ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2019; 15(11):2296-2307. doi:10.7150/ijbs.33735 This issue Cite
Research Paper
Co-expression network analysis of lncRNAs and mRNAs in rat liver tissue reveals the complex interactions in response to pathogenic cytotoxicity
1. Faculty of Health Sciences, University of Macau, Taipa, Macau
2. State Key Laboratory of Respiratory Disease, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
*These authors contributed equally
Abstract
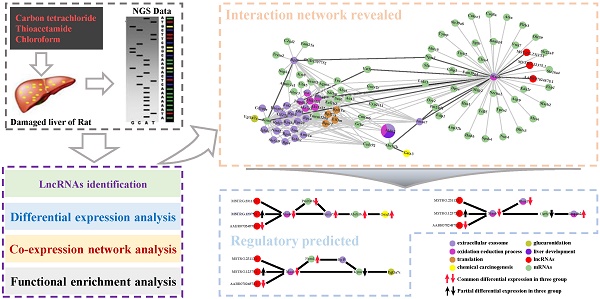
Liver is one of the most vital organs to maintain homeostasis because of its peculiar detoxification functionalities to detoxify chemicals and metabolize drugs and toxins. Due to its crucial functions, the liver is also prone to various diseases, i.e., hepatitis, cirrhosis and hepatoma, etc. Additionally, long non-coding RNAs (lncRNAs) has emerged as key regulators which are found to play important roles in transcription, splicing, translation, replication, chromatin shaping and post translational modification of proteins in living cells. However, the underlying mechanisms of biological processes mediated by lncRNA remain unclear. Here, with the aim of disclosing potential lncRNAs implicated in the biological processes in liver in response to cytotoxicity, we performed a co-expression network analysis based on the transcriptome data of the damaged liver tissue of Rattus norvegicus induced by three cytotoxic compounds (carbon tetrachloride, chloroform and thioacetamide). Our analysis unveils that many biological processes and pathways were collectively affected by the three cytotoxic compounds, including drug metabolism, oxidation-reduction process, oxidative stress, glucuronidation, liver development and flavonoid biosynthetic process, etc. Also, our network analysis has identified several highly conserved lncRNA-mRNA interactions participating in those correlated processes and pathways, implying their potential roles in response to the induced cytotoxicity in liver. Our study provides new insights into lncRNA-mRNA regulatory mechanisms in response to pathogenic cytotoxic damaging in liver and facilitates the development of lncRNA-oriented therapies for hepatic diseases in the future.
Keywords: LncRNAs, interaction network, cytotoxicity, liver disease
