10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(2):97-117. doi:10.7150/ijbs.5.97 This issue Cite
Review
The Fascinating World of RNA Interference
1. Department of Biosciences, Jamia Millia Islamia (A Central University), New Delhi - 110 025, India.
2. Plant Molecular Biology Group, International Centre for Genetic Engineering and Biotechnology, Aruna Asaf Ali Marg, New Delhi - 110 067, India.
Received 2008-7-3; Accepted 2008-11-2; Published 2009-1-15
Abstract
Micro- and short-interfering RNAs represent small RNA family that are recognized as critical regulatory species across the eukaryotes. Recent high-throughput sequencing have revealed two more hidden players of the cellular small RNA pool. Reported in mammals and Caenorhabditis elegans respectively, these new small RNAs are named piwi-interacting RNAs (piRNAs) and 21U-RNAs. Moreover, small RNAs including miRNAs have been identified in unicellular alga Chlamydomonas reinhardtii, redefining the earlier concept of multi-cellularity restricted presence of these molecules. The discovery of these species of small RNAs has allowed us to understand better the usage of genome and the number of genes present but also have complicated the situation in terms of biochemical attributes and functional genesis of these molecules. Nonetheless, these new pools of knowledge have opened up avenues for unraveling the finer details of the small RNA mediated pathways.
Keywords: siRNA, miRNA, piRNA, 21-U RNA, dicer, argonaute, mirtron.
Introduction
The basic research in molecular biology started with DNA, a molecule accommodating all the information required to generate new organism of its own kind. This was followed by an era of studies on proteins, the molecules conferring functionality to the cell. However, RNA largely remained ignored as a meager intermediate of the molecules carrying information and performing functions. On realization of the immense regulatory potential of these species recently, there has been a spurge in the study of the biology of RNA. Though different classes of RNA viz. mRNA, tRNA and rRNA had been identified earlier, the discovery of small RNAs (~19-30 nts), which once thought to be the degradation products of larger RNA molecules, led to the establishment of an independent class of RNA. This class of RNAs is now considered to govern diverse cellular processes across the eukaryotic kingdom.
The discovery of double stranded RNA (dsRNA) mediated transgene silencing by Fire et al. [1] captivated the focus of many, that consequently triggered, in a relatively short period, the emergence of specific classes of small RNAs viz., small interfering RNAs (siRNAs) [2, 3, 4] and microRNAs (miRNAs) [5, 6]. In spite of being different, these molecules have overlapping requirement of dsRNA as precursor and association with Dicer [7, 8, 9] and Ago-subfamily proteins [10, 11, 12, 13]. With the advent in the field, different types of siRNAs e.g. trans-acting small interfering RNAs (tasiRNAs) [14, 15], repeat-associated small interfering RNAs (rasiRNAs) [16, 17] and scan RNAs (scnRNAs) [18, 19] were discovered. These small RNAs are described in detail later in the text.
Two newly discovered small RNAs, viz. piwi-interacting RNAs (piRNAs) [20, 21] and 21-U RNAs [22] have been shown to regulate various cellular pathways and behaving in a manner analogous to the earlier defined miRNA and siRNA species. While mi- and si-RNAs (including its classes) have been identified both in plants and animals, the presence of piRNAs and 21-U RNAs is, so far, known to be limited to animals alone.
The small RNA biogenesis involves the following major steps: (a) The genomic locus is transcribed by pol II / pol III/ pol IV enzymes leading to the formation of double stranded RNA structure [23, 24, 25]; (b) These dsRNAs are sequentially acted on by RNase III type endonucleases (Drosha and Dicer) to generate duplex RNAs of size range 19-28 nts that are unwound by Argonaute proteins; (c) The single stranded mature small RNAs thus formed act as guide molecules to multi protein complex called RNA Induced Silencing Complex (RISC) or RNA Induced Transcriptional Silencing Complex (RITS) [26, 27], the category being dependent on the downstream effect; (d) The whole process culminates with either the cleavage or the translational repression of the homologous message(s) that is determined by the degree of complementarity. Alternatively, the RISC induces transcriptional silencing of corresponding locus by recruiting specific proteins [27].
The discussion below focuses on the biogenesis and the functions of four classes of small RNAs viz. siRNA, miRNA, piRNA and 21-U RNA. Along with the small RNA pool characterized in algae, the newly emerging pathway for miRNA biogenesis viz. the mirtron pathway is also highlighted.
Short-interfering RNAs (siRNAs)
siRNAs are ~20-24 nt long regulatory molecules that besides protecting cell from intrusion of any exogenous nucleic acid (like viruses), are involved in maintaining genome integrity by silencing transcription from undesired loci (retrotransposon, repeat sequences).
siRNA biogenesis
The foremost requirement for siRNA generation is a long double-stranded (ds) RNA molecule. These dsRNAs are formed from any transcription event generating messages with complementary sequences or by some enzymatic activity capable of converting RNA from single strand to double strand (Figure 1, steps a, b). The prime molecule in siRNA generation is Dicer, an RNaseIII type endonuclease. Animals usually encode a single type of Dicer to generate various classes of small RNAs with exceptions of Drosophila and C. elegans each encoding two dicers [28]. Plants on the other hand require multiple dicers. For example, Arabidopsis and Rice possess four and six different Dicer Like proteins (DCLs), respectively. In Arabidopsis, DCL 2, 3, 4 are involved in the generation of different siRNA species while DCL1 is solely responsible for miRNA biogenesis. The activity of each dicer produces siRNAs of characteristic length, e.g. DCL2, DCL3 and DCL4 generate 22, 24 and 21 nt long siRNA species, respectively [29, 30, 31]. Studies carried on Dicer mutants, in both plants and fungi, have allowed us to conclude that these proteins are functionally redundant. The functional redundancy among dicers has been studied in finer details in Arabidopsis, where different combinations of dicer mutants were analyzed on the basis of the size of siRNAs [32, 33]. The siRNA species associated with specific dicer were detected even when the parent dicer was debilitated by mutation. In such a case other dicer(s) may take over the function of the parental dicer, albeit with reduced activity. For instance, in Arabidopsis, DCL1 can produce 21 nt long tasi- RNAs in dcl2dcl3dcl4 mutant and DCL2 can generate tasi-RNAs in dcl4 mutant. Another interesting outcome of these mutant studies was the observation of the existence of functional hierarchy among dicers. This was exemplified by the work on dcl2 and dcl4 mutants, both capable of generating viral siRNAs. However, Arabidopsis dcl4 mutant analysis, where viral siRNA accumulation was observed although to a lesser extent, clearly demonstrated that the efficacy of DCL4 to generate viral siRNAs was significantly higher compared to that of DCL2 [32, 33]. Analysis of the data available till date strongly suggests that the hierarchy among dicers probably confers stringency to the siRNA mediated pathway.
Another important protein involved in siRNA biogenesis in plants, fungi and C. elegans (but not humans) is RNA dependent RNA polymerase (RdRP). The major function of this protein is to generate secondary siRNAs, a step termed signal amplification in siRNA pathway. RdRP can recognize aberrant RNA molecules to produce dsRNAs either in a primer dependent or an independent manner. The dsRNA molecules thus formed are later cleaved by downstream dicer activity.
Dicer, in general, possess six domains viz., DExH Helicase, DUF283, PAZ, RNase IIIa, RNase IIIb and RNA Binding Domain (RBD, Figure 2, a). The crystal structures for few individual domains have been solved providing us opportunity to predict model for dicer activity. The role played by each domain is mentioned in the figure 2 along with the expanded form of the abbreviation used. RBD recognizes the duplex RNA structure while the PAZ domain binds to the 3′-2nt overhangs of the cleaved RNA substrate [34]. Dicer has been proposed to act as a single processing centre, where the RBD and the PAZ domains participate in association with RNA molecule and assist both the RNase III domains to come close to form an intra-molecular dimer [35]. These RNase III domains juxtapose in a manner to cleave the ~21 nt duplex RNA molecules from the ds RNA precursors.
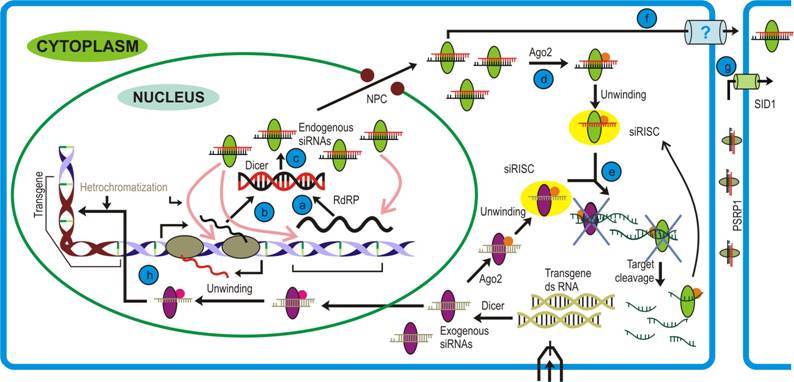
siRNA pathway: Precursor dsRNA are generated by either (a) RNA dependent RNA polymerase (RdRP) activity on aberrant transcripts or (b) transcript having full or partial complementarity. (c) These are recognized and processed by nuclear Dicer (different from the one involved in microRNA pathway) and this siRNA-Dicer complex is then exported to cytoplasm. (d) The siRNA- Dicer complex then recruits Argonaute that unwind the duplex to form si-RISC/RITS. (e) Transcripts bearing complementary sequences to guide siRNA strand are cleaved by RNase activity of Argonaute2. (f) To confer immunity, siRNAs-Dicer complex may also traffic in systemic fashion (g) that is achieved by Systemic RNA Interference-Defective (SID-1; in animals)/ Phloem Small RNA binding protein-1 (PSRP1; in plants). (h) The exogenous siRNA pathway follows parallel to endogenous pathway, but differs in the fact that the cytoplasmic Dicer generates the siRNA duplexes. The RITS complex lead to transcriptional gene silencing that involves various proteins.
The general domain organization of (a) Dicer and (b) Argonaute proteins. The functions of few domains that have been predicted through crystal structures and mutant analysis are mentioned.
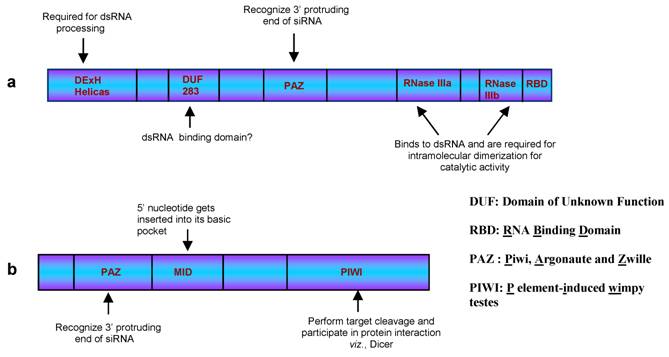
The duplex siRNAs are then unwound by helicase activity of Argonaute (Figure 1, step d), a protein recruited by Dicer. This protein has three well characterized domains namely, PAZ, MID and PIWI domains (Figure 2, b). Both, animals and plants encode multiple Argonautes that participate in different RNAi pathways [36]. Although not all the Argonaute proteins are characterized, the function of few members have been elucidated. For example, AGO2 and AGO1 are prominent members of si- and miRISC, respectively, in the plants and animals [37]. In Arabidopsis, AGO4 plays a crucial role in generating rasi-RNAs that participate in RNA-directed DNA methylation [37], AGO6 is involved in DNA methylation and transcriptional gene silencing [38] while AGO7 has been shown to participate in both tasi-RNA and long siRNAs biogenesis [39]. Similar to dicer, evidences suggest for the functional redundancy among these proteins. This was observed in Arabidopsis ago1 and ago10 mutant where both were found to share similar phenotypes [40, 41]. Further support came from the studies demonstrating AGO10 performing similar function in ago1 mutant. Here, AGO10 was shown to cleave the PHB transcripts under miR165 guidance and thereby establishing the leaf polarity in the ago1 mutants.
Together with the accessory proteins, Argonaute senses the thermal stability of duplex siRNA ends and initiates unwinding from the end with relatively lower thermal energy [42, 43, 44]. Of the two strands, one that is retained with the protein complex (siRISC) is called guide strand while the other (passenger strand) is destined to undergo degradation by exonucleases [45, 46]. In Arabidopsis, the single stranded mature siRNAs are methylated by HEN1 activity thereby rendering stability to these [47]. The single stranded small RNA loaded onto effector complex can be considered analogous to the prokaryotic restriction enzymes that act against any foreign nucleic acid. However, unlike the prokaryotic cellular defense mechanisms which are mediated by restriction enzymes, the small RNAs in eukaryotes can regulate even when the foreign DNA is transcribed to RNA, thus providing a molecular basis to the fact that eukaryotes are evolutionary superior to prokaryotes. On attaining the double-stranded conformation the guide strand of si-RISC activates Argonaute, the RNase activity of which acts specifically on the target sequence at the position complementary to 10 and 11 nt counting from the 5′ end of the siRNA (Figure 1, step e).
Classes of siRNAs
Depending on the nature of loci and biogenesis of dsRNA precursor different versions of siRNAs have been identified:
(i) Trans-acting short interfering RNAs (tasiRNA) are ~21 nt long small RNAs that require endogenous transcript as template [48, 49] that are converted to dsRNA by RNA dependent RNA polymerase (RdRP) activity and subsequently requires the downstream activity of DCL4 and AGO7 to generate functional tasiRNAs [50]. Animals like humans, flies etc., which lack RdRP, are devoid of these small RNA species. Tasi-RNAs resemble miRNAs both in size and function and are involved in targeting non-identical mRNAs. It has been demonstrated that miRNA primed transcripts recruit RdRP that consequently generate tasiRNAs, thereby setting an example of small RNAs mediated regulation of other small RNAs. For instance, miR390 binds to and induces the RdRP activity on primary transcripts and convert them to long dsRNA [50]. In Arabidopsis, the six tasiRNA genes are present that target Auxin Response Factors (ARFs) and MYB transcription factor [15, 51]. One of the recently identified tasiRNA locus, TAS4, has been demonstrated to generate siRNA that targets the transcript at a site which is different from the miR828 cleavage site [51]. This indicates towards the possibility of parallel evolution of tasiRNA, miRNA and their common target in plants.
In an alternate pathway, RdRP can also act on aberrant transcripts (usually viral transcripts) converting them to dsRNA and this mechanism is likely to be responsible in preventing cell from any erroneous transcription event that might affect cellular integrity.
(ii) Repeat-associated short interfering RNAs (rasi-RNAs) are ~24-26 nt long products of DCL3 activity on dsRNAs formed during unchecked transcription event, usually retro-transposon loci [52, 53]. These loci are generally methylated which prevent transcription through such regions. Like tasiRNA, these also require RdRP for amplifying small RNA pool. Rasi-RNAs play important role(s) during gametogenesis in flies, worms and mammals by modulating the chromatin status, and silencing viral transcripts by recruiting histone modifying proteins (Figure 1, step h) [20, 54-56].
(iii) Scan RNA (scn RNA), another type of relatively long (~29 nts) siRNAs have been reported from protozoan Tetrahymena thermophila. This organism exhibits nuclear dimorphism differing by ~15% at the sequence level. During conjugation, scn RNAs derived from micro-nucleus are generated (reproductive nuclei) and eliminate corresponding loci from its own genome while giving birth to macro-nucleus. This phenomenon requires Argonaute like Twi1 protein, and seems to be an ultimate form of RNA interference wherein organism can efficiently utilize small RNA to produce modified versions of genome from the existing ones [18, 19].
(iv) Long siRNAs (lsiRNAs) constitute the more recently introduced class of siRNAs that are 30-40 nt in length and are induced in response to bacterial infection or growth conditions [39]. Discovered in Arabidopsis, the generation of lsiRNAs require DCL1, DCL4 and AGO7 proteins and depend on other established members of both siRNA and miRNA pathway e.g. RDR6, HYL1, HEN1 etc. One of the lsiRNAs targets a protein that confers resistance against bacterial infection. Interestingly, these lsiRNAs unlike other siRNAs are believed to mediate target degradation by a mechanism previously known in animals but not in plants.
Systemic nature of silencing
siRNAs are believed to be a primitive form of immune response evoked against any foreign nucleic acid molecule. Therefore, by corollary, they ought to emanate from the production site to confer rapid cellular defense (Figure 1, steps f, g). This hypothesis is supported from the genetic studies carried out in animals, where import of siRNAs into cells has been demonstrated through a membrane protein called Systemic RNA Interference - Defective (SID-1) [57]. Their work on the ectopic expression with cells lacking SID1 shed light on the probable mechanism of the phenomenon. To their interest, they found that SID-1 mediated uptake of dsRNAs was length dependent, with larger molecules (~500 bps) being imported at a faster rate than the smaller (~30 bps) molecules. Moreover, observation that siRNA import is not affected by either cold treatment to cells or ATP depletion suggested towards a passive diffusion mode of this uptake. How the SID-1 protein discriminates between siRNA and miRNA while importing, still remains an open question to the researchers.
Phloem, a vascular tissue in land plants, has been implicated in the distribution of sugars, nutrients and other biomolecules across the plant. Recently, Yoo et al. [58] provided evidence for another class of molecules that are mobilized through this route i.e. small RNA (Figure 1, step g). Studies with phloem sap of different plant (rich in sap) species like cucurbits, yucca, and lupin revealed the presence of si- and miRNAs. Further they discovered a novel protein, Phloem Small RNA binding protein-1 (PSRP1), which was functionally similar to SID1 in animals and binds to the small RNA species. This was confirmed by studying viral coat protein silenced and non-silenced lines where authors could find accumulation of Coat Protein (CP) siRNAs in the silenced but not in the non-silenced lines. Nonetheless, the significance of miRNA transport across the phloem still remains to be elucidated. However, the PSRP1 protein is not conserved among the plants.
Although the systemic nature of silencing is a well-accepted phenomenon, the underlying mechanism is still ill-defined and demands efforts to resolve the differences between the animal and plant proteins reported till date.
Functions of siRNAs
The functionality of siRNA is the consequence following its binding to target sequences and this is governed by a critical region within the siRNA sequence called “seed region” [59]. The ribonucleotides encompassing the 2-7 positions (with reference to 5′ end) of siRNA constitute the “seed region” and are critical to confer siRNAs their target specificity. It is through the “seed region” that RISC lands onto, anneals and consequently brings about target cleavage/repression. Since siRNAs bind to the sequences from which they are derived, they are not under any kind of selection pressure. It may be noted that although the seed region is important in target recognition, the complementarity in other region of siRNA is critical during the cleavage event.
siRNAs have been involved in almost all possible nucleic acid regulatory pathways like target cleavage [60, 61], transcriptional gene silencing [52, 62-64] and DNA elimination [18, 19]. Moreover, lsiRNAs from plants have been shown to behave functionally similar to animal miRNAs where the siRNA binding rather than the cleavage leads to the decapping of target transcripts [39]. The exonuclease (XRN4) then acts on decapped mRNAs and bring about target cleavage. siRNAs with lesser complementarity has been demonstrated to suppress the targets at translation level [65].
Proteins (other than DCL and AGO) involved in small RNA biogenesis and their downstream function(s).
| Organisms | RNAi players known | Location & Function | Refs. |
|---|---|---|---|
| Animals | Drosha | Nuclear; RNase III type enzyme that binds dsRNA with characteristic structures and generates pre-miR forms by cleaving pri-miRs. | 66 |
| DGCR8/Pasha | Nuclear; dsRNA binding protein assists Drosha function | 67 | |
| PIWI | Nuclear; Ago subfamily protein generating piRNAs | 68, 69 | |
| MILI | Nuclear; Ago subfamily protein generating piRNAs | 70 | |
| Exportin5 /RanGTPase | Nucleo-cytoplasmic; Transports pre-miR to cytoplasm | 71, 72 | |
| Swi6/ HP1 | Nuclear; Heterochromatin formation | 53, 73 | |
| Chp1/ Twi | Nuclear; Heterochromatin formation | 74 | |
| Plants | PSRP1 | Cytoplasmic; Binds and Transports small RNAs across phloem | 58 |
| HYL1 | Nuclear; Interacts with DCL1 and confer stability to miR precursors | 75 | |
| HEN1 | Nuclear; protects duplex small RNAs by 3′ end methylation | 76 | |
| HASTY | Nuclear-membrane; export of duplex small RNAs to cytoplasm | 77 | |
| SDE3/ RDR | Nucleo- cytoplasmic; Performs catalysis of ds long RNA generation that can initiate different RNAi pathways | 78, 79 | |
| Serrate | Nuclear; Binds to pri-miRs in association with DCL1 and HYL1 and helps in processing. | 80 | |
| C. elegans | TUDOR-SN | Cytoplasmic, ds RNA binding putative helicase | 81 |
| SID1 | Transmembrane protein, responsible for systemic nature of RNAi | 57 | |
| RDE-4 | Cytoplasmic; Interact with Dicer1, R2D2-like protein | 82 | |
| Drosophila | VIG, Fmr1 | Cytoplasmic; component of RISC | 83 |
| Loquacious | Cellular; dsRNA binding protein, associates with dicer and participates in miRNA maturation and believed to play crucial role in maintaining germ-line stem cells | 84 | |
| R2D2 | Cytoplasmic; binds to siRNAs with Dicer help in processing and confers asymmetry to siRNAs. | 85 | |
| Armitage | Cytoplasmic; Arabidopsis SDE-3 homolog that acts as helicase during cleaving process. | 44 | |
| Pimet | Nuclear; Homolog of Arabidopsis HEN1 methyltransferase | 86 |
MicroRNAs (miRNAs)
The discovery of microRNA dates back to 1993, when Lee et al. [5] elucidated the function of a non-coding transcript in C. elegans, the expression of which varied spatio-temporally and the mutants showed developmental abnormalities. These genes were later recognized as precursor molecules of yet another important class of endogenous small RNA viz. microRNA that, unlike siRNAs, target messages different from that of the parent. MicroRNA genes constitute ~1% of the total coding genes [87-89] and form the largest class of regulatory molecules.
MicroRNA biogenesis
Micro RNA biogenesis is now believed to be operative by more than one pathway, as described below:
Canonical miRNA pathway: MiRNAs are ~19-23 nt long single-stranded RNAs generated from single-stranded transcript having local-hairpin structure (Figure 3, step a) [6, 89]. These transcripts are generated by pol II and therefore possess 5′ cap and 3′ poly-A tail hallmarks [90, 91]. Unlike the maturation of large RNA classes that occurs in the nucleus, miRNA maturation begins in the nucleus and terminates in the cytoplasm. In animals, the nuclear processing initiates with the endonucleolytic activity of Drosha, an RNase III enzyme, which in association with Pasha (92), recognizes and generates the stem- loop structure (pre-miR) from the pri-miR (Figure 3, step b). Mediated by Exp-5 and Ran-GTPase [93-95], pre-miR are transported out to cytoplasm where these are acted upon by Dicer (Figure 3, step c). Except for the different proteins that participate in miRNA pathway, plants follow almost similar miRNA biogenesis (Table 2). However, an important difference between the miRNA biogenesis pathways of plants and animals lies in the fact that in plants the DCL1 acts on the pre-miRs in the nucleus (Figure 3) [96].
In Arabidopsis and Drosophila, the duplex small RNAs are acted upon by methyl transferases that add methyl group at the 2′-hydroxyl residues of the terminal ribose sugars. Such end-modification protects these RNA species from any kind of degradation or uridylation [47, 97]. Duplex miRs are then unwound by Argonaute1, a prominent member of miRNA- RISC assembly (miRNP) [11], generating mature miRs (Figure 3, step d). The mi-RISC/ miRNP thus formed, under the influence of miRNA and the associated protein(s) achieves its function or follow different fates.
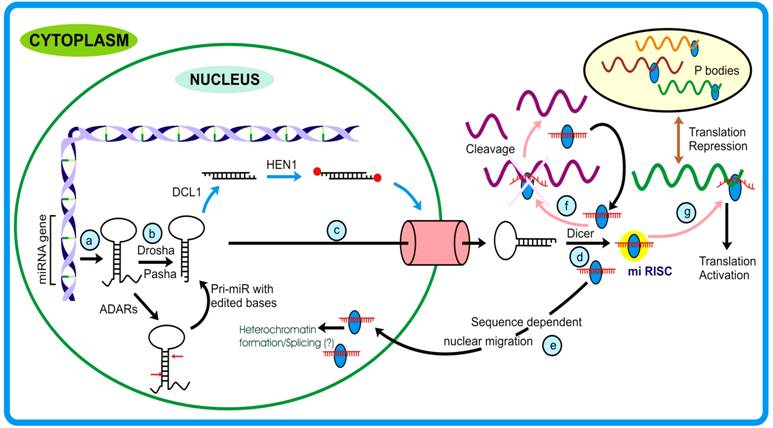
MicroRNA pathway: (a) After being transcribed, the pri-miRNAs stem-loop structure is acted upon by (b) Drosha (that also confers to miRNA strand and target specificity) and generates pre-miRNA. Sometimes, these precursors are edited by Adenosine Deaminase Acting on RNA (ADARs) at specific positions (generally +4 and +44) changing adenine to inosine. In plants, the DCL1 generates miR duplex in the nucleus that is methylated at terminal bases by HEN1. (c) These are then transported to cytoplasm with the assistance of Exportin-5/ HASTY. From here (d) Dicer comes into play (in animals) and generates miRNA duplexes that will be incorporated into micro Ribo-Nucleo-Protein (mi-RNP) complex. After the removal of passenger strand mature miRNA then guides the functional protein complex to the targets. (e) In mammals, miRNAs bearing nuclear signal sequences can traffic back to the nucleus. Depending upon the proteins associated with miRNA leads to either (f) cleavage of target mRNA or modulate the translation turnover by (g) translation activation or repression of respective mRNAs. The repressed mRNAs are transferred to structures called P-bodies.
Comparison between siRNAs and miRNAs among plants and animals.
| Animals | Plants | |
|---|---|---|
| siRNAs | Usually, single dicer involved in all types of siRNA generation | Different dicers required in Arabidopsis (4), Rice (10) |
| Redundancy at functional level not observed | Major proteins (dicer, argonaute) are functionally redundant | |
| Systemic spread requires SID-1 protein | Systemic spread requires PRSP1 (cucurbits) and SNF2 (Arabidopsis) protein | |
| Target cleavage, DNA methylation | Target cleavage, DNA methylation | |
| siRNAs can participate in genomic DNA elimination | No such role attributed here | |
| miRNAs | Generally, target repression | Generally, target cleavage |
| More than one miR can reside on pri-miR | Strictly one miR from one pri-miR | |
| Target various mRNAs | More biased towards TF transcripts | |
| Multiple miRNA binding sites per target | Usually single with one exception | |
| More than one miRNA can bind target | No report | |
| Duplex miRs are formed in cytosol | Duplexes miRs are formed inside nucleus | |
| Mature miRNAs can be trafficked back to nucleus | No such validated report | |
| Pri- or pre-miR are subjected to editing | No such phenomena observed | |
| Repressed mRNAs are stored in special organelles called P-bodies | No such structure observed |
Mirtrons, an emerging concept of microRNA biogenesis: In addition to the canonical miRNA pathway, animals have been shown to follow yet another mode of miRNA biogenesis where intron sequences can produce miRNAs. The hypothesis had emerged through analyses derived from pyro-sequencing of the small RNA pool from Drosophila S2 cell lines where some miR and miR* reads were mapped to the intronic regions. Such miRNAs originating from introns were termed mirtrons [98, 99]. Fourteen mirtrons from Drosophila and four mirtrons from C. elegans have been identified so far. Given that both protein-coding and non-coding genes possess introns, it is predicted that >80% of miRNA are derived from such sites [100, 101].
Introns denote the region of the transcript, generally flanked by few conserved nucleotide residues, which are removed during RNA processing. The conserved GU-AG along with other sequences brings multiple proteins mediating the removal of the intron from the transcript. The characteristic 2′-5′ phosphodiester bond formed within the intron during splicing result in the formation of lariat like structures. Such structures are acted upon by lariat de-branching enzyme to release the single stranded RNA that is consequently degraded by various nucleases present. However, intron sequences having potential to form hairpin like structures might recruit proteins of miRNA pathway viz., Dcr1.
Mirtron generation deviates from the canonical miRNA pathway mainly in the non-requirement of DROSHA/DGCR8 proteins that remove the sequences flanking stem region of pri-miR (Figure 4) [102, 103]. The unavailability of such sequences leads mirtronic pri-miRs to bypass the DROSHA step. This finding is contrary to the previous observation where Drosha was shown to act on intron prior to its splicing [101]. The knock down experiments with different transcripts involved in canonical miRNA biogenesis viz. Dicer1, Loquacious and Argonaute1, clearly supports that mirtron pathway do not require these proteins for their precursor generation. Moreover, the observation that Dcr-2 and RDR-2 (proteins participating in siRNA generation) knock down do not influence the expression of mirtrons, further ruled out any connection existing between the mirtron and siRNA biogenesis. The expression studies demonstrated that such introns can independently give rise to mature miRNAs, as is the case with miRNA genes. Moreover, like miRNAs, mirtrons also require AGO1 for maturation and follow cell-type specific expression [98].
Schematic diagram to depict differences between mirtron and canonical miRNA generation. Introns that assume foldback structures are recognized and cleaved by DROSHA. These stem-loop lariats are then acted upon by Lariat debranching enzyme that cleaves the phosphor-diester bond formed during splicing event. The pri- miR thus formed joins the mainstream miRNA flux, before making exit to cytoplasm.
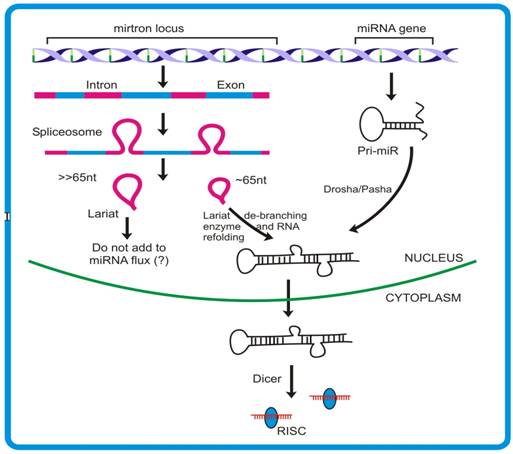
The splicing rules guided by the protein complex spliceosome, mediates appropriate release of the structure, termed lariat [104, 105]. These lariats are subsequently effected by another enzyme, called lariat de-branching enzyme [106] which provides an opportunity for this intronic RNA to assume a fold-back structure. The 3′-2 nt overhang of a mirtron forms a substrate for Exportin-5 mediated nuclear exit. The pre-mirtrons thus produced then merge into the mainstream pre-miR pool and follows similar pathway as that of canonical miRNA. In spite of the fact that Drosha is required for mirtron generation, the experiments with Drosha knock down did not show complete inhibition of pre-mirtron biogenesis. Combining the models proposed by Okamura et al. [98], Ruby et al. [99] and Kim et al. [101], it appears that probably both the models are functional and contribute independently to the miR flux.
Though these studies have revealed an alternative pathway for miRNA generation, the underlying intrinsic details of the mechanism, e.g., whether large lariat-turned-miR generating RNA can be substrate for DROSHA/PASHA, are yet to be unraveled. Interestingly, though the mirtrons have not yet been identified in plants and other organisms, the presence of introns of pre-miR length directs to the possibility of similar pathway being operative.
Since the introns are not subjected to selective pressure it is logical to assume that they are unlikely to preserve their sequences. In light of the above, the mirtron concept might explain the species specificity alongwith the rapid evolution that is observed in miRs. Why an alternate pathway for increasing miR flux has been specifically evolved, poses another interesting puzzle to solve.
MicroRNA functions
MicroRNAs show high tissue-specific and temporal expression and are believed to have evolved to take intensive care of developmental pathways that can be achieved by translation suppression (occurring mainly in animals) or target cleavage (occurring mainly in plants) [107, 108, 109]. However, there are exceptions to the general functions assigned to animal and plant miRNAs. For example, in animals, miR-196 governs the cleavage of HOXB transcript [110], and in plants, e.g. Apetala, a transcription factor, is translationally repressed by miR-172 [111]. Recently microRNAs have been elucidated to play critical role in conferring immunity to both animals and plants [112].
The target-miRNA recognition, like in case of siRNA, is initiated by seed region sequence. In animals however, the target transcript may possess more than one miRNA recognition site, allowing some miRNAs to bind target at multiple locations in proximity. This probably enhances the silencing effect in a cumulative manner and also confers redundancy to the phenomena, thereby making it more stringent [89, 113]. Plants miRs, on the other hand, have single target binding sites through which they achieve the target fate. However, complex relationship exist between plants miRs and their targets as most of the target transcripts falls under the category of transcription factors and can thus regulate many downstream processes. Interestingly, miRNAs are found to negatively regulate the expression levels of prime RNAi enzymes, viz. dicer and Argonaute (114, 115). This adds another layer of intricacy to the regulatory network achieved by these molecules.
In animals, studies suggest that miRNA binding promotes either deadenylation or decapping of the target which is probably achieved by interaction of RISC associated proteins with cap or poly-A tail associated proteins [107, 116, 117]. However, questions like how miRNA binding employs altogether different mechanisms (translational suppression or activation) is poorly understood. Recent studies have elucidated that miRNAs can bring about translational activation and a probable mechanism has been proposed [118, 119]. Previous studies by Pillai et al. (Figure 3, step g) [107] had hinted on the possible cross- talk existing between RNAi and translation activation machinery. These authors showed that AGO2 binding, which is guided by miRNAs (let7 in this case), can relieve the translation arrest of corresponding messages. Lending strong support to these findings, two back-to-back publications from same lab have shed light on our understanding of miRNA and associated proteins in exerting target translation upregulation [118, 119]. The observation that expression of TNF-α, a clinically important protein, vary significantly under starved and nutrient sufficient conditions allowed these authors to hypothesize on the involvement of ARE elements on the mRNA in its stabilization or directly activating translation from the mRNA. Interestingly, the authors found that these AREs bear sequences complementary to miRNAs. Further investigation revealed that two proteins, AGO2 and FXR1 associates, under guidance of miRNAs land onto these sites and exclusively during starved (low glucose) conditions [119]. Importantly, the authors observed similar phenomenon with other miRNAs as well, that led them to hypothesize that miRNAs generally act as repressors during active cellular growth while they tend to behave like translation activator when the conditions are limiting for cell growth. These studies demonstrating the association of miRNAs in translation activation of target mRNAs have provided an altogether new dimension to the functional attributes of miRNAs.
Moreover, as against normal localization, microRNAs have been demonstrated to be channeled back to the nucleus in a sequence-dependent manner (Figure 3, step e) [120]. This phenomenon allows miRNAs to extend their regulatory roots into other territory, namely the nucleus, thereby providing them an opportunity either to target many under-processed transcripts or bringing about silencing of genomic region(s). Interestingly, this was speculated by Bao et al. [121] where they studied Arabidopsis PHB and PHV mutants that govern the organ polarity and are regulated by miR-165 through degradation pathway. The authors observed that mutants resistant to miR degradation had significantly reduced extent of methylation when compared to wild plants and that there was no impact of other RNAi machinery proteins as a consequence. Studies from Hwang et al. [120] and Bao et al. [121], although with different systems, clearly support the notion that miRNAs play role(s) in RNAi mediated gene silencing.
In plants, the miRNA expression pattern is believed to be driven by multiple interacting feedback loops that involve various phytohormones, in particular auxins and gibberellins [122, 123]. Phytohormones regulate transcription of various genes by binding to cis elements and these transcripts possess sites for certain miRNAs. In contrast, transcription of some miRNAs is directly regulated by phytohormones. Such intricate tuning between miR and phytohormones is central to several biological processes.
Modulation of miRNA target range
Animal miRNA, as a general rule, binds to the 3′ UTR region of the target [59, 124, 125] while in case of plants, binding occurs in the coding region [126]. Usually, no compromise (maximum of single mismatch) is accepted in the seed region which otherwise would modulate the target range altogether. Recently another aspect pertaining to gene regulation has come to light in the form of miRNA editing [127]. Nuclear localized editing enzymes Adenosine deaminase acting on RNA (ADARs) [128, 129], edits adenosine to inosine at specific positions on the long dsRNAs (Figure 3). The over-expression of human ADARs reveals that two isoforms (ADAR 1 and 2) are involved in miRNA editing in vivo. Although the editing efficiency of ADAR1 was found to be more than ADAR2, the latter, unlike ADAR1, performed editing at only one particular position on the precursor [130]. This changes the seed region sequence and ultimately modulates the target range of the parent precursors [131]. Such editing of miRNA precursors (both pri- or pre-miRNA) may explain why some miRNAs elicit tissue specificity.
Besides the editing, the miRNA biding to its target can be prohibited by the presence of flanking sequences that are also strikingly conserved and might serve as docking site for certain proteins. The well known HuR proteins (the Hu family of proteins was identified as target antigens in a paraneoplastic neurological syndrome, viz., Hu syndrome) and recently reported Dnd1 (Dead End 1), an RNA binding protein, are among these class of proteins [132, 133]. Both HuR and Dnd1 proteins bind to the AU- rich elements (ARE) in the 3′-UTR and modulate the miRNA function. Dnd1 has been demonstrated to relieve miR122 repression on p27 in human cell lines and similar effect was seen with zebrafish miR430. These results clearly suggest the possibility of other post-transcriptional mechanisms being operative during the modulation of miRNA functionality.
P-bodies as the storage house of repressed messages
MicroRNA-repressed transcripts in animals are engulfed into dynamic vesicles called P (Processing)-bodies (GW1 or cytoplasmic bodies, Figure 3, step g) that carry out active mRNA degradation via nonsense mediated decay and gene silencing [134, 135]. The translationally suppressed mRNA, via interaction between one of the RISC member (AGO1) and the P-body proteins (GW1 and AIN1) gain entry into these structures [136, 137]. These bodies act as storage sites for translationally suppressed mRNA that are released when required and can actively translate [138]. P-bodies have been shown to be associated with various components of translation machinery (except ribosomes) and RNAi components like AGO1, GW182, miRNAs and CCR4 [138-140]. In spite of having homologs for P-bodies associated proteins, such structures are yet to be discovered in plants but some similar nuclear foci called Cajal bodies have recently been reported from Arabidopsis [141]. However, the lsiRNA mediated silencing mechanism that overlaps with P-bodies pathway suggests a similar pathway operating in plants.
Evolution of miRNA targets and sequences
Deep sequencing of Arabidopsis small RNA pool revealed an interesting fact that miRNA genes emerge and are lost frequently along evolution. Inverted duplications are attributed to the birth of new miR genes [142, 143]. MiRNAs face constant selection pressure and loss of any nucleotide especially within seed region of miRNA or complementary sequence on the target itself may increase their vulnerability [144]. On the contrary, considering the facts that > 80% of the miRNAs are generated from introns and that intron sequences are not amenable to stringent selection pressure, it seems likely that only a small fraction of miRNAs can be effected by such phenomenon. Moreover, a number of mRNAs are present among the transciptome, the sequence of which differs with that of existing miRNA targets by one or only a few nucleotides. All these observations together hints on the dynamic changes in the miRNA and its target range that can be achieved by introduction of few nucleotides in either's sequence.
In both plants and animals, some miRNA families are conserved across species while others are species-specific [145-147] which suggests that both target and miRs are evolving in parallel.
Small RNAs from unicellular eukaryote
Research carried out by two independent groups has revealed that contrary to popular beliefs, miRNAs are not restricted merely to multi-cellular organisms. Unicellular alga, Chlamydomonas reinhardtii, has been shown to generate various types of previously described small RNAs in multi-cellular forms i.e. mi- and siRNAs [148, 149]. Sequence analysis of small RNA pool read outs indicated that majority of these are 21-nt long with preference for uridine at 5′ terminal. Most of the sequences studied fall on unique genomic regions (including intergenic, protein coding, non-protein coding, and repetitive loci) thereby making the pool more complex.
Schematic representation of the pri-miRNA transcripts of (i) animal, (ii) plant, and (iii) alga showing differences in the miRNA biogenesis. Note that not all animal and algal pri-miRNAs follow this structural representation but it holds for plants where single miRNA resides within pri-miRNA stem.
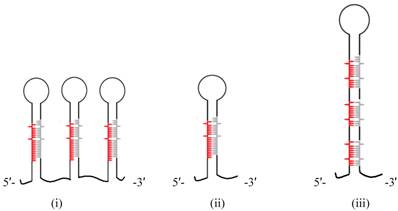
Few (~4) sequences dominated the pool hinting on the absolute requirement of those species and were later found to reside in proximity of each other. Surprisingly, no significant homology was found among known miR sequences. Of the sRNA pool that falls in the intergenic and intronic regions, Zhao et al. [149] looked for the flanking region and selected those capable of generating stem-loop structures. Approximately 200 such loci were predicted by the algorithm. Unlike plant and animal miRs, significant proportions of these sequences were lying in intronic regions. Careful analysis of the stem loop structures revealed interesting fact about these unicellular miR precursors. They can generate more than one mature miR thus making them different from multicellular pri-miR precursors (Figure 5). Although in animals there are loci co-transcribing multiple pri-miRs, each generating single mature miR, plants however, always produce pri-miR with single miR with one exception where a single pre-miR generates more than one miR [150].
Target prediction and function of algal miRs
Targets with diverse biological functions have been predicted and the binding sites were predominant in the protein-coding regions with few falling in Untranslated Regions (UTRs) as well. Interestingly, most of the targets belong to the Flagella- Associated Proteins (FAPs), suggesting that miR might transduce signals received by flagella. Unlike plants, transcription factors were under-represented as targets of algal miRs. However, paucity of annotated alga genome forbids concluding the functional bias of the miR targets and the target prediction per se.
In view of the presence of two AGO proteins and a Dicer, it is logical to assume that alga must be utilizing them for pathway(s) similar to RNAi, which indeed was found to be the case. In order to determine whether the downstream mode of function of algal miRs is similar to that in plants or animals, the biochemical activity was tested using different eluted protein fractions. The target cleavage studies support the notion that algal miR are more akin to plant miRs in terms of function [148]. Further, resistance to ß-elimination confirms the close similarity between algal miR with that of higher plants. Expression profiling of randomly selected miR demonstrated cell type specific expression, suggesting their role in developmental and tissue patterning as is the case with animal and plant miRs.
Algal siRNA species
After categorizing microRNAs, analysis of remainder of sequences characterized those as endo siRNAs. These sequences mapped to protein-coding and intergenic regions and similar to plant ta-siRNAs, were found to be phased relative to each other reflecting that algal small RNAs show more resemblance to plants than animals. But, the paucity of evidence for the presence of protein possessing activity similar to RNA-dependent RNA polymerase (RdRP), raises doubt on the similarity in the mechanisms of their biogenesis. Besides, sequences originating from repeat regions were also obtained, but significantly low reads of such species raise suspicion about their need in transposon silencing. However, the possibility of these RNAs acting as guide to modulate chromatin status cannot be completely refuted. Similar to the scan RNAs [18], these species of siRNA may accumulate during reproductive stage that needs careful evaluation.
Collectively, the analysis of small RNA pool from alga as well as other organisms suggests that these species have evolved independently in all the three existing lineages viz., plants, animals and algae to provide suitable cellular milieu when required during critical events like development, cell death, etc.
Small RNA as therapeutic and diagnostic agents
In view of their diverse roles, attempts were made to exploit the practical applications of si- and miRNAs as therapeutic and diagnostic molecules. Attributing function to a gene has been a great and incessant challenge for the past decade of research. This was revolutionized by the knock-down of almost all the desired messages in vivo using designed siRNAs and allowed researchers to extensively study various pathways including signaling, in both animals and plants. Besides, siRNAs has been shown to be successfully used as anti-viral and anti-cancer agents [151]. One major advantage of utilizing siRNAs relies on its selective (sequence specific) effects. The biggest hurdle encountered in the process was the siRNA delivery that was soon resolved with the advent of delivery molecules like plasmid vectors, transfection agents, etc. The delivery of siRNAs was further improved significantly by modification of these molecules by adding some moieties like 2′-O methyl, phosphothiorates, etc. that increase the stability of small RNAs [152, 153]. This was followed by a series of studies demonstrating the potent knock down of pathogenesis related viral and endogenous undesired transcripts through siRNAs, in different animal models. For instance, Soutschek et al. [154] demonstrated the knock down of apoB messages at the desired site in mice model while Hu et al. [155] used siRNAs to clear EWS-FLI1 transcripts that are upregulated in particular sarcoma. The advancements in siRNA mediated viral resistance against different diseases were achieved almost in parallel. For example, inhibition of Herpes Simplex Virus 2 [156], Hepatitis B virus [157] and Respiratory Syncitial Virus [158] infection are representative examples. Studies to achieve similar resistance are under progress in case of other important viruses like HIV, EBV, etc.
Similar to animals, in plants too, viruses are the major targets where siRNA therapy is being utilized with considerable success to gain resistance. Transgenics generating siRNAs against a particular transcript of a pathogen has been demonstrated to confer increased resistance compared to wild-types, when challenged with viruses. Considering the facts that miRNAs are functionally equivalent to siRNAs in plants and that they are involved in different developmental processes, it seems logical to capitalize their potential in plant system. Interestingly, artificially designed miRNAs (that are not present endogenously) are rather being much more frequently used in plants nowadays both to confer pathogen resistance and study developmental pathways [159-161].
However, in animals, knock down of genes via miRNA cannot be achieved due to lack of complete homology between miRNA and its target(s). Nonetheless, miRNAs can be successfully utilized for diagnostic purposes in animals. In view of their temporal and spatial expression, it is apparent that deregulation of miRs might lead to diseased state. Evidently, the miRNA profilings have been shown to be better indicators of many diseases, especially cancers, where strategies to cure rely on the early disease detection [162]. There is growing evidence that certain cancerous tissues express miRNAs in altered fashion thus supporting miRNAs as promising therapeutic agents [163-165]. In this regard miRNAs can be considered as tumor-suppressors [166]. Similarly, certain other diseased states were also found to be linked with modulated expression levels of miRNAs. For instance, reduced levels of miR-375 in diabetes [167], miR-133 in cardiac hypertrophy [168], and miR-122 and miR-143 in obesity [169] have been reported. Several studies clearly indicate that host encoded microRNAs act as anti-viral molecules targeting pathogen transcripts. Recently, miRNAs associated with viral infection have come into light and the tissue restricted miR expression might explain the viral tissue biasedness. This is supported by occurrence of low levels of miR-122 in HCV infection [170] and miR-32 in Foamy virus infection [171]. Moreover, some viruses (e.g. EBV, HIV, etc.), especially herpes viruses, have been demonstrated to encode miRNAs that confer virulence to the pathogen [172, 173]. These studies provide us with realistic situation during intricate host-pathogen interaction. Interestingly, antagomiRs (amiR) targeting tumor associated transcripts have been utilized in curing animal tumors, thereby promising new opportunities to tackle a number of animal diseases.
Recent attempts to utilize small RNAs for gene therapy for diseases like macular degeneration, Parkinson's disease, etc. are also underway and hopes are high to achieve success in the field. The insights gained from the applications of si- and miRNAs would significantly contribute towards our understanding of eukaryotic functional genomics and diverse biological pathways.
The new nodes of regulatory network
More recently, the discovery of two new species, viz., piRNA and 21-U RNA, has led to the addition to the existing list of small RNA classes. These were assigned to different small RNAs from the previously established classes on the basis of their origin and biogenesis. Nonetheless, they share some overlapping features shown by the previous small RNA classes as well.
Piwi-interacting RNAs (piRNAs)
Argonaute family of proteins is a well-established member of executor RNAi complex [12] and is highly conserved amongst various species [69]. Based on amino acid sequence homology, argonaute family has been categorized into two subclades viz., AGO and PIWI (P element-induced wimpy testes). While the AGO members are ubiquitous and associate with both the existing classes of small RNAs, i.e. siRNA and miRNA, the expression of PIWI proteins is restricted to germline cells alone. Genetic studies revealed that mutants for these proteins lead to male sterility suggesting its probable role in spermatogenesis [174, 68]. However, the exact correlation between the over-expression of PIWI and gametogenesis remained unanswered till recently. The independent works of Lau et al. [175], Aravin et al. [176], Grivna et al. [21] and Girard et al. [69] showing that PIWI proteins associate with ~25-31 nt RNA species which are germline-specific have added new dimensions to our knowledge about the varied nature of small RNA world.
While Lau et al. [175] and Watanabe et al. [55] focused on purifying the protein complexes followed by cloning of the associated small RNAs, as mentioned above. Cloning and sequencing of the germline small RNA population revealed that majority of the piRNA sequences were mapped to the genomic regions previously thought to be non-transcribed, while others corresponded to intergenic, exonic, intronic and repeat regions. These piRNAs were found to fall in two distinct size categories (24-28 nt and 29-31 nt). These are dispersed throughout the genome and reside in clusters ranging from 1 to 100 kilobases each generating 10 to 4500 piRNAs, eliciting strong bias to either DNA strand. Characterization of the protein complexes revealed the presence of two proteins viz. PIWI and Rec Q1 [175]. However, one cannot rule out the possibility of other proteins being loosely bound to the complex which would have subsequently lost during the process. This protein-RNA complex is termed piwi interacting RNA complex (piRC). The presence of RecQ1 as complex constituent was confirmed through its intrinsic ATP-dependent helicase activity.
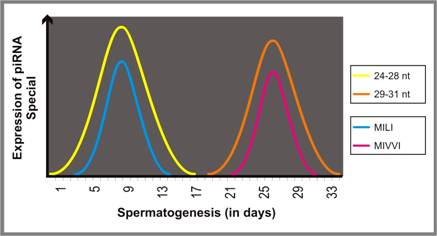
Expression studies have clearly demonstrated that MILI and MIWI (mouse orthologs of PIWI) follow different temporal patterns: while the expression of MILI lasts till pachytene, MIWI expresses till round spermatid stage. Intriguingly, the expression of two different populations of piRNA (24-28 nt and 29-31 nt) follow similar temporal pattern, indicating that each of these proteins might interact with specific piRNA species [177] (Figure 6).
Schematic representation of time-point specific expression each piRNA species follows during spermatogenesis.
In Drosophila, another protein, Pimet (piRNA methylase), has been demonstrated to be involved in piRNA biogenesis. This protein is homologous to Arabidopsis HEN1, a methyltransferase, and mediates methylation at the 2′-O of 3′ ends of piRNAs. However, unlike HEN1, this protein transfers methyl group to single stranded piRNAs [86]. Interestingly, the in vitro experiments with recombinant Pimet showed that it participates in methylation of piRNA but not miRNAs and this biasedness is conferred by interaction with Aubergine (Aub) protein.
piRNA biogenesis
Based on the mapping analysis of piRNA sequences onto genome it is postulated that these piRNA precursors are derived either from the non-overlapping transcripts generated from divergent promoter [175] or from a promoter giving rise to long single stranded RNA. The former postulate was based on the analysis that few piRNA clusters possess gap of few hundred base pairs between transcripts emerging from opposite DNA strands. However, this could not explain the generation of piRNAs from unidirectional promoters.
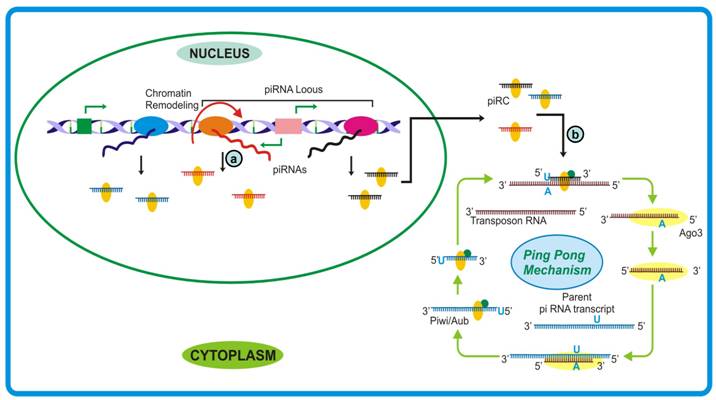
A better understanding of piRNA biogenesis has emerged from the studies conducted by Gunawardane et al. [178] and Brennecke et al. [179] where they proposed a mechanism parallel to secondary siRNA generation, termed as ping-pong model. It was observed that sense piRNAs associate with AGO3 while antisense associate with Piwi/Aub and are complementary till first 10 bases. Also, the 5′ end of antisense piRNA was observed to have strong preference for uridine base and that of sense piRNAs for adenine at position 10. According to the model, the piR-Piwi/Aub complex generated from piRNA cluster binds to target transcript (usually a transposon sequence) and cleaves between 10 and 11 bases. Subsequently, AGO3 binds and guide the cleaved transcript to the piRNA cluster transcript where it follows the hallmark endonucleolytic cleavage after 10th residue (usually adenine). This feed forward loop can rapidly generate sufficient piRNA to take care of any aberrant transcription event especially from retroelements (Figure 7). The cycle can be regulated by sensing reduced production of target transcript and consequently the piRNAs themselves. Whether the piRNA species in an organism are generated following the ping-pong model or by other pathways similar to the biogenesis of si- and miRNAs remains an open question. Moreover, as trigger-target availability is an absolute requirement for the activation of piRNA mediated defense, how these small RNAs take care of recently evolved selfish transposable elements or whether their evolution follows the birth of selfish sequences, still remains obscure.
piRNA biogenesis pathway. (a) Usually a polycistronic transcript, driven by mono- or bidirectional promoter, generates piRNAs by an unknown mechanism. Since the precursor lacks any tendency to achieve double-stranded form, the piRNA biogenesis seemed to be different from other small RNAs. (b) The biogenesis requires template to catalyze generation of desired small RNAs which further cleave corresponding target messages with another set of proteins. These piRNA may either regulate genome organization by checking transposon mobility or move to cytoplasm to take care of cognate messages either by cleaving or stabilizing them.
Functional attributes of piRNAs
Previous studies have shown that PIWI performs multiple functions ranging from epigenetic programming and repression of transposition to post transcriptional regulation [68, 174, 180]. However, in contrast to negative PTGS regulation of si- and miRNAs, piRNAs promote stability of target mRNA and probably enhance the translation as well. Having loci spread throughout the genome, the most important role that could be conferred upon piRNAs would be the patronage of their respective loci [18]. However, in view of the ability of piRC to cleave the cognate transcript [21], the involvement of piRNA at post-transcriptional level cannot be overruled.
21U-RNAs
In an attempt to redefine the small RNA profile in C. elegans, Ruby et al. [22] encountered a novel class of small RNAs, viz., 21U-RNAs. In all the reads analyzed, these molecules were found to be exactly 21 nucleotides long with uridine at its 5′ end. Of the ~5454 sequences obtained, majority were mapped to two major regions on chromosome IV, with few reads lying in between the major regions.
Biogenesis of 21U-RNAs
Though not much has been explored about the factors associated with the biogenesis of these species, the biochemical assays performed with 21-U RNAs have provided us with some of their characteristic features. It was elucidated that these species are sensitive towards alkaline hydrolysis and phosphatase treatment and their capacity to act as substrate for RNA ligase confirms these to be RNA molecules. Similar to small RNAs in plants and rasiRNAs in flies, 21U-RNAs also seemed to be modified at either 2´ or 3´ oxygen [22]. Extrapolating such resemblance to the functionality of these entities suggest that they might play some role(s) in chromatin reorganization and genome stability. In the absence of any evidence for the existence of dsRNA precursor, the biogenesis of 21-U RNAs seems to be dependent on some factor(s), which could sense the uridine residue as the reference point to count the bases. 21U-RNAs show no particular strand biasedness and majority were mapped to intergenic or intronic regions. Because authors used mixed-stage libraries, they could not conclude, as to which stage these species starts accumulating most, which would provide some clues of their functionality. The presence of 21U-RNAs during L1 and dauer stages suggests their role during worm development.
Motifs for 21U-RNA transcription
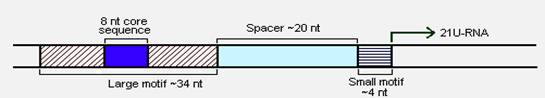
Taking closer look at the sequences flanking 21U-RNAs Ruby et al. [22] predicted two upstream elements, large and small motifs. While the large motif are ~34 nt long with 8-nucleotide core consensus sequence CTGTTTCA, the small motifs were ~4 nt long having YRNT as the core sequence. These two motifs were separated by linker sequences of ~19-27 bps in all the cases (Figure 8). Further analysis revealed that these motifs were highly conserved, suggesting for the requirement of these sequences during the transcription. In contrast, the 21U-RNA sequences were not at all conserved even within the same species.
Each 21U-RNA is transcribed autonomously suggests that they are independent genes and that 5′ flanking sequences may act as promoter clearly supports the hypothesis (Figure 9). The consideration of the above fact would dramatically influence the current scenario where it is believed that there are approximately 25,000 genes, at least in worms (these small RNAs are not yet reported in other organisms). This number may increase up to 1.5 times the existing figure.
General structure of 21U-RNA locus
21U-RNA pathway. Dictated by their own promoters, the independent transcripts are made that may involve specific factors to sense the terminal U residue and the 21 nt, thereby releasing mature 21-U RNAs.
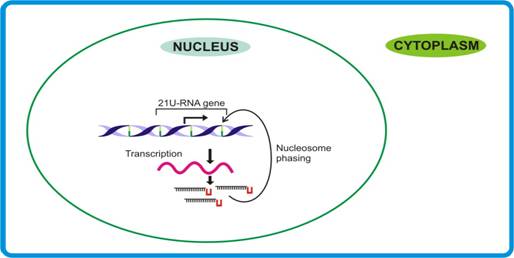
Function of 21-U RNAs
Considering the fact that 21-U RNA sequences show no homology with any transcript point towards their possible role in genome stability. However, in view of the earlier findings that sRNAs responsible for genome stability generally are of ≥24 nt, the possible role of 21U-RNAs (size 21nt) in genome stability remains doubtful. Moreover, since 21U-RNAs seemed to undergo maturation in the nucleus itself, their likely involvement in splicing cannot be over-ruled.
Prokaryotic RNAi
Though the prokaryotes encode few proteins having domains similar to proteins that participate in RNAi pathways, there are still no reports supporting this prediction. The sequence analysis of bacteria suggested that these organisms incorporate sequences from the parasite genome called Clustered regularly interspaced short palindromic repeats (CRISPR). These sequences, from foreign genome, are multiple noncontiguous direct repeats with spacer sequences and contribute resistance to the bacteria via RNAi pathway [181, 182]. The incorporation of spacer sequences is critical in achieving “adaptive” immunity which is evident from the studies challenging bacterial strains with phages [182]. Interestingly, the presence of spacers alone was not found to be sufficient rather their arrangement within genome is critical. But how these sequences help bacteria to gain resistance and the underlying mechanisms are yet to be explored and which will provide us important information regarding bacterial evolution.
Comparison between different small RNA species.
| siRNAs | miRNAs | piRNAs | 21U-RNAs | |
|---|---|---|---|---|
| Length (in nts) | 21-24 | ~22 | 25-31 | 21 |
| Requirement of dsRNA precursor | Yes | Yes | No | No |
| Genomic location | Dispersed throughout | Dispersed throughout | Discrete loci | Chromosome IV |
| Frequency (in %) of 5′ U Monophophate | ~80% | ~76% | ~94% | 100% |
| Location of Biogenesis | Cytoplasm/Nucleus | Nucleus and Cytoplasm | Nucleus? | Nucleus |
| Nature of gene | Autonomous /clustered | Autonomous | Tightly clustered | Autonomous |
| Proteins strictly associated with biogenesis (animals) | Dcr 2, AGO2 | Dcr 1, AGO1, Drosha/ Pasha, Exportin-5 | Piwi/ Aubergine, AGO3 | ? |
| Detected in | All eukaryotes studied | All eukaryotes but S. cerevisiae | Worms, Zebrafish mammals | C. elegans, C. briggsae |
| Expression | All tissues | Every tissue but few shows tissue specificity | Male germ line cells | All tissues |
| Downstream effects | Target cleavage, Chromatin remodeling, Translation repression, Genome reorganization | Translation repression, Target cleavage, Chromatin remodeling? | Genome organization, Enhances translation and mRNA stability | Nucleosome phasing |
| 3´ end modification | Yes | Yes | Yes | Yes |
| Mode of transcription | Divergent but partial overlapping Convergent | Autonomous | Divergent | Autonomous |
| Strand biasedness | Yes | Yes | High | Yes |
| Selection pressure | No | High | No | No |
| Nature of transcript | Polycistronic | Polycistronic/ Monocistronic | Polycistronic | Monocistronic |
| Potential tool without adverse effects | Yes | In plants | No | No |
Conclusions
The discovery that genomic regions, previously thought to be untranscribed, generates huge amount of small RNAs that participates actively in genome regulation allows us to redefine the concept of what regions shall be annotated as able-to-transcribe and also tempt us to ponder what actually is C-value paradox. That these piRNAs and 21U-RNA sequences are not conserved suggest that evolution has taken place even at the lowest classification level.
However many question still remains unanswered, for example, what could be the evolutionary significance of having autonomously expressed sequences on chromosome IV? How could the birth of such highly diversified transcripts be explained? What is the significance of mirtron pathway other than adding to miRNA flux? Can we expect similar pathways operating in prokaryotes? It would be interesting to determine the order in which all the four existing classes of small RNA have evolved. Moreover, with more reports supporting the RNAi like pathway in prokaryotes, we hope of connecting the missing links during the evolution of the phenomenon.
In future we expect diverse forms of these entities getting added to the list to get a complete picture of how organisms differ at the regulatory level and what roles do these small RNA play to achieve the goal. The discovery of diverse hitherto unknown phenomenon involving small RNAs tempts us to explore these enigmatic species. Though rigorous efforts for about more than a decade have gathered significant information on small RNAs, scientists are still trying to unleash the complete mechanisms underlying the origin, biogenesis and functions of this small RNA world.
Acknowledgements
We sincerely thank Dr Arun Kumar Sharma, Department of Plant Molecular Biology, University of Delhi, South Campus, New Delhi for valuable suggestions in manuscript preparation. One of the authors ARN is thankful to CSIR, Government of India, for financial Support.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Fire A, Xu S, Montgomery M. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806-11
2. Hamilton A, Baulcombe D. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950-2
3. Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188-200
4. Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21(17):4671-9
5. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843-54
6. Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13(10):807-18
7. Ketting RF, Fischer SE, Bernstein E. et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654-9
8. Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in the RNA interference and germline development in Caenorhabditis elegans. Science. 2001;293(5538):2269-71
9. Myers JW, Jones JT, Meyer T, Ferrell JEJr. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol. 2003;21:324-8
10. Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA. 2000;97(21):11650-4
11. Hutvagner G, Zamore PD. A microRNA in a multiple turnover RNAi enzyme complex. Science. 2002;297(5589):2056-60
12. Liu J, Carmell MA, Rivas FV. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437-41
13. Meister G, Landthaler M, Patkaniowska A. et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185-97
14. Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/RDE1/ RDR6 are required for juvenile development and the production of trans-acting siRNAs in arabidopsis. Genes Dev. 2004;18(19):2368-79
15. Vazquez F, Vaucheret H, Rajagopalan R. et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16(1):69-79
16. Aravin AA, Lagos-Quintana M, Yalcin A. et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5(2):337-50
17. Pélisson A, Sarot E, Payen-Groschêne G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81(4):1951-60
18. Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004;18(17):2068-73
19. Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci USA. 2004;101(6):1679-84
20. Saito K, Nishida KM, Mori T. et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the drosophila genome. Genes Dev. 2006;20(16):2214-22
21. Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709-14
22. Ruby JG, Jan C, Player C. et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127(6):1193-207
23. Lee Y, Kim M, Han J. et al. MicroRNA genes are transcribed by RNA polymerase II. The EMBO J. 2004;23(20):4051-60
24. Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human miRNAs. Nat Struc & Mol Biol. 2006;13(12):1097-101
25. Onodera Y, Haag JR, Ream T. et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120(5):613-22
26. Noma K, Sugiyama T, Cam H, Verdel A, Zofall M. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36(11):1174-80
27. Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9(2):315-27
28. Lee YS, Nakahara K, Pham JW. et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117(1):69-81
29. Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchial action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;303(5783):68-71
30. Blevins T, Rajeswaran R, Shivaprasad PV. et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34(21):6233-46
31. Xie Z, Allen E, Wilken A, Carrington JC. Dicer-like 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102(36):12984- 9
32. Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15(16):1494-500
33. Henderson IR, Zhang X, Lu C. et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38(6):721-5
34. Yan KS, Yan S, Farooq A. et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426(6965):468-74
35. Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human dicer and bacterial RNase III. Cell. 2004;118(1):57-68
36. Okamura K, Ischizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA- directed RNA cleavage pathways. Genes Dev. 2004;18(14):1655-66
37. Zilberman D, Cao X, Johansen LK, Carrington JC, Jacobsen SE. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol. 2004;14(13):1214-20
38. Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26(6):1691-701
39. Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007;21(23):3123-34
40. Lynn K, Fernandez A, Aida M. et al. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126(3):469-81
41. Moussain B, Haecker A, Laux T. ZWILLE buffers meristem stability in Arabidopsis thaliana. Dev Genes Evol. 2003;213(11):534-40
42. Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209-16
43. Schwarz DS, Hutvagner G, Du T. et al. Assymetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199-208
44. Tomari Y, Du T, Haley B, Schwarz DS. et al. RISC assembly defects in the drosophila RNAi mutant armitage. Cell. 2004;116(6):831-41
45. Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607-20
46. Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123(4):621-9
47. Yu B, Yang Z, Li J. et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932-5
48. Talmor-Neiman M, Stay R, Klipcan L, Kobi B, Baulcombe DC, Arazi T. Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 2006;48(4):511-21
49. Fahlgren N, Montogomery TA, Howell MD. et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16(9):939-44
50. Montgomery TA, Howell MD, Cuperus JT. et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133(1):128-41
51. Rajagopal R, Vaucheret H, Trejo J, Bartel DP. A diverse evolutionary fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20(24):3407-25
52. Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double stranded RNA. EMBO J. 2000;19(19):5194-201
53. Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320-4
54. Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20(10):2536-44
55. Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol. 2001;11(10):747-57
56. Pal-Bhadra M, Leibovitch BA, Gandhi SG. et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669-72
57. Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301(5639):1545-7
58. Yoo BC, Kragler F, Varkonyi-Gasic E. et al. A systemic small RNA signaling system in plants. Plant Cell. 2006;16(8):1979-2000
59. Lewis B, Burge C, Bartel D. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15-20
60. Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293-6
61. Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20(23):6877-88
62. Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110(6):701-11
63. Aufsatz W, Mette MF, van der Winden J, Matzke AJ, Matzke M. RNA directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16499-506
64. Volpe TA, Kidner C, Hall IM. et al. Regulation of heterchromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833-7
65. Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17(4):438-42
66. Han J, Lee Y, Yeom KH. et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016-27
67. Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14(23):2162-7
68. Cox DN, Chao A, Lin H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127(3):503-14
69. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199-202
70. Kuramochi-Miyagawa S, Kimura T, Ijiri TW. et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839-49
71. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011-6
72. Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185-91
73. Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319(5859):94-7
74. Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol. 2005;25(6):2331-46
75. Han MH, Goud S, Song L, Fedoroff N. The arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101(4):1093-8
76. Yu B, Yang Z, Li J. et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932-5
77. Bollman KM, Aukerman MJ, Park MY. et al. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development. 2003;130(8):1493-504
78. Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14(4):857-67
79. Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102(1):152-7
80. Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. EMBO Reports. 2006;7(10):152-8
81. Caudy AA, Ketting RF, Hammond SM. et al. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425(6956):411-4
82. Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12(5):807-18
83. Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16(19):2491-6
84. Forstemann K, Tomari Y, Du T. et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3(7):e236
85. Liu Q, Rand TA, Kalidas S. et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301(5641):1921-5
86. Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H & Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2'-O-methylation of Piwi- interacting RNAs at their 3' ends. Genes Dev. 2007;21(13):1603-8
87. Grad Y, Aach J, Hayes GD. et al. Computational and experimental identification of C. elegans microRNAs. Mol Cell. 2003;11(5):1253-63
88. Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299(5612):1540
89. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281-97
90. Cai X, Hagedorn C, Cullen B. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957-66
91. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376-85
92. Lee Y, Ahn C, Han J. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415-9
93. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011-6
94. Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95-8
95. Shibata S, Sasaki M, Miki T. et al. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34(17):4711-21
96. Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in arabidopsis. Proc Natl Acad Sci USA. 2005;102(10):3691-6
97. Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in arabidopsis. Curr Biol. 2005;15(16):1501-7
98. Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in drosophila. Cell. 2007;130(1):1-12
99. Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83-6
100. Rodriguez A, Griffith-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(104):1902-10
101. Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26(3):775-83
102. Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380-5
103. Han J, Lee Y, Yeom KH. et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887-901
104. Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367-409
105. Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: (ed.) Gesteland RF, Cech TR, Atkins JF. The RNA World, 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 1999:525-560
106. Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA. 1998;4(9):1096-110
107. Pillai RS, Bhattacharyya SN, Artus CG. et al. Inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309(5740):1573-6
108. Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15(11):2730-41
109. Palatnik JF, Allen E, Wu X. et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425(6955):257-63
110. Yekta S, Shih I, Bartel DP. MicroRNA-directed-cleavage of HOXB8 mRNA. Science. 2004;304(5670):594-6
111. Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022-5
112. Navarro L, Dunoyer P, Jay F. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312(5772):436-9
113. Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363-4
114. Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-like1 (DCL1) in arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13(9):784-9
115. Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22(1):129-36
116. Giraldez AJ, Mishima Y, Rihel J. et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75-9
117. Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & Dev. 2007;21(15):1857-62
118. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931-34
119. Vasudevan S, Steitz J. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128(6):1105- 18
120. Hwang HW, Wentzel EA Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315(5808):97-100
121. Bao N, Lye KW, Barton MK. MicroRNA binding sites in arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004;7(5):653-62
122. Guo HS, Xie Q, Fie JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17(5):1376-86
123. Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131(14):3357-3365
124. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3(3):e85
125. Enright AJ, John B, Gaul U. et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1
126. Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16(17):1616-26
127. Blow MJ, Grocock RJ, van Dongen S. et al. RNA editing of human microRNAs. Genome Biol. 2006;7(4):R27
128. Keegan LP, Brindle J, Gallo A, Leroy A, Reenan RA, O'Connell MA. Tuning of RNA editing by ADAR is required by drosophila. EMBO J. 2005;24(12):2183-93
129. Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucl Acid Res Mol Biol. 2005;79:299-338
130. Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10(8):1174-7
131. Kawahara Y, Zinshteyn B, Sethupathy P. et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315(5815):1137-40
132. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111-24
133. Kedde M, Strasser MJ, Boldajipour B. et al. RNA-Binding Protein Dnd1 Inhibits MicroRNA Access to Target mRNA. Cell. 2007;131(7):1273-86
134. Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300(5620):805-8
135. Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165(1):31-40
136. Liu J, Rivas FJ, Wohlscjelgel J. et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7(12):1261-6
137. Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7(7):719-23
138. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111-24
139. Eystathioy T, Jakymiw A, Chan EK. et al. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9(10):1171-3
140. Behm-Ansmant I, Rehwinkel J, Doerks T. et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20(14):1885-98
141. Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the cajal body. Proc Natl Acad Sci USA. 2007;104(13):5437-42
142. Allen E, Xie Z, Gustafson AM, Sung G, Spatafora JW. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36(12):1282-90
143. Maher C, Stein L, Ware D. Evolution of arabidopsis microRNA families through duplication events. Genome Res. 2006;16(4):510-9
144. Fahlgren N, Howell MD, Kasschau KD. et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2(2):e219
145. Sunkar R, Zhu JK. Novel and stress regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16(8):2001-19
146. Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766-70
147. Berezikov E, Guryev V, van de Belt J. et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21-4
148. Molna´r A, Schwach F, Studholme DJ, Thueneman EC, Baulcombe DC. MiRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447(7148):1126-9
149. Zhao T, Li G, Mi S. et al. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21(10):1190-203
150. Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101(34):12753-8
151. Lu PY, Xie FY, Woodle MC. siRNA-mediated antitumorigenesis for drug target validation and therapeutics. Curr Opin Mol Ther. 2003;5(3):225-34
152. Braasch DA, Jensen S, Liu Y. et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42(26):7967-75
153. Elmén J, Thonberg H, Ljungberg K. et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33(1):439-47
154. Soutschek J, Akinc A, Bramlage B. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173-8
155. Hu HM, Zielinska-Kwiatkowska A, Munro K. et al. EWS/FLI1 suppresses retinoblastoma protein function and senescence in Ewing's sarcoma cells. J Orthop Res. 2008;26(6):886-93
156. Palliser D, Chowdhury D, Wang QY. et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439(7072):89-94
157. Morrissey DV, Lockridge JA, Shaw L. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotech. 2005;23(23):1002-7
158. Zhang W, Yang H, Kong X. et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat Med. 2005;11(1):56-62
159. Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18(5):1121-33
160. Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18(5):1134-51
161. Qu J, Ye J, Fang R. Artificial microRNA-mediated virus resistance in plants. J Virol. 2007;81(12):6690-9
162. Lu J, Getz G, Miska EA. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834-8
163. Calin GA, Dumitru CD, Shimizu M. et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524-9
164. Lee EJ, Gusev Y, Jiang J. et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120(5):1046-54
165. Zhao Y, Ransom JF, Vedantham V. et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA1-2. Cell. 2007;129(2):303-17
166. Lee YS, Datta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025-30
167. Poy MN, Eliasson L, Krutzfeldt J. et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226-30
168. Care A, Catalucci D, Felicetti F. et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613-8
169. Esau C, Davis S, Murray SF. et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Met. 2006;3(2):87-98
170. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577-81
171. Pfeffer S, Zavolan M, Grässer FA. et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734-6
172. Lecellier CH, Dunoyer P, Arar K. et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308(5721):557-60
173. Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337(4):1214-8
174. Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2(6):819-30
175. Lau NC, Seto AG, Kim J. et al. Characterization of the piRNA complex from Rat Testes. Science. 2006;313(5785):363-7
176. Aravin A, Gaidatzis D, Pfeffer S. et al. A novel class of small RNAs binds to MILI protein in mouse testes. Nature. 2006;442(7099):203-7
177. Watanabe T, Takeda A, Tsukiyama T. et al. 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20(13):1732-43
178. Gunawardane LS, Saito K, Nishida KM. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315(5818):1587-90
179. Brennecke J, Aravin AA, Stark A. et al. Discrete small RNA-generating loci as master regulators of transposon activity in drosophila. Cell. 2007;128(6):1089-103
180. Houwing S, Kamminga LM, Berezikov E. et al. A Role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129(1):69-82
181. Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7
182. Barrangou R, Fremaux C, Deveau H. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2006;315(5819):1709-12
Author contact
![]() Correspondence to: Dr. Qazi Mohd. Rizwanul Haq., Department of Biosciences, Jamia Millia Islamia (A Central University), New Delhi-110025, India. Tel: +91 11 26981717 Ext. 3412, Fax: +91 11 26980229, E mail: qmrhaq.biac.in
Correspondence to: Dr. Qazi Mohd. Rizwanul Haq., Department of Biosciences, Jamia Millia Islamia (A Central University), New Delhi-110025, India. Tel: +91 11 26981717 Ext. 3412, Fax: +91 11 26980229, E mail: qmrhaq.biac.in

 Global reach, higher impact
Global reach, higher impact