTGACGGCTGTTGCC; apela sgRNA1: AGCAGCAGATACAGCGGG, apela sgRNA2: CAGCAGCAGCAGATACAG, apela sgRNA3: AGACTCGACTCTCCTCAC, apela sgRNA4: GTGATGCTCAGGGTGGTT. The specific primers for checking genome editing efficiency were as the following: aplnra_F: 5_CTGCTCAAGAAGGACTCAAAGCC_3, aplnra_R: 5_GTGGACGATGGCGAGGTAG_3; apln_F: 5_CGCACTGAAGAGCAAACAGTC_3, apln_R: 5_CATGCAGAAGTCGGCAAGTAATT_3; apela_F: 5_GGATTTCTACAGTCCGTTAC_3, apela_R: 5_CTCGAATCGTTTGCCTCATG_3.
Microscope
To examine the KV morphogenesis, the heart and liver of the living embryos in transgenic lines, the embryos were laid in the proper holes made in the Agar plate (1.5% of Low Melting-point Agar (LMP, Sigma)) or directly laid on the Agar plate. In situ hybridized, immunostained embryos were mounted in the 100% glycerol for imaging. Images for living embryos and for in situ experiments were captured at room temperature using AxioVision4 software (Carl Zeiss, Inc.). Cilia images were captured by using LAS AF software (Leica), a x20/0.70 N.A. HC Plan Apochromat air objective (Leica).
Abbreviations
hpf: hours post fertilization; dpf: days post- fertilization; WISH: whole-mount in situ hybridization; qPCR: quantitative PCR; LR patterning: left-right patterning; GPCP: G protein coupled receptor; MPCs: myocardial progenitor cells; DFCs: dorsal forerunner cells; KV: Kupffer's vesicle; LPM: lateral plate mesoderm; KO: Knock Out.
Supplementary Material
Supplementary figures.
Acknowledgements
We are most grateful to Dr. Yi Feng's lab to host most of this work in University of Edinburgh, and to the members in Yi Feng's lab. We are also grateful to Dr. Ian C Scott and Dr. Qiang Wang for the gift plasmids.
Author contributions statement
S.H, C.Z, Z.G designed the experiments. C.Z, Y.Z, M.L, ZG, and S.H performed the experiments, K.C, Y.W, M.Y, X.Z, H.Z, H.T, M. L, C. L, B. S, Y. F, S. L and X.Y discussed the results and commented on the manuscript, S.H, C.Z, Z.G, F.Y, Y.W wrote the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFA0106600), National Natural Science Foundation of China (No. 31741091, 31470080, 31201092, 81573154, 81773432,), Royal Society K.C. Wong Foundation (NF140476) and Scientifc Research Fund of Sichuan Provincial Education Department (18Z021).
Competing Interests
The authors have declared that no competing interest exists.
References
1. O'Dowd BF, Heiber M, Chan A. et al. A human gene that shows identity with the gene encoding the angiotensin receptor is located on chromosome 11. Gene. 1993;136:355-360
2. Tatemoto K, Hosoya M, Habata Y. et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun. 1998;251:471-476
3. Charo DN, Ho M, Fajardo G. et al. Endogenous regulation of cardiovascular function by apelin-APJ. Am J Physiol Heart Circ Physiol. 2009;297:H1904-1913
4. Farkasfalvi K, Stagg MA, Coppen SR. et al. Direct effects of apelin on cardiomyocyte contractility and electrophysiology. Biochem Biophys Res Commun. 2007;357:889-895
5. Kuba K, Zhang L, Imai Y. et al. Impaired heart contractility in Apelin gene-deficient mice associated with aging and pressure overload. Circ Res. 2007;101:e32-42
6. Sonmez A, Celebi G, Erdem G. et al. Plasma apelin and ADMA Levels in patients with essential hypertension. Clin Exp Hypertens. 2010;32:179-183
7. D'Aniello C, Lonardo E, Iaconis S. et al. G protein-coupled receptor APJ and its ligand apelin act downstream of Cripto to specify embryonic stem cells toward the cardiac lineage through extracellular signal-regulated kinase/p70S6 kinase signaling pathway. Circ Res. 2009;105:231-238
8. Pitkin SL, Maguire JJ, Kuc RE. et al. Modulation of the apelin/APJ system in heart failure and atherosclerosis in man. Br J Pharmacol. 2010;160:1785-1795
9. Hashimoto T, Kihara M, Imai N. et al. Requirement of apelin-apelin receptor system for oxidative stress-linked atherosclerosis. Am J Pathol. 2007;171:1705-1712
10. Chun HJ, Ali ZA, Kojima Y. et al. Apelin signaling antagonizes Ang II effects in mouse models of atherosclerosis. J Clin Invest. 2008;118:3343-3354
11. Tucker B, Hepperle C, Kortschak D. et al. Zebrafish Angiotensin II Receptor-like 1a (agtrl1a) is expressed in migrating hypoblast, vasculature, and in multiple embryonic epithelia. Gene Expr Patterns. 2007;7:258-265
12. Chng SC, Ho L, Tian J. et al. ELABELA: a hormone essential for heart development signals via the apelin receptor. Dev Cell. 2013;27:672-680
13. Scott I C, Masri B, D'Amico LA. et al. The g protein-coupled receptor agtrl1b regulates early development of myocardial progenitors. Dev Cell. 2007;12:403-413
14. Pauli A, Norris ML, Valen E. et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science. 2014;343:1248636
15. Nornes S, Tucker B, Lardelli M. Zebrafish aplnra functions in epiboly. BMC Res Notes. 2009;2:231
16. Deshwar AR, Chng SC, Ho L. et al. The Apelin receptor enhances Nodal/TGFbeta signaling to ensure proper cardiac development. Elife. 2016:5
17. Paskaradevan S, Scott IC. The Aplnr GPCR regulates myocardial progenitor development via a novel cell-non-autonomous, Galpha(i/o) protein-independent pathway. Biol Open. 2012;1:275-285
18. Zeng XX, Wilm TP, Sepich DS. et al. Apelin and its receptor control heart field formation during zebrafish gastrulation. Dev Cell. 2007;12:391-402
19. Kim JD, Kang Y, Kim J. et al. Essential role of Apelin signaling during lymphatic development in zebrafish. Arterioscler Thromb Vasc Biol. 2014;34:338-345
20. Karpinich NO, Caron KM. Apelin signaling: new G protein-coupled receptor pathway in lymphatic vascular development. Arterioscler Thromb Vasc Biol. 2014;34:239-241
21. Kang Y, Kim J, Anderson JP. et al. Apelin-APJ signaling is a critical regulator of endothelial MEF2 activation in cardiovascular development. Circ Res. 2013;113:22-31
22. Speder P, Petzoldt A, Suzanne M. et al. Strategies to establish left/right asymmetry in vertebrates and invertebrates. Curr Opin Genet Dev. 2007;17:351-358
23. Raya A, Izpisua Belmonte JC. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat Rev Genet. 2006;7:283-293
24. Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303-2316
25. Huang S, Ma J, Liu X. et al. Geminin is required for left-right patterning through regulating Kupffer's vesicle formation and ciliogenesis in zebrafish. Biochem Biophys Res Commun. 2011;410:164-169
26. Essner JJ, Amack JD, Nyholm MK. et al. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247-1260
27. Wang G, Cadwallader AB, Jang DS. et al. The Rho kinase Rock2b establishes anteroposterior asymmetry of the ciliated Kupffer's vesicle in zebrafish. Development. 2011;138:45-54
28. Caron A, Xu X, Lin X. Wnt/beta-catenin signaling directly regulates Foxj1 expression and ciliogenesis in zebrafish Kupffer's vesicle. Development. 2012;139:514-524
29. Lai SL, Yao WL, Tsao KC. et al. Autotaxin/Lpar3 signaling regulates Kupffer's vesicle formation and left-right asymmetry in zebrafish. Development. 2012;139:4439-4448
30. Manning DK, Sergeev M, van Heesbeen RG. et al. Loss of the ciliary kinase Nek8 causes left-right asymmetry defects. J Am Soc Nephrol. 2013;24:100-112
31. John LB, Trengove MC, Fraser FW. et al. Pegasus, the 'atypical' Ikaros family member, influences left-right asymmetry and regulates pitx2 expression. Dev Biol. 2013;377:46-54
32. Ellertsdottir E, Ganz J, Durr K. et al. A mutation in the zebrafish Na,K-ATPase subunit atp1a1a.1 provides genetic evidence that the sodium potassium pump contributes to left-right asymmetry downstream or in parallel to nodal flow. Dev Dyn. 2006;235:1794-1808
33. Helker CS, Schuermann A, Pollmann C. et al. The hormonal peptide Elabela guides angioblasts to the midline during vasculogenesis. Elife. 2015:4
34. Liu J, Zhou Y, Qi X. et al. CRISPR/Cas9 in zebrafish: an efficient combination for human genetic diseases modeling. Hum Genet. 2017;136:1-12
35. Wu R S, Lam II, Clay H. et al. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev Cell. 2018;46:112-125 e114
36. Shankaran SS, Dahlem TJ, Bisgrove BW. et al. CRISPR/Cas9-Directed Gene Editing for the Generation of Loss-of-Function Mutants in High-Throughput Zebrafish F0 Screens. Curr Protoc Mol Biol. 2017;119:31.9.1-31.9.22
37. Norris ML, Pauli A, Gagnon JA. et al. Toddler signaling regulates mesodermal cell migration downstream of Nodal signaling. Elife. 2017:6
38. Huang S, Ma J, Liu X, Zhang Y. et al. Retinoic acid signaling sequentially controls visceral and heart laterality in zebrafish. J Biol Chem. 2011;286:28533-28543
39. O'Carroll AM, Lolait SJ, Harris LE. et al. The apelin receptor APJ: journey from an orphan to a multifaceted regulator of homeostasis. J Endocrinol. 2013;219:R13-35
40. Yang P, Read C, Kuc RE. et al. Elabela/Toddler Is an Endogenous Agonist of the Apelin APJ Receptor in the Adult Cardiovascular System, and Exogenous Administration of the Peptide Compensates for the Downregulation of Its Expression in Pulmonary Arterial Hypertension. Circulation. 2017;135:1160-1173
41. Roberts EM, Newson MJ, Pope GR. et al. Abnormal fluid homeostasis in apelin receptor knockout mice. J Endocrinol. 2009;202:453-462
42. Tay HG, Schulze SK, Compagnon J. et al. Lethal giant larvae 2 regulates development of the ciliated organ Kupffer's vesicle. Development. 2013;140:1550-1559
43. Dasgupta A, Merkel M, Clark MJ. et al. Cell volume changes contribute to epithelial morphogenesis in zebrafish Kupffer's vesicle. Elife. 2018:7
44. Neugebauer JM, Amack JD, Peterson AG. et al. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651-654
45. Matsui T, Thitamadee S, Murata T. et al. Canopy1, a positive feedback regulator of FGF signaling, controls progenitor cell clustering during Kupffer's vesicle organogenesis. Proc Natl Acad Sci U S A. 2011;108:9881-9886
46. Aquilina-Beck A, Ilagan K, Liu Q. et al. Nodal signaling is required for closure of the anterior neural tube in zebrafish. BMC Dev Biol. 2007;7:126
47. Frisca F, Colquhoun D, Goldshmit Y. et al. Role of ectonucleotide pyrophosphatase/phosphodiesterase 2 in the midline axis formation of zebrafish. Sci Rep. 2016;6:37678
48. Burdine RD, Grimes DT. Antagonistic interactions in the zebrafish midline prior to the emergence of asymmetric gene expression are important for left-right patterning. Philos Trans R Soc Lond B Biol Sci. 2016;371:1710
49. Xu J, Chen L, Jiang Z. et al. Biological functions of Elabela, a novel endogenous ligand of APJ receptor. J Cell Physiol. 2018;9:6472-6482
50. Szokodi I, Tavi P, Foldes G. et al. Apelin, the novel endogenous ligand of the orphan receptor APJ, regulates cardiac contractility. Circ Res. 2002;91:434-440
51. Huang S, Xu W, Su B. et al. Distinct mechanisms determine organ left-right asymmetry patterning in an uncoupled way. Bioessays. 2014;36:293-304
52. Roxo-Rosa M, Jacinto R, Sampaio P. et al. The zebrafish Kupffer's vesicle as a model system for the molecular mechanisms by which the lack of Polycystin-2 leads to stimulation of CFTR. Biol Open. 2015;4:1356-1366
53. Zhang J, Jiang Z, Liu X. et al. Eph/ephrin signaling maintains the boundary of dorsal forerunner cell cluster during morphogenesis of the zebrafish embryonic left-right organizer. Development. 2016;143:2603-2615
54. Gokey JJ, Ji Y, Tay HG. et al. Kupffer's vesicle size threshold for robust left-right patterning of the zebrafish embryo. Dev Dyn. 2016;245:22-33
55. Dasgupta A, Amack JD. Cilia in vertebrate left-right patterning. Philos Trans R Soc Lond B Biol Sci. 2016:371
56. Matsui T, Bessho Y. Left-right asymmetry in zebrafish. Cell Mol Life Sci. 2012;69:3069-3077
57. Lu H, Ma J, Yang Y. et al. EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev Cell. 2013;24:543-553
58. Huang CJ, Tu CT, Hsiao CD. et al. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30-40
59. Kimmel CB, Ballard WW, Kimmel S R. et al. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253-310
60. Carl M, Bianco IH, Bajoghli B. et al. Wnt/Axin1/beta-catenin signaling regulates asymmetric nodal activation, elaboration, and concordance of CNS asymmetries. Neuron. 2007;55:393-405
61. Varshney GK, Pei W, LaFave MC. et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015;25:1030-1042
Author contact
![]() Corresponding author: Sizhou Huang, Email: huangyuy1027edu.cn; Tel: (086) 28-6273-9327; Fax. (086)28-62739298.
Corresponding author: Sizhou Huang, Email: huangyuy1027edu.cn; Tel: (086) 28-6273-9327; Fax. (086)28-62739298.
Received 2018-9-20
Accepted 2019-3-12
Published 2019-5-11
