10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2008; 4(6):338-344. doi:10.7150/ijbs.4.338 This issue Cite
Review
Inducible SOS Response System of DNA Repair and Mutagenesis in Escherichia coli
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawinskiego 5a, 02-106 Warszawa, Poland
Received 2008-8-20; Accepted 2008-9-17; Published 2008-9-23
Abstract
Chromosomal DNA is exposed to continuous damage and repair. Cells contain a number of proteins and specific DNA repair systems that help maintain its correct structure. The SOS response was the first DNA repair system described in Escherichia coli induced upon treatment of bacteria with DNA damaging agents arrest DNA replication and cell division. Induction of the SOS response involves more than forty independent SOS genes, most of which encode proteins engaged in protection, repair, replication, mutagenesis and metabolism of DNA. Under normal growth conditions the SOS genes are expressed at a basal level, which increases distinctly upon induction of the SOS response. The SOS-response has been found in many bacterial species (e.g., Salmonella typhimurium, Caulobacter crescentus, Mycobacterium tuberculosis), but not in eukaryotic cells. However, species from all kingdoms contain some SOS-like proteins taking part in DNA repair that exhibit amino acid homology and enzymatic activities related to those found in E. coli. but are not organized in an SOS system. This paper presents a brief up-to-date review describing the discovery of the SOS system, the physiology of SOS induction, methods for its determination, and the role of some SOS-induced genes.
Keywords: SOS response, DNA repair, DNA mutations, error-prone repair, mutagenic DNA polymerases
1. Historical Overview
After it was recognized that genes are composed of DNA (Oswald T. Avery, 1940), numerous experiments were performed to explore the chemical properties of DNA, mostly by treating bacteria and bacteriophages with a variety of agents and chemicals, like UV light, mitomycin C (MC), etc. Consequently, a growing list of bacterial mutants showing new and unusual properties was obtained, and their properties were subsequently determined.
The hypothesis of SOS response was developed on the basis of the following data: (i) The observation by Jean Weigle in l953 [1] that reactivation of UV-irradiated phage λ greatly increased when the irradiated phages were plated on previously irradiated E. coli host cells. This phenomenon was later termed W- or Weigle- reactivation [2]; (ii) Induction of prophage λ and lysis of bacteria (transformation from lysogenic to lytic development) when E. coli bacterial lysogens were UV irradiated [3 - 6], and (iii) Observation of filamentous growth of E. coli B cells in response to UV irradiation, suggesting a relation between the arrest of cell division, the mechanisms of λ prophage induction and UV-induced mutation. [7]. These data led Miroslav Radman to conclude that in E. coli there is a DNA repair system dependent on the LexA and RecA proteins that is induced when DNA is severely damaged and its synthesis arrested and its induction of this system is connected with induction of mutations. Radman named it "SOS repair" and "SOS replication" after an international telegraph (or optical) distress signal “SOS” in the Morse alphabet (three dots, three dashes, three dots).
The SOS hypothesis of Miroslav Radman was initially put forward in an unpublished letter sent to numerous researchers in l970, which was subsequently published only in1974 [2]. Evelyn Witkin hypothesized earlier that formation of filaments and prophage induction in irradiated E. coli B cells could have a related mechanism. The original letter of Radman and the Witkin's early paper, regarded as the basis for the discovery of the SOS response phenomenon, were recently reprinted in a paper by Bryn A. Bridges [8]. Further work along this line confirmed and developed this hypothesis. Systems resembling in some respect the SOS response described in E. coli were later found to operate in eukaryotic cells as well, but the bacterial and in eukaryotic responses are in fact substantially different [9].
2. Mechanism of SOS Induction: Role of RecA* Coprotease and LexA Repressor Protein
The recA and lexA genes were the first to be recognized as being involved in SOS induction. Mutations in these genes make cells highly sensitive to UV irradiation. The 27 kDa LexA and the 36 kDa RecA proteins were previously known as recombination proteins operating in the sexual life and genetic exchange of bacteria [10]. Presently, it is known that RecA protein also participates in genetic DNA exchange, in recF, recO, recR, recN and ruvABC-dependent recombinational DNA repair [11], and, together with LexA protein, plays a major role in the regulation of the SOS response. The down- and up-regulation of the SOS-induced genes is basically an interplay of two proteins, LexA repressor and RecA* where LexA is a transcriptional repressor protein, and RecA* is a coprotease aiding the autocatalytic selfcleavage of LexA [12-14].
Agents capable of inducing the SOS response system are, e.g., UV-radiation, MC, methyl methane sulfonate (MMS), and many other chemicals that disrupt DNA, arrest DNA synthesis, and cell division, and lead to accumulation of single stranded (ss) DNA. The level of RecA protein in bacterial cells (like that of UvrD helicase II) is very high. The RecA protein has a strong tendency to form nucleoprotein filaments on ssDNA, and a much weaker one with broken, double stranded (ds) DNA [15, 16]. This probably protects DNA against destruction, and is required for every aspect of RecA activity. The assembly of RecA on ssDNA proceeds in the 5'-3' direction at a ratio of 1 molecule RecA per 3 DNA bases, and requires dATP or ATP, but no ATP-ase activity. The disassembly, in contrast, requires hydrolysis of ATP to ADP and proceeds much more slowly than the assembly. RecA assembled on ssDNA acquires a coprotease activity, RecA*, which facilitates the self-cleavage of LexA protein resulting in derepression of SOS-regulated genes. LexA protein has a weak auto-cleavage activity, but its cleavage and derepression of the SOS genes occur only in the presence of the RecA* coprotease.
Each of the SOS-induced damage-inducible (din) or sos genes has near its promoter/operator site a specific 20-nucleotide-long “SOS-box" (also named, LexA-box) to which the LexA repressor protein is bound, preventing RNA polymerase binding and gene expression [12 , 14, 16, 17]. The SOS box has a palindromic structure suggesting that the LexA repressor binds as a dimer, as was later confirmed [18]. The role of the RecA* coprotease in SOS-induced cells therefore is: 1. to assist in the cleavage of LexA protein (202 amino acids) at the Ala84-Gly85 site, which causes derepression of SOS-genes [19, 20]; 2. to cleave the CI repressor of λ lambda phage, which transforms the phage from a lysogenic to a lytic form [6, 12]; 3. to process UmuD → UmuD' by nicking UmuD at the Cys24-Gly25 site [19, 20] which is a prerequisite for the assembly of the SOS-induced mutagenic DNA polymerase V (Pol V) consisting of UmuD'2C. The rate limiting step of Pol V synthesis is UmuD→ UmuD' processing, which occurs much more slowly than the self-cleavage of LexA. The role of Pol V in mutagenesis is translesion synthesis (TLS) across the damage in template DNA, enabling DNA replication, frequently at the cost of fidelity leading to mutation [21]. All these proteins, the CI repressor of λ phage and LexA repressor, UmuD, PolB/DinA (Pol II) and DinB (Pol IV) proteins are homologous within their carboxy-terminal domains, and all are encoded by din (sos) genes regulated under SOS response.
Induction of the SOS response proceeds until 45-60 min after treatment of bacteria with SOS inducing agents and then abruptly ceases. Within this time most of the lesions have been repaired. The timing of the derepression of individual din genes depends on the strength of the LexA repressor binding with the SOS box and on the ease with the LexA repressor is detached from a particular SOS-box.
3. Detection of SOS -Induced Genes
3.1. By din::lacZ formation and β-galactosidase assay
The SOS response was studied earlier by testing the increase in din genes expression either from the natural genes, or by using a reporter gene construct e.g., fusion a putative din promoter with promoter-less lacZ gene encoding β-galactosidase. Graham Walker and coworkers [14, 22] were the first to employ for this task a defective phage, Mu1d(Ap,lacZ) constructed by Casadaban and Cohen [23], which easily inserts randomly into the chromosome of E. coli K12 and creates a mutation. This phage bears a promoter-less lacZ gene, so that β-galactosidase is not expressed. However, when the Mu phage is by chance integrated under the promoter of a din gene forming a functional din::Mu-1d(Ap,lacZ) operon, β-galactosidase is synthesized in response to DNA damage.
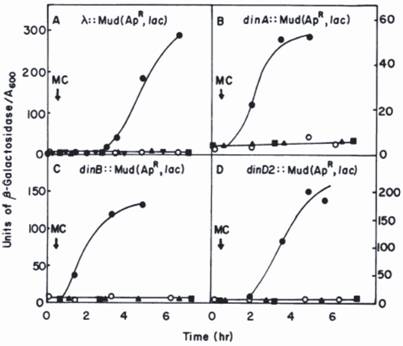
Since hydrolysis of β-galactosidase substrate (o-nitrophenyl-β-galactoside) forms yellow colonies, those bearing a din::Mu-lacZ fusion are easily selected on agar plates; the precise level of β-galactosidase expressed in response to DNA damage can then be accurately measured in liquid medium (see Fig.1 for details). In this way more than ten novel din-genes were detected and most of them subsequently identified.
Kinetics of induction of β-galactosidase in din::lacZ fusion strains by mitomycin C (MC) [14]. The din::lac fusions were generated by the insertion of the Mu d1(Ap, lac) bacteriophage into the E. coli chromosome. The λ::Mu d1(Ap lac) derivative was generated by an insertion of Mu d1(Ap lac) in the λ phage into E. coli chromosome. Symbols: o, untreated fusion strain; ●, fusion strains plus MC; lexA(Ind-) derivatives of the fusion strain plus MC; ■, recA (Def) derivatives of the fusion strain plus MC; ▼, a pKM280-containing derivative of the of the λ:: Mu d1(Ap lac) strain plus MC. Reprinted from [22] with author's permission. Two of genes, dinA and dinB were subsequently identified as polB (Pol II) and Pol IV, respectively [24, 25, 43].
Recently, a new method has been elaborated to measure SOS gene expression and promoter activity of the SOS-genes (e.g., recA, lexA, umuDC) by using a plasmid bearing an SOS promoter to be investigated fused to the reporter gene gfp-encoding green fluorescent protein (GFP) [26, 27]. This allows one to measure the promoter activity of SOS genes in a single bacterial cell, as well as localization and duration of the SOS induction. It appears that induction of the SOS genes does not proceed as a single event, but follows in several repeatable steps whose modulation depends on the SOS-inducing dose, the level of damage in the DNA, and the UmuD' protein accumulation. This method opens a new way to measuring the dynamics of the SOS response.
3.2. By search for SOS boxes: Determination of heterology index (HI)
Progress in DNA sequencing and knowledge of the characteristic elements of SOS gene sequences enabled direct computational searches for SOS-inducible genes. When 33% of E. coli chromosomal DNA had been sequenced, Lewis et al. localized by sequence analysis and quantitative DNA binding experiments six novel potentially LexA-regulated genes, and named them sosA-F [17]. For two of those, sosC and sosD, the authors confirmed experimentally that they strongly bound purified LexA repressor.
Subsequently, by comparing the sequences of SOS-boxes from 19 din genes known at the time (including sosA-sosF), they established that the consensus SOS box sequence is a perfect palindrome, TACTG(TA)5CAGTA [see also 14]; and on the basis of the theory of Berg and von Hippel they calculated mathematically for each of the SOS-boxes a heterology index (HI). This index indicates the deviation of an SOS box from the consensus and, when its value is low the gene is tightly suppressed, and when its value is high it is more easily de-repressed. At an HI greater than 15 the LexA repressor does not bind to the SOS-box [28]. Hence, HI value is a measure of the relative strength of LexA repressor binding to a given SOS box, and is responsible for the variation in derepression potential.
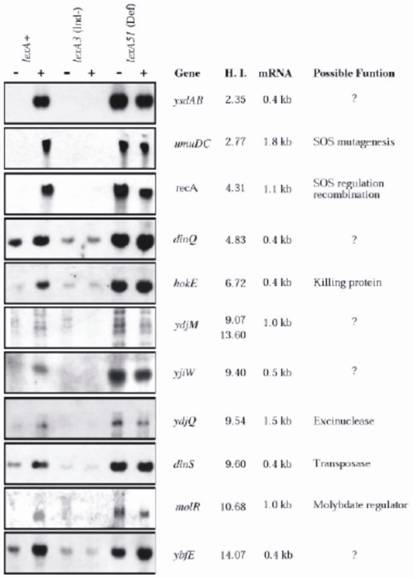
When the DNA sequence of the entire E. coli K12 chromosome has been determined Fernandez de Henestrosa et al [28] localized, by searching for potential SOS-boxes associated with open reading frames, 69 potential SOS-boxes with an HI value ≤ 15, including all previously known ones and seven novel. The new genes were subsequently analyzed for their ability to be expressed upon MC treatment, for the length of the expressed mRNA, and the HI values (Fig. 2). The analyses were conducted in three isogenic E. coli strains differing in the lexA allele: lexA+(wild type) SOS-inducible, SOS non-inducible lexA3(Ind-), and the constitutively expressed lexA51(Def) allele. Potential functions of the new and old genes were further characterized; and discussed. The results confirmed that each of the new SOS box-containing genes was indeed LexA-dependent gene, and its expression was induced by MC only in the lexA+ strain; otherwise, they were either not expressed (lexA3), or fully expressed (lexA51), regardless of whether the bacteria were MC-treated or not (see Fig 2 for details).
From the nucleotide sequence of the ydjQ gene (alternative names b1741 sosD) it was deduced that it encodes the 295 amino acid-long protein that shares significant homology with the N -terminal half of UvrC protein [28]. The UvrABC-excinuclease (consisting of UvrA, UvrB and UvrC proteins) was known to be involved in nucleotide excison repair (NER) which removes bulky adducts or structure-affecting lesions (e.g., pyrimidine dimers, pyrimidine (6-4) photoproducts, and abasic sites) from modified DNA [29]. It was also known that the N-part of UvrC incises ssDNA initially at the 3' side of the lesion, and then the C-part of UvrC incises the DNA at the 5' side of the lesion. Recently, Moolenaar et al, renamed the YdjQ protein as Cho (after UvrC homolog) and examined its enzymatic activity [30]. They confirmed that Cho protein incises UvrB-DNA preincision complexes only at the 3'- side of the lesion; however, some lesions in DNA that were very poorly incised by the UvrC protein were very efficiently incised by Cho protein. So, the Cho protein greatly increases the substrate range in DNA repair by the NER system. The uvrA, uvrB, and ydjQ (but not uvrC) genes are SOS-induced genes.
Northern analysis of E. coli genes that appear to be regulated by LexA. RNA was extracted from three isogenic strains that differed in lexA gene: RW118 (lexA+), RW434 (lexA[Ind-]), and RW542 (lexA51[Def]). RNA was obtained from undamaged cells (-) and from cells that had been exposed to mitomycin C (5 µg ml-1) (+) for 30 min before extraction. The previously identified LexA-regulated recA and umuDC genes were used as positive controls. The genes are depicted according to their ascending heterology index (HI). The sizes of the mRNA transcript and the possible functions of the genes are also indicated See ref. [28] for more details. (By courtesy of Blackwell Science).
3.3. Localisation of din genes by microarray techniques
The microarray technique allows a great number of genes to be monitored in one experiment. Courcelle et al., [31] used microarrays containing amplified E coli DNA chromosomal fragments with open reading frames from 4101 genes (95.5% of the total) to measure the expression of all the genes in UV-irradiated and non-irradiated SOS-inducible (lexA+) and non-inducible lexA1(Ind-) strains. In the UV irradiated lexA+ strain the authors identified 17 newly LexA-dependent SOS-induced genes, in addition to the 26 known beforehand; therefore the total number of SOS-inducible genes in E. coli is probably 43. In the same publication the authors established that the ssb gene coding for an ssDNA- binding protein is not SOS-inducible, as has been thought previously. They also observed a number of genes whose expression increased (usually not above twofold) in UV-irradiated cells but which were not regulated by LexA protein. They noted that protein transcripts from many genes unregulated by LexA were reduced after UV-irradiation, and concluded that these transcripts were probably either damaged or degraded by UV. They also identified thirty genes having potential SOS box-like structures, but which were not LexA- regulated.
4. Mechanism and Specificity of LexA Repressor Binding to SOS Boxes
It is believed that the sequences of all potential SOS boxes in the E. coli chromosome have been identified. Some examples of SOS boxes that bind (A) or not bind (B and C) LexA repressor, together with HI values and the number of mismatches (NM) are shown in Table 1. NM denotes the number of positions in SOS boxes deviating from a perfect palindrome. Both the number and the pattern of mismatches may be key for the specificity the LexA protein binding to each individual SOS box. It seems that this hypothesis is a good explanation for specificity and different strength of LexA protein binding to sequences of SOS boxes. But this should be confirmed.
It can be seen that generally, when the SOS-boxes have low HI values, between 2.7 and 12, they can bind LexA repressor (Table 1A); and when the HI is above 16.4 (Table 1B) they apparently are unable to bind LexA repressor. However, in some cases (shown in part C) such as the yigN (alternative name sosB), and dinJ (sosA) genes, the SOS boxes fail to bind LexA repressor despite their moderate HI values (9.27 and 7.06, respectively) [28].
Potential SOS boxes of genes that bind or do not bind LexA repressor.
| Gene | SOS box sequence | HI* | NM** |
|---|---|---|---|
| Consensus | TACTGTATATATATACAGTA | 0 | |
| A. Genes whose SOS boxes bind LexA and are regulated by LexA represor | |||
| recA | TACTGTATGAGCATACAGTA | 4.31 | 1 |
| umuDC | TACTGTATATAAAAACAGTA | 2.77 | 2 |
| uvrB | AACTGTTTTTTTATCCAGTA | 6.11 | 5 |
| polB | GACTGTATAAAACCACAGCC | 12.09 | 5 |
| lexA1 | TGCTGTATATACTCACAGCA | 6.34 | 4 |
| lexA2 | AACTGTATATACACCCAGGG | 8.32 | 6 |
| B. Genes whose potential SOS boxes do not bind LexA but re not LexA regulated | |||
| intE | GGCTGCTGAAAAATACAGAA | 16.04 | 7 |
| ymfI | TTCTGTACCAGAAAACAGTT | 15.48 | 8 |
| ymfM | AGCTGCAGGAGCATGCAGCA | 19.32 | 3 |
| lit | TGATGACAGAGTGTCCAGTG | 20.32 | 8 |
| C. Genes whose SOS boxes do not bind LexA in spate of low HI value | |||
| yigN | AACTGGACGTTTGTACAGCA | 9.27 | 5 |
| dinJ | AGCTGAATAAATATACAGCA | 7.06 | 3 |
Potential SOS boxes (sequence on coding strand) that bind (A), or do not bind (B and C) LexA repressor.
HI* denotes heterology index; NM** denotes the number of mismatches in SOS boxes deviating from a perfect palindrome. The lack of LexA repressor binding despite a relatively low HI value (section C) testifies that there is no direct correlation between them [28]. Anyhow, it indicates that the HI value cannot be the only indicator of the ability of an SOS box to bind LexA. The number of mismatches in the palindromic SOS boxes in each of the sections is similar, and does therefore not determine LexA binding ability with the SOS boxes. Data in parts A and C are from ref. [28], those in part B are from ref. [31].
5. Characteristics of Some SOS-Induced Genes
The SOS- response genes are found scattered throughout the E. coli chromosome as single genes situated in single operons. Six of them, umuDC (the source of Pol V), ruvAB (catalyzing branch migration in Holliday structures), and ysdAB (of unknown function) are encoded by pairs of genes forming an operon. Generally, only one SOS-box is present in one operon. The exceptions are the lexA and ydjM (b1728) genes that contain two SOS-boxes each (separated by one and two bases, respectively) and recN containing three SOS boxes. The sequences of the SOS boxes in one gene are different [28]. In the case of ydjM, two dimeric LexA repressors bind cooperatively to each SOS box, and as estimated, both of them are functional [28]. The sequences of the SOS-boxes in one gene differ by 2 to 4 bases. How and why the extra SOS-boxes in genes arise, and how they influence the gene expression potential are questions that remain to be answered.
6. The Time Required for Derepression of SOS-Induced Genes
The time scale for gene derepression and synthesis of the SOS-induced proteins varies for individual genes. The most rapidly derepressed genes (<1 min after SOS induction) include: lexA encoding LexA repressor protein (quickly degraded in SOS induced cells), uvrAB, cho and uvrD involved in NER repair, ruvAB taking part in recombinational DNA repair, polB and dinB encoding Pol II and Pol IV, respectively [24, 25, 43], and dinI, whose product inhibits processing of UmuD to UmuD' [32]. The UmuD' protein is necessary for the synthesis of the mutagenic Pol V (UmuD'2C) [21]. Therefore, the DinI protein retards synthesis of Pol V. The expression of the recA and recN genes encoding recombination and recombinational repair proteins, takes place 5 min after SOS induction, while that of sulA (old name, sfiA) and umuDC occurs at the latest stage of SOS induction [11]. SulA protein is an inhibitor of cell division causing filamentous growth of cells and prolonging the time during which the cellular DNA may be repaired.
7. Copy Numbers of din Genes Encoding Proteins
RecA and UvrD belong to the most abundantly synthesized proteins. Their numbers at a constitutive level are, respectively, about 10,000 and 8,000 copies per cell and increase 10-fold after SOS induction [11]. The RecA protein binds to ssDNA and probably protects it against uncontrolled destruction. UvrD, DNA helicase II, participates in dam-instructed mutHLS-dependent mismatch repair (MMR) [33], and takes part in UvrABC- and Cho-dependent (NER) repair by displacing the damage-containing ssDNA from the repaired DNA strand [29, 30]. The numbers of protein molecules synthesized in uninduced vs. SOS-induced cells are as follows: 20:250 for UvrA; 250:1000 for UvrB; 40:300 for DNA Pol II, and 250:2500 for DNA Pol IV (11, 34). UmuD protein is expressed, at 180 molecules per uninduced, and at 2400 molecules per lacked functional LexA repressor cell; there is 200 UmuC molecules per SOS-induced cell and no Pol V (< 15 molecules) in uninduced cell [35, 16].
8. Mutagenic SOS-induced DNA Polymerases
In E. coli, apart from the constitutively synthesized DNA replicating Pol III there are three potentially mutagenic DNA polymerases whose synthesis is increased (Pol II and Pol IV) or occurs only in SOS-induced cells (Pol V). Among these, Pol II is the only DNA polymerase that possesses a 3'-5' exonuclease proofreading activity and it is the least error-prone; its role also includes the recovery of degraded DNA at replication forks [36]. Both pol II and pol IV appear in the early stages of SOS induction, and Pol V in its final stage [11, 28, 16]. Pol V is the most error-prone enzyme and the most important one for mutagenicity of the SOS-induced cells.
In E. coli defective in umuD(C) almost no mutations are induced after UV-irradiation [37] and the mutations induced by MMS treatment are greatly reduced [38, 39]. The major mutagenic lesions formed in DNA after UV irradiation are TT-cis-syn cyclobutane dimers and CT or TT (6-4) photoproducts [14, 16], while after MMS treatment, 3-methyladenine (3meA), apurinic sites and 1meA and 3meC (in alkB-mutant cells) are found [37, 38] predominate. Both UV and MMS, like many other mutagens, are SOS inducers and hence although the damaging lesions are different, the SOS-inducing signal must be common; it is generally accepted that SOS-inducing signals are RecA/ssDNA filaments formed on accumulating ssDNA in the cells when DNA synthesis is arrested. Some of the premutagenic lesions require mutagenic DNA polymerases to lead to mutations, while others do not. However which SOS-induced, mutagenic DNA polymerase is required depends on the type of lesion [40-42].
9. Conclusion
The hypothesis of SOS response was astonishing, fruitful and inspiring. We gathered much information regarding the metabolism of DNA, the expression of the SOS-induced genes, and their functions. Yet, new ideas are still forthcoming.
Acknowledgements
I am greatly thankful to Professor Graham C.Walker, Roger Woodgate and Blackwell Science Publisher for reprinting permissions.
Conflict of Interest
The author has declared that no conflict of interest exists.
References
1. Weigle JJ. Induction of mutation in a bacterial virus. Proc Natl Acad Sci U S A. 1953;39(7):628-636
2. Radman M. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis. In: (ed.) Sherman S, Miller M, Lawrence C, Tabor WH. Molecular and Environmental aspects of mutagenesis. Springfield IL: Charles C Thomas publisher. 1974:128-142
3. Borek E, Ryan A. The transfer of irradiation-elicited induction in a lysogenic organism. Proc Natl Acad Sci U S A. 1958;44(5):374-347
4. Hertman I, Luria SE. Transduction studies on the role of a rec+ gene in the ultraviolet induction of prophage lambda. J Mol Biol. 1967;23(2):117-133
5. Defais M, Fauquet P, Radman M, Errera M. Ultraviolet reactivation and ultraviolet mutagenesis of lambda in different genetic systems. Virology. 1971;43(2):495-503
6. Craig R. Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein-directed cleavage of phage lambda repressor. J Biol Chem. 1981;256(15):8039-8044
7. Witkin EM. Ultraviolet-induced mutation and DNA repair. Annu Rev Microbiol. 1969;23:487-514
8. Bridges BA. Error-prone DNA repair and translesion DNA synthesis II: The inducible SOS hypothesis. DNA Repair (Amst). 2005;4(6):725-739
9. Eller MS, Asarch A, Gilchrest BA. Photoprotection in human skin-A multifaceted SOS response. Phtochem & Photobiol. 2008;84:339-349
10. Clark AJ, Margulies AD. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc Natl Acad Sci U S A. 1965;53:451-459
11. Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63(4):751-813
12. Horii T, Ogawa T, Nakatani T, Hase T, Matsubara H, Ogawa H. Regulation of SOS functions: purification of E. coli LexA protein and determination of its specific site cleaved by the RecA protein. Cell. 1981;27(3 Pt 2):515-522
13. Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29(1):11-22
14. Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48(1):60-93
15. Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J Mol Biol. 1999;288(3):391-401
16. Schlacher K, Pham P, Cox MM, Goodman MF. Roles of DNA polymerase V and RecA protein in SOS damage-induced mutation. Chem Rev. 2006;106(2):406-419
17. Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol. 1994;241(4):507-523
18. Thliveris AT, Little JW, Mount DW. Repression of the E coli recA gene requires at least two LexA protein monomers. Biochimie. 1991;73(4):449-456
19. Burckhardt SE, Woodgate R, Scheuermann RH, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci U S A. 1988;85(6):1811-1815
20. Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85(6):1806-1810
21. Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, Goodman MF. UmuD'(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci U S A. 1999;96(16):8919-8924
22. Kenyon CJ, Walker GC. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980;77(5):2819-2823
23. Casadaban MJ, Cohen SN. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979;76(9):4530-4533
24. Bonner CA, Hays S, McEntee K, Goodman MF. DNA Polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:7663-7667
25. Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RPP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4(2):281-286
26. Friedman N, Vardi S, Ronen S, Alin M, Stavans J. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol. 2005;3(7):e238
27. Krishna S, Maslov S, Sneppen K. UV-induced mutagenesis in Escherichia coli SOS response: a quantitative model. PLoS Comput Biol. 2007;3(3):e41
28. Fernandez de Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35(6):1560-1572
29. Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58(3):317-29
30. Moolenaar GF, van Rossum-Fikkert S, van Kesteren M, Goosen N. Cho, a second endonuclease involved in Escherichia coli nucleotide excision repair. Proc Natl Acad Sci U S A. 2002;99(3):1467-1472
31. Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158(1):41-64
32. Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. Inhibition of Escherichia coli RecA coprotease by Din I. EMBO J. 1998;17(11):3207-3216
33. Cooper DL, Lahue R.S, Modrich P. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101-133
34. Kim S.-R, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal din genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Cell. 1999;4(2):281-286
35. Woodgate R, Ennis DG. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991;229(1):10-16
36. Rangarajan S, Woodgate R, Goodman MF. A phenotype for enigmatic DNA polymerase II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci USA. 1999;96(16):9224-9229
37. Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156(2):121-131
38. Sledziewska-Gojska E, Janion C. Alternative pathways of methyl methanesulfonate-induced mutagenesis in Escherichia coli. Mol Gen Genet. 1989;216(1):126-31
39. Nieminuszczy J, Sikora A, Wrzesinski M, Janion C, Grzesiuk E. AlkB dioxygenase in preventing MMS-induced mutagenesis in Escherichia coli: effect of Pol V and AlkA proteins. DNA Repair (Amst). 2006;5(2):181-188
40. Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19(22):6259-6265
41. Fuchs RP, Fujii S, Wagner J. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem. 2004;69:229-264
42. Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Molec Biol. 2007;42:373-397
43. Iwasaki H, Nakata A, Walker GC, Shinagawa H. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol. 1990;172(11):6268-6273
Author contact
![]() Correspondence to: Prof. Celina Janion, celinawaw.pl
Correspondence to: Prof. Celina Janion, celinawaw.pl

 Global reach, higher impact
Global reach, higher impact