ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2019; 15(7):1383-1395. doi:10.7150/ijbs.33773 This issue Cite
Research Paper
P11 Loss-of-Function is Associated with Decreased Cell Proliferation and Neurobehavioral Disorders in Mice
Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan 430072, P. R. China
Abstract
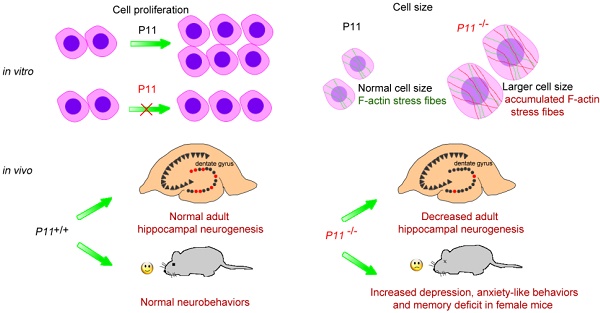
Although depression is associated with anxiety and memory deficit in humans, the molecular mechanisms of the complication remain largely unknown. In this study, we generated P11 knockout mice using CRISPR/Cas9 technology, as well as P11 knockout MEF cell lines, and confirmed depression-like phenotype. We observed that knockout of P11 in MEFs led to a decreased cell proliferation compared with P11+/+ MEFs. Moreover, P11 knockout resulted in a larger cell size, which resulted probably from accumulated F-actin stress fibers. The number of proliferating cells was decreased in the hippocampus of P11 KO mice. We observed anxiety-like disorder in addition to depression phenotype in the knockout mice. In addition, knockout of P11 led to memory deficit in female mice, but not in males. These data indicated that P11 is involved in regulating cell proliferation and cell size. The molecular associations of depression behavior with anxiety and memory deficit suggested a potential approach to improve therapeutic intervention through P11 in these disorders.
Keywords: P11, depression, anxiety, memory deficit, cell proliferation, cell size
Introduction
Major depressive disorder (MDD), also simply known as depression, affects approximately 3% of the world's population (216 million people) [1].The lifetime incidence of major depressive disorder is higher in women than in men [2], and patients with depression have a tendency of suicide. The most common time of depression onset is in a person's mid-20s [3]. In mental illnesses, anxiety disorders are specifically prevalent. Their onset age is often very young (at 11 yrs) [4]. There is considerable overlap of symptomatology between these two disorders. There are many anxious patients who present with concurrent depression-like symptoms and anxiety-like symptoms are often present in many depressed patients [5]. These symptoms can persist and affect life of patients, if treated not timely [6]. Despite several decades of intense studies, our understanding of molecular etiology of the disorders is still limited.
P11, also known as S100A10, belongs to S100 family, a family of small acid Ca2+-binding proteins in vertebrates. In mammals, S100a genes form a cluster on a single chromosome, which could duplicate onto separate chromosomes in teleost fish species [7, 8]. P11 consists of two EF-hands separated by a central small region, and the EF-hand at the C-terminal is vital for its target binding [9-12]. Unlike other members, P11 is Ca2+ insensitive because of essential amino acid replacements in its EF-hand Ca2+-binding loops that keep the protein in a permanently active status [13, 14]. P11 is expressed ubiquitously [15, 16], especially in brain regions that are implicated in the pathophysiology of depression, including the nucleus accumbens, cerebral cortex, prelimbic cortex and hippocampus[17-22]. P11 usually exists in the form of annexin II-P11 heterotetramer (AIIt) [23, 24]. P11 plays important roles in depression together with several neurotransmitter receptors, such as 5-hydroxytryptamine (5-HT1B) receptor, 5-HT4 receptor and metabotropic glutamate receptor 5 (mGluR5) [25-29]. Recent studies indicated that P11 plays roles in both depression and Parkinson's disease [30-32]. In addition, P11 contributes to the clinical hemorrhagic phenotypes of acute promyelocytic leukemia as a plasminogen receptor [33]. Dysregulation of P11 was also involved in cancers [34, 35], fatty livers [36], cocaine reward and cannabis dependence [37, 38], and Diarrhea-predominant Irritable Bowel Syndrome [39]. These studies suggested that P11 is multifunctional in regulation of development of various diseases in humans, in addition to its major roles in depression.
Neurogenesis was first detected in the dentate gyrus of young rats by injection with thymidine-H3 [40]. In humans, adult neurogenesis even persisted into the eighth decade of life, although quiescent stem cell pools and angiogenesis declined [41]. However, another study showed that the number of proliferating progenitors and young neurons in the dentate gyrus decreased sharply after birth and neurogenesis were scarcely observed in teenagers [42]. In adult humans, many questions concerning neurogenesis in hippocampus remain unanswered [43], particularly, evolutionary divergence in brain exists between humans and rodents. Treatment of antidepressant agent fluoxetine can increase cell proliferation in hippocampus of wild-type mice. However, no difference between fluoxetine-treated and control P11 KO mice was observed [44], indicating an association of P11 with cell proliferation. Although roles of P11 in many activities and processes have been well documented, a direct and precise function for P11 in cell proliferation and its possible contribution to psychological disorders remain elusive.
To explore the functions of P11 in cell proliferation and its potential contribution to depression, memory deficit and anxiety, in this study, we generated P11 knockout mice, as well as P11 knockout MEF cell lines, using CRISPR/Cas9 technology. By comparing cell proliferation of P11+/+ and P11-/- MEFs, we found that ablation of P11 leads to a decreased cell proliferation. Moreover, P11 knockout resulted in a larger cell size, compared with that of wild type, which resulted probably from accumulated F-actin stress fibers. Moreover, BrdU staining in the hippocampus showed a decrease in the number of proliferating cells in P11 KO mice. Then we observed anxiety-like disorder in addition to depression-like phenotype in P11 knockout mice. Besides, knockout of P11 led to memory deficit in female mice, while not in males. These findings are valuable for understanding of the roles of P11 in the neurological disorders.
Materials and methods
Animals
C57BL/6 wild-type mice were purchased from Wuhan University Center for Animal Experiment (Wuhan, China) and Shanghai Biomodel Organism Science & Technology Development (Shanghai, China). All animal experiments and methods were performed in accordance with the relevant approved guidelines and regulations, as well as under the approval of the Ethics Committee of Wuhan University (wdsky0201703).
Antibodies
The following primary antibodies were used: anti-P11 (AF2377, R&D Systems, Minneapolis, USA), anti-β-actin (20536-1-AP, Proteintech Group, Chicago, USA) and Bromodeoxyuridine (BrdU) antibody (GTX128091, GeneTex, Texas, USA). The following second antibodies were used: peroxidase-conjugated affinipure rabbit anti-goat IgG (H+L) (SA00001-4, Proteintech Group), peroxidase-conjugated affinipure goat anti-rabbit IgG (H+L) (SA00001-2, Proteintech Group) and TRITC-conjugated goat anti-rabbit IgG (H+L) (T-2769, Thermo Fisher, Massachusetts, USA).
Plasmid constructs
LentiCRISPR-v2 plasmid was a gift from Dr. Feng Zhang of Broad Institute of MIT and Harvard. LentiCRISPRv2-p11-gRNA was constructed as described previously [45, 46]. Briefly, P11-gRNAs were designed according to CRISPR Design Tool (http://crispr.mit.edu/) and synthesized with BsmBI sticky end, then annealed and inserted into the BsmBI (Fermentas, Vilnius, Lithuania) digested lentiCRISPR-v2 plasmid. For constructing lentiviral vector MSCV- P11, the P11 coding sequence was amplified using wild-type MEF cDNA as the template, and then digested with XhoI and EcoRI (Fermentas), and cloned into MSCV PIG(18751, Addgene, Cambridge, USA). Primer sequences are described below (restriction enzyme recognition sites shown underlined):
F(5'-3'): AATCTCGAGATGCCATCCCAAATGGAGCAC; R(5'-3'): AATGAATTCCTACTTCTTCCCCTTCTGCT.
Cas9 expression vector (pST1374-N-NLS-flag-linkerCas9) for in vitro transcription (44758) and PUC57-sgRNA expression vector (51132) were obtained from Addgene. P11-gRNAs were designed according to CRISPR Design Tool (http://crispr.mit.edu/). Synthesized oligos for gRNA expression were denatured at 95 °C for 5 minutes and annealed at room temperature, then cloned into linearized PUC57-sgRNA expression vector.
Generation of P11 knockout mice
Cas9 expression vector was linearized by AgeI enzyme (Fermentas). Cas9 mRNA was transcribed using mMESSAGE mMACHINE T7 kit (AM1344, Thermo Fisher). PUC57-sgRNA expression vector was linearized by DraIII (Fermentas) and in vitro transcribed using MEGAshortscript T7 kit (AM1354, Thermo Fisher). The transcribed mRNA and gRNAs were further purified using MEGAclear Transcription Clean-Up Kit (AM1908, Thermo Fisher) for embryo microinjection.
Fertilized eggs were collected from the oviducts of C57BL/6 mice. Both transcribed gRNAs (100 ng/ ml) and Cas9 mRNA (150 ng/ml) were mixed and microinjected into male pronuclei of fertilized eggs in M2 medium (M7167, Sigma, St. Louis, USA). The embryos were transferred to the uterus of pseudo- pregnant mice, from which viable founders were obtained.
Lentivirus generation and infection
To generate lentivirus as described before [47], the 293T cells seeded in 100-mm plate were transfected with lentiCRISPRv2-P11-gRNA or MSCV- P11 and lentiviral packaging vectors (pRSV-Rev (12253, Addgene), pMD2.G (12259, Addgene) and pCMV-VSV-G (8454, Addgene) or pCL-Eco (12371, Addgene)) using Lipofectamine 2000 (11668019, Thermo Fisher) according to the manufacturer's instructions. The cells were incubated for 48 hrs and the supernatants were filtered through 0.45 μm filters and used directly to infect MEFs (mouse embryonic fibroblasts). Puromycin (A1113802, Thermo Fisher) was used to screen cell lines stably expressing Cas9-P11-gRNA or P11.
Preparation of primary MEFs, cell culture and transfection
Primary MEFs were obtained from embryos at 13.5 days post-coitum (dpc). The cells at passage 3 were used for subsequent experiments. HEK293T cells were obtained from China Center for Type Culture Collection (3115CNCB00266, CCTCC, Wuhan, China). Wild-type MEF cells were a gift from Dr. William J. Craigen of Baylor. The cells were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM; SH30022.01B, HyClone, Beijing, China) containing 10% fetal bovine serum (FBS; SV30087.02, HyClone) in 6-well plates at 37 °C in 5 % CO2. For transfection, Lipofectamine 2000 (8µl) (11668019, Thermo Fisher) was used in each well.
Western blot analysis
Western blots were performed as previously described [47]. Briefly, protein extracts from cell lines or tissues were separated using 10% Tricine-SDS- PAGE and transferred onto a 0.22 μm PVDF membrane (NK0414, Roche Diagnostics). After incubation overnight with the antibodies anti-P11 (1:1000 dilution) or anti-β-actin (1:2000 dilution), the membranes were washed in TBST (20 mM Tris-HCl pH7.5, 150 mM NaCl, 0.1% Tween 20) five times, incubated with peroxidase-conjugated second antibody for 1 h, and then washed five times at 37 °C. A Super Signal Chemiluminescent Substrate system (34080, Pierce, Rockford) was used to detect the signals.
Cell growth assay
Cell growth was evaluated using a real-time cell analysis (RTCA) system (xCELLigence RTCA DP, Roche, Penzberg, Germany). Briefly, background values were taken from the blank culture of 50µl medium in the E-16 plates. RTCA (Software 2.0) was used to calibrate the plates. The cells (~2,000/well) were plated with fresh medium to a final volume of 150 µl. Afterwards, the E-plate was located at RTCA cradle. The impedance signals were recorded every 15 min until 120 hrs.
Flow cytometry
For cell size measurement, cells were trypsinized, washed with ice-cold PBS, and their forward scatter values were measured by flow cytometry (CyAn ADP, Beckman Coulter, Brea, USA). The forward scatter (FSC-A) values of cells were analyzed by Modfit Software (Verity Software House, Turramurra, Australia).
Cell proliferation was analyzed by BrdU incorporation using anti-BrdU antibody, PE conjugated (339812, Biolegend, San Diego, USA). Briefly, cells at the logarithmic growth phase were cultured with 10 µM BrdU (B23151, Thermo Fisher) for 1 h, then harvested for ice-cold (-20 °C) 70% ethanol fixation for at least 2 hrs. For BrdU staining, cells were treated with 2 N HCl to denature DNA, and then neutralized with 0.1 mol/L Na2B4O7. Then, the samples were incubated with anti-BrdU antibody (5ul per million cells) in PBS/5% BSA for 20 minutes at room temperature in the dark. After three washes in PBS, the nuclei were stained with DAPI (C1002, Beyotime, Haimen, China). Stained cells were analyzed by flow cytometry (CyAn ADP, Beckman Coulter).
Eosin staining and analysis
Briefly, cells were plated onto glass slides in 24-well plates and subjected to eosin staining 24 hrs later. The stained cells were examined under a bright light microscope using 200 x magnification, and captured multiple images from selected eosin-stained slides. At least, twenty-two cells were selected randomly for analysis. All the analyses were performed in a double blind manner.
F-Actin staining by phalloidin
Phalloidin-iFluor 647 (ab176759, Abcam, Cambridge, UK) was used according to the manufacturer's instruction. In brief, cells were fixed with 4% paraformaldehyde and then permeabilized in 0.1% Triton X-100. The samples were then incubated with phalloidin conjugate working solution at room temperature for 30 min. The nuclei were stained with Hoechst (C1026, Beyotime, Haimen, China). Images were captured by confocal fluorescence microscopy (FV1000, Olympus, Tokyo, Japan).
BrdU staining and immunofluorescence
4-month-old mice were intraperitoneally injected with BrdU quartic daily (50 mg/kg). 24 hrs later, the mice were deeply anesthetized and perfused with PBS, then 4% paraformaldehyde (PFA). The brain samples were further fixed in 4% PFA overnight at 4 °C and saturated with 30% sucrose. The samples were embedded in OCT medium (Tissue-Tek, Miles, 4583) and frozen at-20 °C. Serial sections (40 µm) were cut through the entire hippocampus with a crystal microtome (CM1850, Leica, Bensheim, Germany), with totally 72 sections from the individual mouse. The sections were permeabilized in 1% Triton X-100 for 15 min, treated with 1 N HCl to denature DNA, and neutralized with 1 mol/L boric acid. After that, the sections were incubated with anti-BrdU antibody (GTX128091) (1:500) in PBS/5% BSA overnight at 4°C, then incubated with TRITC-conjugated goat anti- rabbit (T-2769) (1:1000) in PBS/5% BSA for 1 h at room temperature. After washes with PBS, the nuclei were stained with Hoechst (C1026, Beyotime). Images were acquired by confocal fluorescence microscopy (FV1000, Olympus). BrdU staining was performed with every sixth section throughout the hippocampus. All BrdU-positive cells in the subgranular zone of the dentate gyrus were counted. The total number of BrdU-positive cells per section was determined and multiplied by 72 to obtain the total number of cells per dentate gyrus.
Off-target assay
CRISPR Design Tool was used to predict the potential off-target sites (http://crispr.mit.edu/). All possible off-target sites were screened by ungapped alignments, allowing for up to 4 mismatches in the target gRNA sequence. In the output, three potential off-target sequences with high scores for every gRNA were selected and PCR amplified using genomic DNA from skin as templates. The PCR products were cloned into pGEM-T Easy Vector (A1360, Promega, Madison, USA) and subjected to sequencing. At least 5 recombinant clones were sequenced.
Tail suspension tests (TST)
Tail suspension tests were performed as previously described [48]. All behavioral experiments, except for sucrose preference, were performed in a quiet room illuminated by a dim light (50 lx) during the daytime. Briefly, 4-month-old mice were suspended from a hook with medical tape attached ∼2 cm from the tail tip for 6 min. The trials were recorded by a video camera positioned directly in front of the mice, and immobility was scored with Ethovision XT 12 (Noldus Information Technology, Wageningen, Netherlands) at a 10% immobility threshold.
Sucrose preference
The sucrose preference was assayed as described before [49]. Briefly, the mice were single housed and given two drinking bottles to choose between solutions water or 1% sucrose. The positions of the two bottles were exchanged 12hrs later to prevent potential side-preference bias. Consumptions of water and sucrose were measured the following 24hrs. Preference for sucrose over water was calculated as the weight ratio of (sucrose / [sucrose + water]).
Forced swimming tests (FST)
The procedure was used as described before [50]. Briefly, swimming sessions were performed by placing mice (N=2/group) for 6 min test in individual transparent beaker (30 cm height x 20 cm diameters) containing 15 cm of water level (23-25 °C). The mice were then removed from the beakers, towel dried and placed in heated cages (37 °C). Each session was scored with Ethovision XT 12 (Noldus Information Technology). A mouse was determined to be immobile when it remained floating in water without struggling or movements sometimes to keep its head over water. The data were analyzed by Ethovision XT 12 (Noldus Information Technology) at a 0.5% immobility threshold.
Open field tests
Open field tests were conducted as described before [51]. Briefly, mice were placed individually in four plexiglas open field chamber (30×30×30 cm). One chamber was subdivided into nine 10 cm×10 cm arenas, including eight outer zone and one inner zone. The movement of each mouse during the 10 min test was recorded by the tracking software Ethovision XT 12 (Noldus Information Technology). Besides, prior to cleaning the chamber, the fecal boli pellets presented in the chamber were counted for further analysis.
Elevated plus maze tests
Anxiety analysis was performed as described before [52]. Briefly, the maze consisted of 2 open arms, 50 × 10 cm (L × W) and 2 closed arms, 50 × 10 × 50 cm (L × W × H) with an open roof. The 2 arms of each type were opposite to each other. The maze was elevated 50 cm from the ground with a video camera fixed above to record the movements of the subject. For the test, each mouse was placed in the center of the maze, facing one of the closed arms and the movement during 5 min was recorded and analyzed by software Ethovision XT 12 (Noldus Information Technology).
Novel object recognition tests
The tests were performed as described before with some modifications [53]. Briefly, the experiments were carried out in an open field box (30 cm × 30 cm × 30 cm). On the test day, each mouse was placed in the box with two objects located in two corners of the same side. The objects were plastic blocks of the same size (9 cm x 5 cm x 9 cm). The time spent exploring each object was recorded during 10 min period. After a break for 2 hrs in their home cage, the mice were placed in the box once again, in which one object used in the training session was replaced with a novel object with different shape and color. The time spent exploring each object was recorded during the subsequent 5 min period. The mice were regarded to be exploring when they were facing, sniffing or biting the object, and were scored by Ethovision XT 12 (Noldus Information Technology). A ratio of the time spent exploring any one of the two objects (during the training session) or the replaced novel one (during the test session) over the total time spent exploring both objects as a preference index, was used to measure memory preference.
Statistical analysis
One-way ANOVA was used to determine significant differences between control and experimental groups, with two-way ANOVA for sexes and genotypes. In all analysis, data are presented as Mean ± SEM from at least 3 independent experiments. *p<0.05, **p<0.01 versus respective control by Student's t test. Statistical analysis was conducted using GraphPad Prism 5 (GraphPad Software, La Jolla, USA). Cell size in eosin staining analysis was calculated using ImageJ software (https://imagej.nih.gov/ij/).
Results
Ablation of P11 leads to a decreased cell proliferation
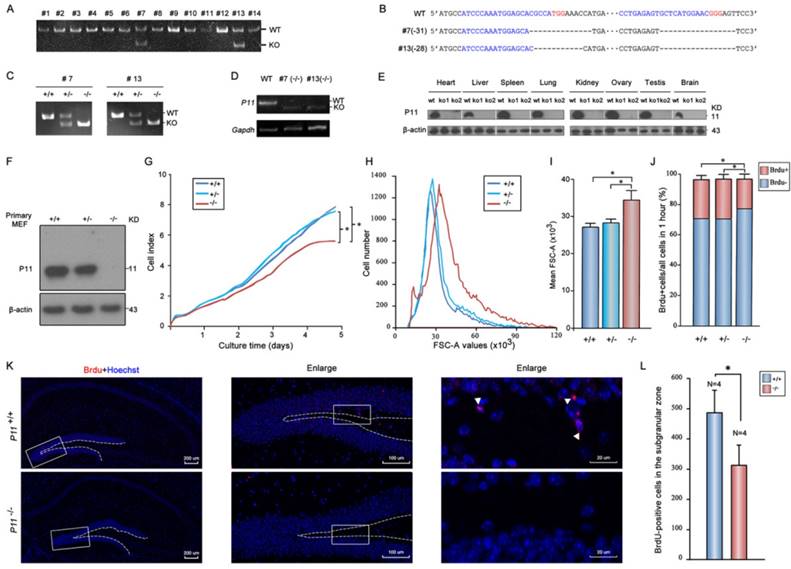
To elucidate the potential effect of P11 on cell proliferation, P11 knockout mice were generated using CRISPR/Cas9 technology. Two gRNAs were designed to target to around translation start codon (Fig. S1A). Two knockout lines (#7 and #13) were obtained (Fig. 1A). Off-target analysis confirmed the validity of genome editing of p11 gene using CRISPR/Cas9 system (Tables S1, S2). Deletions of 31 bp (#7) and 28 bp (#13) led to frameshift mutations (Fig. 1B, Fig. S1B). Homozygote mutant offsprings (P11-/-) were produced by intercross of the P11+/- mice (Fig. 1C). Although the mRNA of its mutant were present in P11 KO mice (Fig. 1D), the P11 protein was undetectable in different tissues of P11 KO mice (Fig. 1E). Consistent with previous reports, P11 is expressed relatively ubiquitously. Particularly, there is an obvious expression of P11 in brain samples of wide-type mice. Genotyping of heterozygous intercrosses revealed that proportions of wild-type, heterozygote and homozygote mutant mice were 28%, 50% and 22% respectively, which is consistent with genetic segregation of the Mendel's Laws (Table 1). The subsequent studies were performed with animals derived from the two independent lines.
Genotype distribution among offsprings of heterozygote intercrosses a
| Mice strains | Total | Genotype number of offsprings b | χ2 | p value | ||
|---|---|---|---|---|---|---|
| +/+ (%) | +/- (%) | -/- (%) | ||||
| # 7 | 170 | 47(27) | 86(51) | 37(22) | 1.2 | p>0.1 |
| #13 | 201 | 59(29) | 97(49) | 45(22) | 2.19 | p>0.1 |
| Total | 371 | 106(28) | 183(50) | 82(22) | 3.17 | p>0.1 |
a Total number of #7 offsprings: 170; Total number of #13 offsprings: 201; Total number of these two strains: 371. b N(P), N indicates the genotype number of each strain or these two strains, P indicates the proportion of each genotype. For example, 47(27) indicated that there were 47 P11+/+ mice among offsprings of #7 heterozygote intercrosses, and the proportion of P11+/+ offsprings was 27%. Expected distribution: 25% +/+, 50% +/- and 25% -/-; p>0.1, χ2 test.
Next, we investigated potential role of P11 in cell proliferation. We compared growth properties of P11+/+, P11+/- and P11-/- MEFs from E13.5 embryos. Real-time cell analysis (RTCA) showed a moderately slower growth rate after deletion of P11 (Fig. 1F, G). We also determined doubling time of the P11+/+, P11+/- and P11-/- MEFs, which was 33, 33 and 35 hours, respectively. More remarkable, primary P11-/- MEFs showed a lower saturation density than controls. Measurement of single MEF size further showed that P11-/- MEFs were significantly larger than wild-type ones and heterozygous cells (Fig. 1H, I). BrdU incorporation showed that depletion of P11 led to a decreased cell proliferation, from 25.77% or 26.07% to 19.32%, compared to those of P11+/+ or P11+/- MEFs (Fig. 1J). These results suggested that P11 knockout decreased cell proliferation and altered cell size.
To further elucidate the potential effect of P11 on cell proliferation in the hippocampus, we performed BrdU staining of the hippocampus in WT and P11 KO mice. A comparison of BrdU-positive cells in the hippocampus showed an decrease in the number of proliferating cells in P11 KO mice (Fig. 1K, L), suggesting a role of P11 in cell proliferation.
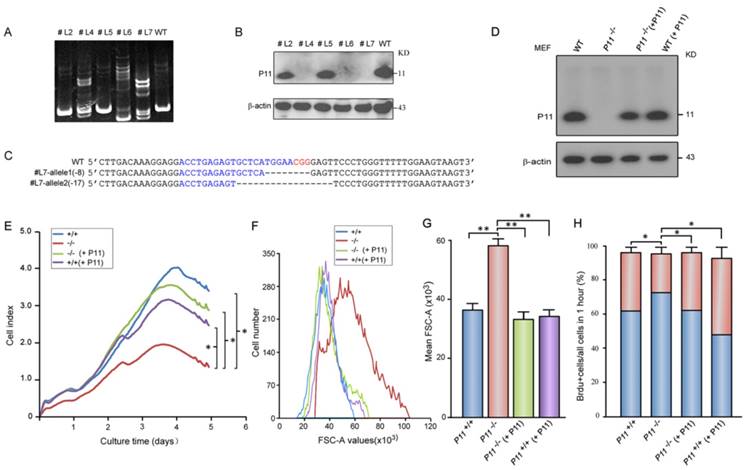
Rescue of growth retardation and cell size after P11 re-expression in P11-/- MEFs
To confirm roles of P11 in cell proliferation and regulation of cell size, we constructed two P11-/- MEF cell lines (Fig. 2A-C, Fig. S1C) and further rescued P11 expression in P11-/- MEF cell lines by P11 over-expression (Fig. 2D). MEF cell line with over-expression of P11 was also constructed (Fig. 2D). Consistently, cell proliferation was rescued after re-expression of P11 in P11-/- MEFs (Fig. 2E). Doubling time of the P11-/- MEFs was rescued from 28 hours in P11-/- MEFs to 17 hours after re-expression of P11. Consistent with the far lower saturation density in P11-/- MEFs, size of single P11-/- MEF was significantly larger than that of wild-type, while P11 re-expression in P11-/- MEFs rescued cell size as large as wild-type MEFs (Fig. 2F, G). Furthermore, BrdU incorporation measurement also demonstrated the rescued cell proliferation (Fig. 2H). These results indicated that P11 promoted cell proliferation and regulated cell size.
Targeted disruption of P11 in mice using CRISPR/Cas9 system and regulation of cell proliferation and morphology by P11. A. PCR analysis of genomic DNA isolated from mice tails of F0 generation. The KO band (~150 bp) can be distinguished from WT band (~180 bp). Primer sequences and PCR conditions are listed in Table S1. B. Nucleotide sequence alignments of P11 mutant alleles of #7 and #13 strains from F2 generation with WT sequence. The deleted sequences are indicated in dash lines. C. PCR analysis to determine genotypes of offsprings from heterozygote intercrosses in two KO strains. The absence of a 182-bp WT band signifies the generation of the P11 KO offsprings. D. RT-PCR analysis of P11 expression in P11 KO mice. Brain extracts from adult mice were subjected to RT-PCR to detect P11 expression. Gapdh was used as a control. E. Western blots of P11 protein expression in different tissues from P11 WT and KO mice. Tissues extracts from adult mice (ko1, #7; ko2, #13; and wt) were subjected to Western blots to detect P11 expression. β-actin was used as a control. F. Western blot analysis of P11. Primary MEFs were derived from 13.5-day-old embryos and cultured at 37 °C in 5 % CO2. β-actin was used as a control. G. Growth kinetics of MEFs from P11+/+, P11+/- and P11-/- embryos. Primary MEFs were cultured in E-Plate 16 at a density of 2×103 cells per well to perform RTCA for 5 days with each of these cells repeated four wells. The impedance value of each well was automatically monitored by the xCELLigence system. The strength of impedance is positively correlated with the number of cells having attached to the electrodes and is recorded as cell index (CI) values. Three independent experiments, n = 4 wells of cells per genotype. Two-way ANOVA was performed. *p < 0.05. H. Cell size of P11+/+, P11+/- and P11-/- MEFs determined by flow cytometry (N=3). Forward scatter (FSC-A) values, as a measure of cell size. Forward scatter intensity was measured by flow cytometry. I. Statistical analysis of the mean FSC-A values of MEFs in (H). Data are from three independent experiments (mean ± SEM, n= 3). One-way ANOVA was performed. *p < 0.05. J. BrdU incorporation analysis. P11+/+, P11+/- and P11-/- MEFs at the logarithmic growth phase were labeled with 10 µM BrdU for 1 h in culture. The cells were then harvested, fixed, and stained with anti-BrdU antibody. Stained cells were analyzed by flow cytometry. Data are from three independent experiments (mean ± SEM, n= 3). One-way ANOVA was performed. *p < 0.05. K. Cell proliferation in the hippocampus in WT and P11 KO mice. Representative micrographs of the hippocampus labeled with the cell proliferation marker BrdU (arrowheads). The image on the right is enlarged from the box area of the image on the left. The subgranular zone of the dentate gyrus was indicated with dotted lines. L. Statistics of BrdU-positive cells in the subgranular zone. *p<0.05, data represent mean ± SEM for indicated groups. One-way ANOVA was performed.
Targeted disruption of P11 in MEF cell lines using CRISPR/Cas9 system and decreased cell proliferation and altered morphology of P11-/- MEFs. A. Detection of CRISPR/Cas9-mediated mutation of P11 locus by PAGE-based genotyping assay. MEF cells were infected with specified P11-gRNA lentivirus. The genomic DNA from the different cell clones was extracted and PCR amplified to test P11 disruption in 12% PAGE gel. Primer sequences and PCR conditions are listed in Table S1. B. Western blot analysis of P11 expression in cellular extracts prepared from the clones shown in (A), β-actin was a control. C. Nucleotide sequence alignments of two P11 alleles of the modified #L7 clone with WT sequence. The deleted sequences are indicated in dash lines. D. Western blot analysis of rescued P11-/- MEFs by over-expression of P11 (P11-/- + P11). P11 knockout cell line was constructed by CRISPR/Cas9. To rescue P11 expression in P11-/- MEF cells, the cells were infected with the MSCV- P11 retroviral vectors, while WT MEFs with over-expression of P11 were used as a control. E. Growth kinetics of P11+/+, P11-/-, P11-/- (+ P11) and P11+/+ (+ P11) MEFs. RTCA was performed as shown in Fig. 1G. Three independent experiments, n = 4 wells of cells per genotype. Two-way ANOVA was performed. *p < 0.05. F. Rescued cell size of P11-/- (+ P11) MEFs determined by flow cytometry (N=3). Forward scatter (FSC-A) values, as a measure of cell size. Forward scatter intensity was measured by flow cytometry. G. Statistical analysis of the mean FSC-A of MEFs in (F). Data are from three independent experiments (mean ± SEM, n= 3). One-way ANOVA was performed. **p < 0.01. H. BrdU incorporation analysis. P11+/+, P11-/-, P11-/- (+ P11) and P11+/+ (+ P11) MEFs at the logarithmic growth phase were labeled with 10 µM BrdU for 1 h in culture. The cells were then harvested, fixed, and stained with anti-BrdU antibody. Stained cells were analyzed by flow cytometry. Data are from three independent experiments (mean ± SEM, n= 3). One-way ANOVA was performed. *p < 0.05.
Ablation of P11 alters MEF morphology
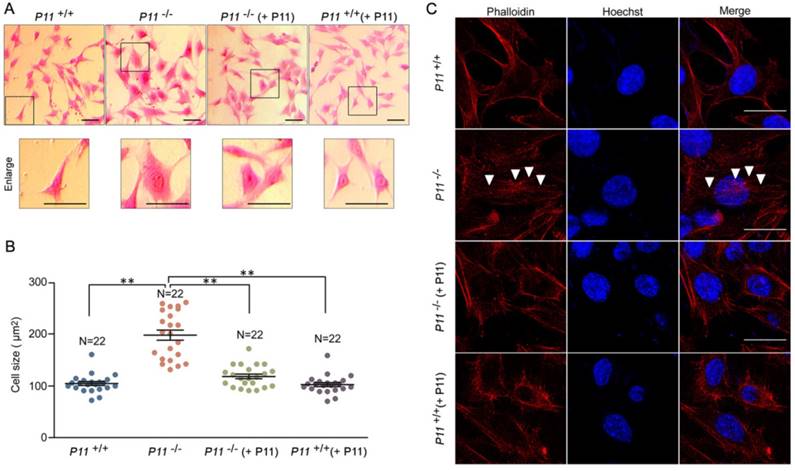
To further observe cell phenotype, we performed eosin staining and cell size analysis of P11+/+, P11-/-, P11-/- (+ P11) and P11+/+ (+ P11) MEFs. Knockout of P11 led to a larger cell size as well as the nucleus, in comparison with wild type, while P11 re-expression in P11-/- MEFs rescued the cell size (Fig. 3A, B). To explore influence of P11 knockout on F-actin cytoskeleton, we performed F-actin staining by phalloidin. Ablation of P11 showed an obvious accumulation of F-actin stress fibers (Fig. 3C). While P11-/- (+ P11) and P11+/+ (+ P11) MEFs shared the similar distribution of F-actin stress fibers as P11+/+ MEFs (Fig. 3C). These results suggested that P11 may regulate MEF morphology through organization of F-actin cytoskeleton.
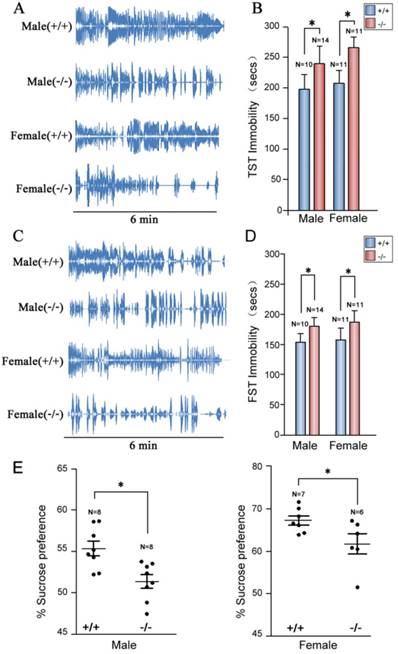
Depression-like disorders in the P11 KO mice
To confirm depression-like behaviors of P11 knockout mice targeted by homologous recombinetion in ES cells [25], we performed tail suspension tests. Consistently, P11 KO mice exhibited a similar depression-like phenotype as the original P11 KO mouse line. Immobility time of P11 knockout mice was significantly higher than wild-types (Fig. 4A, B), as well as the performance in forced swimming tests (Fig. 4C, D). Sucrose preference tests further confirmed this depression-like phenotype, in which P11 knockout mice did not show an obvious sucrose preference than wild-types (Fig. 4E). These data confirmed depression-like behaviors in P11 knockout mice produced by CRISPR/Cas9 technology.
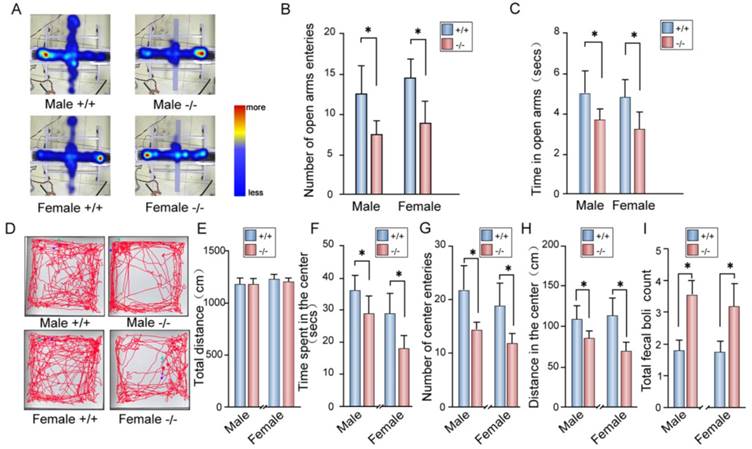
Anxiety-like disorders in the P11 KO mice
Depression is often accompanied by anxiety in patients [54]. Thus, we measured anxiety behavior in P11 KO mice. In elevated plus maze tests, P11 KO mice displayed less entrance into the open arms than wild-type mice (Fig. 5A, B), while KO mice spent less time in the open arms and more time in the closed arms than wild-type mice (Fig. 5C).
Ablation of P11 alters MEF morphology. A. Eosin staining of MEF cell lines, P11+/+, P11-/-, P11-/- (+ P11) and P11+/+ (+ P11). Cell size of P11 knockout cells is larger than WT MEFs, which was rescued after re-expression of P11. Dashed boxes showed enlarged area. Enlarged cells were shown in lower panel. Scale bar, 30 µm. B. Statistical analysis of cell size in (A). Cell size was calculated using ImageJ software. One-way ANOVA was performed. **p < 0.01. C. Phalloidin staining of F-Actin filaments of MEF cell lines. Accumulated F-Actin filaments were observed in P11 knockout cell line. P11+/+, P11-/-, P11-/- (+ P11) and P11+/+ (+ P11) MEFs were subjected to phalloidin staining of F-Actin filaments (red). The nuclei were stained by Hoechst (blue). White arrowheads highlighted F-Actin filaments. Scale bar, 30 µm.
We further tested anxiety-like phenotype in P11 KO mice by open field tests. The P11 KO mice spent less time in the center of the open field arena and more time in the margin area than wild-type mice (Fig. 5D-F), while there is no difference in overall hyperactivity as measured by the horizontal distance traveled when compared to wild-type mice (Fig. 5E). Consistently, P11 KO mice entered into the center less (Fig. 5G) and traveled a less distance in the center (Fig. 5H) than wild-type mice. Furthermore, P11 KO mice had more counts of fecal boli than wild-type mice (Fig. 5I). These data showed an increased anxiety in P11 KO mice.
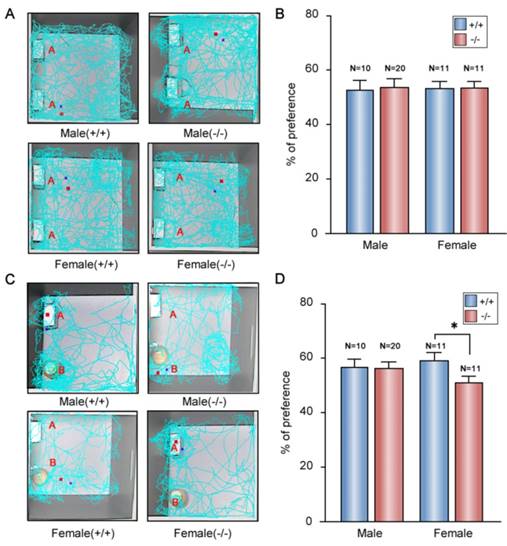
Memory deficit in the P11 KO females
Memory deficit were observed in patients with depression and anxiety-like disorders [55]. An obvious impairment of memory performance was also observed in genetic rat model of depression [56]. Thus, we performed the novel object recognition tests to evaluate memory characterization of the P11 KO mice. In the tests, we used a ratio of the time spent exploring novel object over the total time spent exploring both objects. In training session, all the mice spent the same time to explore the two identical objects in the open field (Fig. 6A, B). In the test session, there was no difference between KO and wild-type males (Fig. 6C, D). However, compared to wild-type females, KO females displayed less novel object exploring behavior (Fig. 6C, D), which indicated a memory deficit in the P11 KO females.
Discussion
Depression, anxiety and memory deficit are often caused by a combination of genetic, environmental, and psychological factors. Molecular mechanisms underlying these disorders remain elusive. In adult humans, many questions concerning neurogenesis in hippocampus also remain unanswered [43]. To explore the functions of P11 in cell proliferation, in the present study, we generated P11 knockout mice by CRISPR/Cas9 technology and observed that ablation of P11 leads to a decreased cell proliferation. P11 knockout resulted in a larger cell size, compared with that of wild type, which resulted from accumulated F-actin stress fibers. Furthermore, BrdU staining in the hippocampus showed a decrease of the number of proliferating cells in P11 KO mice. We also explored the contribution of P11 to depression, memory deficit and anxiety. We observed that anxiety was accompanied by depression when P11 was knockout in mice. Particularly, P11 knockout resulted in memory deficit in female mice, in addition to depression and anxiety. These findings suggest that there is an intrinsic and molecular correlation of these disorders linked by P11, which probably regulates neurogenesis by cell proliferation and morphology. These findings are valuable for understanding of the roles of P11 in several neurological disorders.
Depression-like phenotypes in P11 KO mice. A. Representative histograms of WT and P11 KO mice in the tail suspension tests within 6 min. Individual histograms showed temporal courses of 'struggle' and 'immobile' episodes. B. Statistical analysis of immobility time during tail suspension tests. Data represent mean ± SEM for indicated number per group, two-way ANOVA was performed. *p < 0.05. C. Representative histograms of WT and P11 KO mice in the forced swimming tests within 6 min. Individual histograms showed temporal courses of 'struggle' and 'immobile' episodes. D. Statistical analysis of immobility time during forced swimming tests (FST). Data represent mean ± SEM for indicated number per group, two-way ANOVA was performed. *p < 0.05. E. Statistical analysis of sucrose preference tests. Consumed sucrose solution or water was analyzed by the weight ratio of (sucrose / [sucrose + water]). Data represent mean ± SEM for indicated number per group, one-way ANOVA was performed. *p < 0.05.
There are two main hypotheses to underlie the etiology of depression-like disorders. Monoamine hypothesis suggests that deficiency of 5-HT and norepinephrine function leads to depression, and neurogenesis hypothesis has been proposed to focus on the downstream events, particularly induced growth of new neurons after antidepressant therapies [57]. Antidepressant therapies effectively prevent the reuptake or metabolism of neurotransmitters, and often take 4-6 weeks for clinical improvement of symptom, which suggest possible neurogenesis during this treatment. Adult hippocampal neurogenesis contributes to modulation of cognitive behaviors, and its dysregulation, especially in the dentate gyrus, triggers psychiatric disorders, such as depression[58]. Consistently, we observed that there is an obvious expression of P11 in brain samples of wide-type mice. BrdU staining in dentate gyrus of P11 KO mice further showed a decrease of the number of proliferating cells. Thus, our results support the neurogenesis hypothesis, which may explain pathogenesis of depression disorder.
Since detected in the dentate gyrus of young rats by injection with thymidine-H3[40], neurogenesis was found to persist into the eighth decade of life in adult humans [41]. Although the number of proliferating progenitors and young neurons in the dentate gyrus decreases sharply after birth, neurogenesis can be also observed in teenagers [42]. In adult humans, many questions concerning neurogenesis in hippocampus remain unanswered [43], because evolutionary divergence in brain exists between human and rodents. In addition, treatment of fluoxetine, an antidepressant agent, showed an increase of proliferating cells in hippocampus of wild-type mice. However, no difference between fluoxetine-treated and control P11 KO mice was observed [44], indicating an association of P11 with cell proliferation. These studies support our findings that P11 probably regulates neurogenesis by cell proliferation and morphology.
Identification of specific molecules that modulate cognitive behaviors would contribute to our better understanding of pathophysiology of psychiatric disorders. Several genes that regulate neurogenesis have been identified, which are associated with anxiety or depression disorders, such as p21Cip1 [59], miR-17-92 cluster [60] and PINK1[61]. In the adult brain, P11 emerged as an important component of 5-HT signaling. Multiple chemical and behavioral factors trigger neurogenic processes. Consistent with this, nerve growth factor (NGF) can regulate P11 expression levels in the pheocytochroma cell line PC12, which suggested a possible role for p11 in the regulation of cell growth [62, 63]. Likewise, NGF induced P11 expression in primary cultured dorsal root ganglia neurons [64]. Brain-derived neurotrophic factor (BDNF) can also increase P11 protein levels in primary hippocampal cells [65], and the antidepressant-like effect of BDNF in rodent models requires P11 [66]. Cell proliferation was attenuated by fluoxetine (antidepressant drug) in the subgranular zone in hippocampus of P11 KO mice, compared with WT mice [44]. Furthermore, P11 was also expressed in fetal cortex, thalamus, hippocampus, and hypothalamus at gestation day 17, and a decreased expression of P11 after exposure of citalopram (antidepressant drug) in uterus led to significantly decreased neurogenesis in relevant fetal brain regions [67]. In addition, down-regulation of P11 with siRNA or microRNA decreased cell proliferation in cancer cell lines [68-70]. Consistent with these, our results demonstrated that P11 can promote cell proliferation and maintain cell size through F-actin filaments, which probably affect hippocampal dendritic morphology and hippocampal cell proliferation both in fetal and adult brain. This speculation needs to be further elucidated.
Another important finding in our study is the molecular link of anxiety comorbid with depression and memory deficit through P11 action. Particularly, P11 knockout resulted in memory deficit in female mice, in addition to depression and anxiety. Actually, female patients of depression are more likely to have anxiety and/or memory deficit, compared with men [2, 71, 72]. The sex difference may result from effects of steroid hormones. There are plenty of sex differences that exist in the hormonal regulation of neurogenesis [73, 74]. Whether P11 is involved in this process needs to be further explored. Although many efforts have been made to explain pathological mechanisms of these disorders [43, 75, 76], molecular link of depression, anxiety and memory deficit remains elusive. Our study provides evidence that P11 contributes to comorbid of depression, anxiety and memory deficit in mouse models. Further evidence should be obtained through exploring hippocampal cell proliferation in human patients.
In summary, we first demonstrated that P11 can promote cell proliferation and maintain cell size through F-actin filaments both in primary MEF and MEF cell lines. BrdU staining in the hippocampus indicated a decrease in the number of proliferating cells in P11 KO mice. Secondly, we observed depression-like phenotype accompanied with anxiety-like phenotype in P11 KO mice, as well as memory deficit in the female KO mice, confirming a molecular association of depression behavior with anxiety and memory deficit through P11. Our study suggested a new approach for better understanding of the effect of P11 on depression, anxiety and memory deficit.
Anxiety-like phenotypes in P11 KO mice. A. Representative images for both P11 WT and KO mice in the elevated plus maze tests. Tracks traveled and time spent of each mouse were recorded for 10 min in each test. Scale bar represents time spent from less (blue) to more (red). Light blue indicates no visit. B-C. Statistical analysis of number of open arms entries (B) and time spent in the open arms (C) of P11 WT and KO mice in the elevated plus maze test. (N =11 female KO mice, 11 female WT mice, 20 male KO mice, 10 male WT mice), *p<0.05, data represent mean ± SEM, two-way ANOVA was performed. D. Representative tracks for both P11 WT and KO mice in the open field tests. Each track represents the total distance traveled by each mouse during the 10 min time period of the test. E-I. Statistical analysis of the distance traveled (E), time spent in the center area (F), number of center entries (G), distance in the center area (H) and total fecal boli count (I) of P11 WT and KO mice in the open field tests. (N =11 female KO mice, 11 female WT mice, 20 male KO mice, 10 male WT mice), *p<0.05, data represent mean ± SEM, two-way ANOVA was performed.
Memory deficit in the P11 female KO mice. A. Representative tracks for both WT and KO mice during the training session (~10 min). Two identical objects (A) are placed in the open field. Each track represents the total distance traveled by each mouse during the 10 min time period of the test. B. Statistical analysis of the preference index of P11 WT and KO mice during the training session. A preference percentage, a ratio of the amount of time spent exploring any one of the two objects over the total time spent exploring both objects. Data represent mean ± SEM for indicated number per group, two-way ANOVA was performed. C. Representative tracks for WT and KO mice during the test session (~5 min). A novel object (B) replaced one familiar object (A). Each track represents the total distance traveled by each mouse during the 5 min time period of the test. D. Statistical analysis of the preference index of P11 WT and KO mice during the test session. A preference percentage, a ratio of the amount of time spent exploring the replaced novel one over the total time spent exploring both objects. *p<0.05, data represent mean ± SEM for indicated number per group, two-way ANOVA was performed.
Abbreviations
KO: knock out; CRISPR: Clustered regularly interspaced short palindromic repeats; MDD: Major depressive disorder; AIIt: annexin II-P11 heterotetramer; 5-HT: 5-hydroxytryptamine; mGluR5: metabotropic glutamate receptor 5; CDS: coding sequence; MEF: mouse embryonic fibroblasts; dpc: days post-coitum; DMEM: Dulbecco's modified Eagle's medium; RTCA: real-time cell analysis; BrdU: bromodeoxyuridine; FSC: forward scatter; NGF: nerve growth factor; BDNF: Brain-derived neurotrophic factor; PAM: protospacer adjacent motif; TST: Tail suspension tests; FST: Forced swimming tests.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank Dr. Feng Zhang of Broad Institute of MIT and Harvard for providing LentiCRISPR-v2 plasmid and Dr. William J. Craigen of Baylor College of Medicine for MEF cells. We appreciate Shanghai Biomodel Organism Science & Technology Development Co. for the assistance in KO mice. This work was supported by the National Natural Science Foundation of China (31571280, 31771487, 31471182 and 31771370) and National Key Technologies R&D Program (2018ZX0800950B).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Vos T, Allen C, Arora M, Barber RM, Bhutta ZA, Brown A. et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. The Lancet. 2016;388:1545-602
2. Belmaker RH, Agam G. Major depressive disorder. N Engl J Med. 2008;358:55-68
3. Kessler RC, Bromet EJ. The epidemiology of depression across cultures. Annu Rev Public Health. 2013;34:119-38
4. Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593-602
5. Stavrakaki C, Vargo B. The relationship of anxiety and depression: a review of the literature. Br J Psychiatry. 1986;149:7-16
6. Wittchen HU. Generalized anxiety disorder: prevalence, burden, and cost to society. Depress Anxiety. 2002;16:162-71
7. Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem Biophys Res Commun. 2004;322:1111-22
8. Shang X, Cheng H, Zhou R. Chromosomal mapping, differential origin and evolution of the S100 gene family. Genet Sel Evol. 2008;40:449-64
9. Kube E, Becker T, Weber K, Gerke V. Protein-protein interaction studied by site-directed mutagenesis. Characterization of the annexin II-binding site on p11, a member of the S100 protein family. J Biol Chem. 1992;267:14175-82
10. Kassam G, Le BH, Choi KS, Kang HM, Fitzpatrick SL, Louie P. et al. The p11 subunit of the annexin II tetramer plays a key role in the stimulation of t-PA-dependent plasminogen activation. Biochemistry. 1998;37:16958-66
11. Poon WY, Malik-Hall M, Wood JN, Okuse K. Identification of binding domains in the sodium channel Na(V)1.8 intracellular N-terminal region and annexin II light chain p11. FEBS Lett. 2004;558:114-8
12. Johnsson N, Weber K. Alkylation of cysteine 82 of p11 abolishes the complex formation with the tyrosine-protein kinase substrate p36 (annexin 2, calpactin 1, lipocortin 2). J Biol Chem. 1990;265:14464-8
13. Gerke V, Weber K. The regulatory chain in the p36-kd substrate complex of viral tyrosine-specific protein kinases is related in sequence to the S-100 protein of glial cells. EMBO J. 1985;4:2917-20
14. Rety S, Sopkova J, Renouard M, Osterloh D, Gerke V, Tabaries S. et al. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat Struct Biol. 1999;6:89-95
15. Saris CJ, Kristensen T, D'Eustachio P, Hicks LJ, Noonan DJ, Hunter T. et al. cDNA sequence and tissue distribution of the mRNA for bovine and murine p11, the S100-related light chain of the protein-tyrosine kinase substrate p36 (calpactin I). J Biol Chem. 1987;262:10663-71
16. Zokas L, Glenney JR Jr. The calpactin light chain is tightly linked to the cytoskeletal form of calpactin I: studies using monoclonal antibodies to calpactin subunits. J Cell Biol. 1987;105:2111-21
17. Warner-Schmidt JL, Schmidt EF, Marshall JJ, Rubin AJ, Arango-Lievano M, Kaplitt MG. et al. Cholinergic interneurons in the nucleus accumbens regulate depression-like behavior. Proc Natl Acad Sci U S A. 2012;109:11360-5
18. Schmidt EF, Warner-Schmidt JL, Otopalik BG, Pickett SB, Greengard P, Heintz N. Identification of the cortical neurons that mediate antidepressant responses. Cell. 2012;149:1152-63
19. Oh YS, Gao P, Lee KW, Ceglia I, Seo JS, Zhang X. et al. SMARCA3, a chromatin-remodeling factor, is required for p11-dependent antidepressant action. Cell. 2013;152:831-43
20. Milosevic A, Liebmann T, Knudsen M, Schintu N, Svenningsson P, Greengard P. Cell- and region-specific expression of depression-related protein p11 (S100a10) in the brain. J Comp Neurol. 2017;525:955-75
21. Seo JS, Wei J, Qin L, Kim Y, Yan Z, Greengard P. Cellular and molecular basis for stress-induced depression. Mol Psychiatry. 2016;26:118
22. Svenningsson P, Kim Y, Warner-Schmidt J, Oh YS, Greengard P. p11 and its role in depression and therapeutic responses to antidepressants. Nat Rev Neurosci. 2013;14:673-80
23. Gerke V, Weber K. Identity of p36K phosphorylated upon Rous sarcoma virus transformation with a protein purified from brush borders; calcium-dependent binding to non-erythroid spectrin and F-actin. EMBO J. 1984;3:227-33
24. Johnsson N, Marriott G, Weber K. p36, the major cytoplasmic substrate of src tyrosine protein kinase, binds to its p11 regulatory subunit via a short amino-terminal amphiphatic helix. EMBO J. 1988;7:2435-42
25. Svenningsson P, Chergui K, Rachleff I, Flajolet M, Zhang X, El Yacoubi M. et al. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77-80
26. Warner-Schmidt JL, Flajolet M, Maller A, Chen EY, Qi H, Svenningsson P. et al. Role of p11 in cellular and behavioral effects of 5-HT4 receptor stimulation. J Neurosci. 2009;29:1937-46
27. Lee KW, Westin L, Kim J, Chang JC, Oh YS, Amreen B. et al. Alteration by p11 of mGluR5 localization regulates depression-like behaviors. Mol Psychiatry. 2015;20:1546-56
28. Guo J, Zhang W, Zhang L, Ding H, Zhang J, Song C. et al. Probable involvement of p11 with interferon alpha induced depression. Sci Rep. 2016;6:17029
29. Egeland M, Warner-Schmidt J, Greengard P, Svenningsson P. Co-expression of serotonin 5-HT(1B) and 5-HT(4) receptors in p11 containing cells in cerebral cortex, hippocampus, caudate-putamen and cerebellum. Neuropharmacology. 2011;61:442-50
30. Marongiu R, Arango-Lievano M, Francardo V, Morgenstern P, Zhang X, Cenci MA. et al. Gene therapy blockade of dorsal striatal p11 improves motor function and dyskinesia in parkinsonian mice. Proc Natl Acad Sci U S A. 2016;113:1423-8
31. Schintu N, Zhang X, Alvarsson A, Marongiu R, Kaplitt MG, Greengard P. et al. p11 modulates L-DOPA therapeutic effects and dyskinesia via distinct cell types in experimental Parkinsonism. Proc Natl Acad Sci U S A. 2016;113:1429-34
32. Green H, Zhang X, Tiklova K, Volakakis N, Brodin L, Berg L. et al. Alterations of p11 in brain tissue and peripheral blood leukocytes in Parkinson's disease. Proc Natl Acad Sci U S A. 2017;114:2735-40
33. O'Connell PA, Madureira PA, Berman JN, Liwski RS, Waisman DM. Regulation of S100A10 by the PML-RAR-alpha oncoprotein. Blood. 2011;117:4095-105
34. Ito Y, Arai K, Nozawa R, Yoshida H, Higashiyama T, Takamura Y. et al. S100A10 expression in thyroid neoplasms originating from the follicular epithelium: contribution to the aggressive characteristic of anaplastic carcinoma. Anticancer Res. 2007;27:2679-83
35. Phipps KD, Surette AP, O'Connell PA, Waisman DM. Plasminogen receptor S100A10 is essential for the migration of tumor-promoting macrophages into tumor sites. Cancer Res. 2011;71:6676-83
36. Liu M, Ge R, Liu W, Liu Q, Xia X, Lai M. et al. Differential proteomics profiling identifies LDPs and biological functions in high-fat diet-induced fatty livers. J Lipid Res. 2017;58:681-94
37. Arango-Lievano M, Schwarz JT, Vernov M, Wilkinson MB, Bradbury K, Feliz A. et al. Cell-type specific expression of p11 controls cocaine reward. Biol Psychiatry. 2014;76:794-801
38. Polimanti R, Meda SA, Pearlson GD, Zhao H, Sherva R, Farrer LA. et al. S100A10 identified in a genome-wide gene x cannabis dependence interaction analysis of risky sexual behaviours. J Psychiatry Neurosci. 2017;42:160189
39. Katsumata R, Shiotani A, Murao T, Ishii M, Fujita M, Matsumoto H. et al. Gender Differences in Serotonin Signaling in Patients with Diarrhea-predominant Irritable Bowel Syndrome. Intern Med. 2017;56:993-9
40. Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319-35
41. Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V. et al. Human Hippocampal Neurogenesis Persists throughout Aging. Cell Stem Cell. 2018;22(e5):589-99
42. Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW. et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018;555:377-81
43. Kempermann G, Gage FH, Aigner L, Song H, Curtis MA, Thuret S. et al. Human Adult Neurogenesis: Evidence and Remaining Questions. Cell Stem Cell. 2018;23:25-30
44. Egeland M, Warner-Schmidt J, Greengard P, Svenningsson P. Neurogenic effects of fluoxetine are attenuated in p11 (S100A10) knockout mice. Biol Psychiatry. 2010;67:1048-56
45. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783-4
46. Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84-7
47. Yuan J, Zhang Y, Sheng Y, Fu X, Cheng H, Zhou R. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11:1081-98
48. Steru L, Chermat R, Thierry B, Simon P. The tail suspension test: a new method for screening antidepressants in mice. Psychopharmacology. 1985;85:367-70
49. Iniguez SD, Warren BL, Parise EM, Alcantara LF, Schuh B, Maffeo ML. et al. Nicotine exposure during adolescence induces a depression-like state in adulthood. Neuropsychopharmacology. 2009;34:1609-24
50. Wieland S, Lucki I. Antidepressant-like activity of 5-HT1A agonists measured with the forced swim test. Psychopharmacology (Berl). 1990;101:497-504
51. Seibenhener ML, Wooten MC. Use of the Open Field Maze to measure locomotor and anxiety-like behavior in mice. J Vis Exp. 2015;6:e52434
52. Mansouri MT, Soltani M, Naghizadeh B, Farbood Y, Mashak A, Sarkaki A. A possible mechanism for the anxiolytic-like effect of gallic acid in the rat elevated plus maze. Pharmacol Biochem Behav. 2014;117:40-6
53. Karasawa J, Hashimoto K, Chaki S. D-Serine and a glycine transporter inhibitor improve MK-801-induced cognitive deficits in a novel object recognition test in rats. Behav Brain Res. 2008;186:78-83
54. Sartorius N, Ustun TB, Lecrubier Y, Wittchen HU. Depression comorbid with anxiety: results from the WHO study on psychological disorders in primary health care. Br J Psychiatry Suppl. 1996;30:38-43
55. Castaneda AE, Tuulio-Henriksson A, Marttunen M, Suvisaari J, Lonnqvist J. A review on cognitive impairments in depressive and anxiety disorders with a focus on young adults. J Affect Disord. 2008;106:1-27
56. Eriksson TM, Delagrange P, Spedding M, Popoli M, Mathe AA, Ogren SO. et al. Emotional memory impairments in a genetic rat model of depression: involvement of 5-HT/MEK/Arc signaling in restoration. Mol Psychiatry. 2012;17:173-84
57. Cunningham KA, Watson CS. Cell cycle regulation, neurogenesis, and depression. Proc Natl Acad Sci U S A. 2008;105:2259-60
58. Kang E, Wen Z, Song H, Christian KM, Ming G-l. Adult Neurogenesis and Psychiatric Disorders. Cold Spring Harb Perspect Biol. 2016;8:a019026
59. Pechnick RN, Zonis S, Wawrowsky K, Pourmorady J, Chesnokova V. p21Cip1 restricts neuronal proliferation in the subgranular zone of the dentate gyrus of the hippocampus. Proc Natl Acad Sci U S A. 2008;105:1358-63
60. Jin J, Kim SN, Liu X, Zhang H, Zhang C, Seo JS. et al. miR-17-92 Cluster Regulates Adult Hippocampal Neurogenesis, Anxiety, and Depression. Cell Rep. 2016;16:1653-63
61. Agnihotri SK, Sun L, Yee BK, Shen R, Akundi RS, Zhi L. et al. PINK1 deficiency is associated with increased deficits of adult hippocampal neurogenesis and lowers the threshold for stress-induced depression in mice. Behav Brain Res. 2019;363:161-72
62. Masiakowski P, Shooter EM. Nerve growth factor induces the genes for two proteins related to a family of calcium-binding proteins in PC12 cells. Proc Natl Acad Sci U S A. 1988;85:1277-81
63. Masiakowski P, Shooter EM. Changes in PC12 cell morphology induced by transfection with 42C cDNA, coding for a member of the S-100 protein family. J Neurosci Res. 1990;27:264-9
64. Okuse K, Malik-Hall M, Baker MD, Poon WY, Kong H, Chao MV. et al. Annexin II light chain regulates sensory neuron-specific sodium channel expression. Nature. 2002;417:653-6
65. Park SW, Nhu LH, Cho HY, Seo MK, Lee CH, Ly NN. et al. p11 mediates the BDNF-protective effects in dendritic outgrowth and spine formation in B27-deprived primary hippocampal cells. J Affect Disord. 2016;196:1-10
66. Warner-Schmidt JL, Chen EY, Zhang X, Marshall JJ, Morozov A, Svenningsson P. et al. A role for p11 in the antidepressant action of brain-derived neurotrophic factor. Biol Psychiatry. 2010;68:528-35
67. King JR, Velasquez JC, Torii M, Bonnin A. Effect of Maternal +/-Citalopram Exposure on P11 Expression and Neurogenesis in the Mouse Fetal Brain. ACS Chem Neurosci. 2017;8:1019-25
68. Yang X, Popescu NC, Zimonjic DB. DLC1 interaction with S100A10 mediates inhibition of in vitro cell invasion and tumorigenicity of lung cancer cells through a RhoGAP-independent mechanism. Cancer Res. 2011;71:2916-25
69. Shan X, Miao Y, Fan R, Qian H, Chen P, Liu H. et al. MiR-590-5P inhibits growth of HepG2 cells via decrease of S100A10 expression and Inhibition of the Wnt pathway. Int J Mol Sci. 2013;14:8556-69
70. Shang J, Zhang Z, Song W, Zhou B, Zhang Y, Li G. et al. S100A10 as a novel biomarker in colorectal cancer. Tumour Biol. 2013;34:3785-90
71. Robichaud M, Dugas MJ, Conway M. Gender differences in worry and associated cognitive-behavioral variables. J Anxiety Disord. 2003;17:501-16
72. Asher M, Aderka IM. Gender differences in social anxiety disorder. J Clin Psychol. 2018;18:22624
73. Koss WA, Frick KM. Sex differences in hippocampal function. J Neurosci Res. 2017;95:539-62
74. Gheorghe A, Qiu W, Galea LAM. Hormonal Regulation of Hippocampal Neurogenesis: Implications for Depression and Exercise. Curr Top Behav Neurosci. 2018:10
75. Balsamo M, Cataldi F, Carlucci L, Fairfield B. Assessment of anxiety in older adults: a review of self-report measures. Clin Interv Aging. 2018;13:573-93
76. Borelli WV, Schilling LP, Radaelli G, Ferreira LB, Pisani L, Portuguez MW. et al. Neurobiological findings associated with high cognitive performance in older adults: a systematic review. Int Psychogeriatr. 2018;18:1-13
Author contact
![]() Corresponding authors: Professors Rongjia Zhou and Hanhua Cheng, College of Life Sciences, Wuhan University, Wuhan 430072, P. R. China, Fax: 0086-27-68756253, E-mail: rjzhouedu.cn, hhchengedu.cn
Corresponding authors: Professors Rongjia Zhou and Hanhua Cheng, College of Life Sciences, Wuhan University, Wuhan 430072, P. R. China, Fax: 0086-27-68756253, E-mail: rjzhouedu.cn, hhchengedu.cn
Received 2019-2-2
Accepted 2019-3-9
Published 2019-5-20
