10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(2):118-127. doi:10.7150/ijbs.5.118 This issue Cite
Research Paper
Differential expression of neurotrophins in postnatal C57BL/6 mice striatum
Unidad de Biomedicina, FES-I, Universidad Nacional Autónoma de México, Av. De Los Barrios # 1, Los Reyes Iztacala, C. P. 54090 Tlalnepantla, México
Received 2008-6-7; Accepted 2009-1-15; Published 2009-1-16
Abstract
Neurotrophin expression in early stages of development is crucial for brain assembly and function. In particular, postnatal expression of neurotrophins has not been well documented in the neostriatum and in general neurotrophins or their receptor mRNA's are normally reported, but not protein expression. In the present study, immunocytochemical expression of BDNF, NT-3 and NT-4/5 was characterized in striatal tissue of C57BL/6 mice at postnatal days 10th (P10), 21st (P21), 42nd (P42) and 80th (P80).
We found that the expression of BDNF diminished along the postnatal time course we evaluated, while staining for NT-4 increased up to age P42 and remained constant, thereafter in the cell's soma. In contrast, NT-3 was first expressed in the neostriatal bundles and later on, in neostriatal cell somas. These results provide information about differences in the spatial and temporal expression of each neurotrophin in the neostriatum during the first 80th postnatal days.
RT-PCR procedures were also carried out to further determine whether protein levels of neurotrophins observed in the neostriatum were under control of gene expression. All neurotrophin mRNAs were expressed and only mRNABDNF was reduced during the postnatal evaluated days.
Differences in temporal expression of neurotrophins may be related to the heterochronic development of neostriatal cell populations, but also with the specificity of each neurotrophin modulating different neuronal targets.
Keywords: neurotrophins, postnatal, BDNF, NT-3, NT4/5, striatum
Introduction
Neurotrophins are a family of trophic factors involved in differentiation and survival of neural cells [1]. Neural Growth Factor (NGF), Brain Derived Neurotrophic Factor (BDNF), Neurotrophin 3 (NT-3) and Neurotrophin 4/5 (NT-4/5) are active in different neuronal groups throughout the embryonic period but sometimes their roles are restricted to a specific developmental stage [2].
Neurotrophin distribution is not homogeneous throughout the nervous system and its expression is low during postnatal period [3]. In mice striatum, neurotrophin expression is important for the establishment of neuronal connections as well as its correct cell functions. Previous studies have established messenger RNA (mRNA) expression of NGF [4], BDNF [5-7], NT-3 [7, 8] and NT-4/5 [9] in neostriatal tissue, although these molecules have not always been detected by Northern blot in fetal or postnatal brain [10]. In addition to the presence of neurotrophin mRNA, expression of neurotrophin receptors have been documented during the early development of the striatum [3, 7, 8, 11-13]. Nonetheless, the time-course of postnatal expression of neurotrophins, has not been well recognized in the striatum. Neurodegenerative disorders affecting the central nervous system are characterized by cell death of specific neuronal populations. Some degenerative diseases afflicting neural system have clear genetic origin [14]; however most of them are sporadic, suggesting that other mechanisms are influencing this outcome. Striatal neurons are particularly sensitive to excitotoxicity [15] and metabolic insults [16]; additionally, trophic factors can supply deficits during the first two postnatal weeks that may worsen cellular harm. As neurodegenerative diseases may be a consequence of abnormal levels of neurotrophins during the first stages of development, a clear description of their expression patterns during the two postnatal months in the normal brain is important to address before evaluating them in striatal dysfunction.
We aim to describe postnatal expression of neurotrophin proteins visualized with immunostaining techniques at P10, P21, P42, and P80 in C57BL/6 mice. To further verify whether neurotrophin protein expression is determined by gene expression, we examined the mRNA levels of each neurotrophin at P10, P21 and P42 in mice striatum.
Materials and Methods
Male C57BL/6 mice (Harlan, México) of 10th, 21st, 42nd, and 80th postnatal days were euthanized with pentobarbital (45mg/kg of body weight) and heart perfused with saline solution (NaCl 0.15 M) followed by phosphate-buffered saline (PBS, 0.1M, pH 7.4) with paraformaldehyde (4%). Brains were dissected and post-fixed in the same fixative for two hours at room temperature, and then transferred to a sucrose solution (30%) until they sank for cryoprotection. The brains were frozen in cold 2-methylbutane. Cryostat coronal sections (30 μm) containing dorsal striatum were cut and collected sequentially into 24-well dishes filled with blocking solution for immunocytochemistry (3% bovine serum albumin, 0.1% triton X-100 and 0.025% sodium azide in PBS).
All animal procedures were carried out with the approval of the animal care committee of the National Autonomous University of México and in agreement with the NIH.
Immunocytochemistry (IHC)
Tissue sections were incubated in blocking serum overnight at 4° C. After that, tissue slices were incubated with primary antibodies against BDNF, NT-3, and NT-4/5 (cat # AB1534SP, AB1517P, AB1519P, respectively, Chemicon International, Inc.), diluted all (1:1000) in blocking buffer and incubated for 24 hours at 4° C. Three washes were done with blocking solution (every 15 min), then tissue slices were incubated for 90 minutes at room temperature with the corresponding biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA, USA) diluted (1:500) in blocking solution. After three washes with PB, slices were incubated with the avidin-peroxidase kit as recommended by the supplier (ABC kit; Vector). Peroxidase activity was revealed and intensified by using 3, 3'-diamobenzidine as a chromogen and nickel chloride according to the protocol provided by the supplier (Peroxidase staining kit; Vector). Finally, the staining was stopped after ~5 minutes by washing the slices with PB. For control IHC staining, the primary antibody was omitted and the tissue sections were processed with the same procedure as described before. Slices were mounted on gelatin-coated slides and cover slipped with Permount (Fisher, Inc).
Quantitative assessment of neurotrophins expression in the striatum
Images of immunostained sections of dorsal neostriatum, from animals of 10 (n=10), 21 (n=10), 42 (n=10), 80 (n=10) days old were captured and quantified using an imaging analyses system (Image J system, NIH). For each mouse, six striatal slices (30 μm thick) spaced at intervals of 180 μm were taken and processed for immunocytochemistry. Afterwards, positive cells of four selected fields (252 μm2, area) were counted in digitized images taken at the level of dorsal striatum. Each image slice was digitized using a computer assisted system consisting of a Nikon Optiphot microscope and a digital camera connected to a Pentium computer equipped with the morphometric software analysis. Contrast and area values were kept constant and intensity threshold was established to eliminate background staining. The number of cells counted per field was averaged per animal and then per group and compared using variance analysis with a significance value of p <0.05. Post hoc Bonferroni´s comparisons were done when significance was observed. Data are expressed in mean ± S.E.M
RNA extraction and Reverse Transcriptase (RT)-Polymerase Chain Reaction (PCR)
Total RNA was isolated from 0.5 g samples of frozen mice striatal tissue. The extraction was carried out with the TRIzol reagent method using the Total RNA Isolation Reagent (Invitrogen, Life Technologies, Inc.). RNA concentration was calculated from the optical density at 260 nm and the purity was determined by 260nm /280nm of absorbance [17].
Total RNA (~300ng) from each sample was reverse transcribed and amplified with polymerase chain reaction with gene-specific for NGF, BDNF, NT-3, NT4/5 and β-actin, using the Super Scrit II kit (Invitrogen, Life Technologies, Inc.). Reverse Transcription was performed with a TECHNE, TC 3000 thermocycler. 2 μl of each RT product obtained was amplified with PCR using the RED Taq DNA Polymerase (Sigma-Aldrich, Inc.) in a final volume of 20 μl and 0.5 μl of each specific neurotrophin primer. The amplification cycles and the primers are specified in the Table 1. For the semi-quantitative analysis of RT-PCR products, β-actin was used as an internal control [18, 19]. Total PCR products were detected by electrophoresis on a 2% agarose gel and stained with 1 μl de SYBR Green. The area and density of PCR product bands were measured by chemio luminescence using a FUJIFILM FLA-5000 scanner and digitized with the Image Reader FLA-5000 V2.1 program. The levels of β-actin mRNA served as an internal standard to ensure equal loading of RNA, then neurotrophin mRNA levels were normalized with the housekeeping gene. The resulting measures were expressed as arbitrary densitometry units and statistically analyzed.
Primers used for the PCR
| Neurotrophin | Sequence | Cycles | Product (bp) | ºC | |
|---|---|---|---|---|---|
| NGF | Sense | 5'-TAGCGTAATGTCCATGTTGT | 35 | 434 | 58 |
| Antisense | 5'-CCCACACACTGACACTGTCA | ||||
| BDNF | Sense | 5'-GAAGAGCTGCTGGATGAGGAC | 35 | 332 | 60 |
| Antisense | 5'-TTCAGTTGGCCTTTTGATACC | ||||
| NT-3 | Sense | 5'-CTCATTATCAAGTTGATCCA | 35 | 312 | 55 |
| Antisense | 5'-CCTCCGTGGTGATGTTCTATT | ||||
| NT-4/5 | Sense | 5'-CCCTGCGTCAGTACTTCTTCGAGAC | 35 | 249 | 65 |
| Antisense | 5'-CTGGACGTCAGGCACGGCCTGTTC | ||||
| β-actin | Sense | 5'-TGGTGGGTATGGGTCAGAAGGACTC | 35 | 266 | 60 |
| Antisense | 5'-CATGGCTGGGGTGTTGAAGGTCTCA | ||||
Primer sequences were designed according to ref [19]. All primers were obtained from Sigma-Alrdrich
Statistical Analysis
Data are presented as mean ± S.E.M values if anything else is indicated. For IHC and RT-PCR experiments groups were compared using variance analysis, p <0.05 were considered to be statistically significant. Post hoc Bonferroni´s comparisons were performed when significance was observed. The statistical analysis was conducted using SigmaStat statistical software for Windows, version 2.03 (Systat Software, Inc.)
Results
Neurotrophin IHC expression
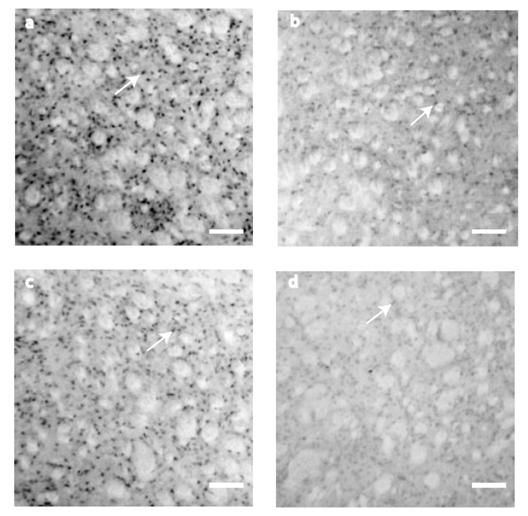
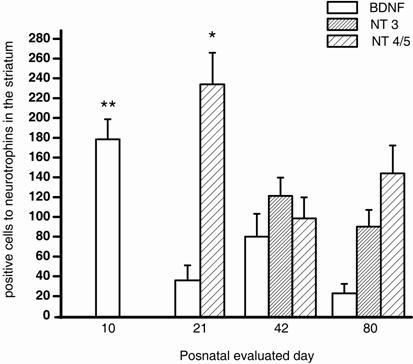
BDNF IHC expression was evaluated at 10th, 21st, 42nd, and 80th postnatal days. BDNF positive cells showed a significant reduction along the postnatal days analyzed (F3=7.907, P<0.001, Fig.1 and Fig. 4). BDNF peak expression was obtained at P10 (178 ± 20 cells), immunostaining was present mainly in somas of ~15 μm of diameter (Fig. 1a). By P21, the number of BDNF positive cells decreased (36 ± 14 cells) significantly (t=4.144, P<0.001, Multiple Comparison Procedures, Bonferroni´s t-test) in comparison with P10 (Fig. 1b). BDNF positive cells (80 ± 23, Fig. 1c) at P42 were significantly different than in P10 (t=2.875, P=0.032, Bonferroni´s t-test). At P80, BDNF positive cells were significantly reduced (23 ± 9) in comparison with P10 (t=4.516, P<0.001, Bonferroni´s t-test). There were no differences between cells detected at P21 vs. P42 (t=1.807, P=0.449, Bonferroni´s t-test); P42 vs. P80 (t=2.331, P=0.135, Bonferroni´s t-test) and P21 vs. P80 (t=0.519, P=1.000, Bonferroni´s t-test; Fig. 1d). There were not cells detected in control sections for IHC (data not shown).
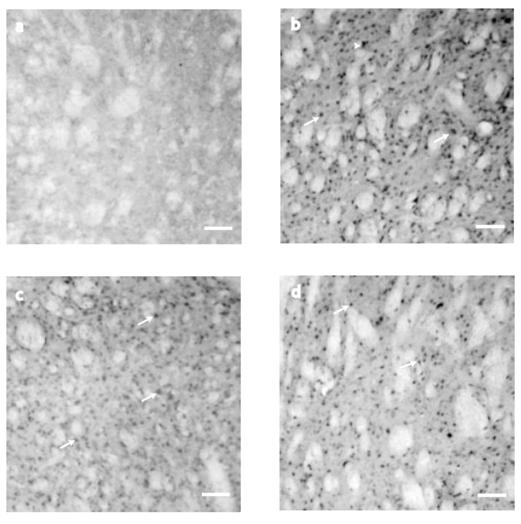
IHC expression of Neurotrophin 4/5 in the striatum evaluated at P10, P21, P42, and P80 displayed significant differences in the postnatal days evaluated (F2=4.860, P=0.011, Fig 2 and Fig. 4). We did not see any NT-4/5 IHC expression at P10 (Fig. 2 a). Nevertheless, NT-4/5 expression increased at P21 (234 ± 30 cells, Fig. 2 b). By P42, NT-4/5 stained cells (98 ± 21) showed a significant reduction (t=3.074, P=0.010, Bonferroni´s t-test) in comparison with P21 (Fig. 2 c). No significant differences were observed at P80 (144 ± 28 cells, Fig. 2d and Fig. 4) in comparison with P21 (t=1.874, P=0.198, Bonferroni´s t-test) or P42 (t=1.329, P=0.568, Bonferroni´s t-test). NT-4/5 immunostaining was mainly somatic in each evaluated age. No cells were detected in control sections for IHC were primary antibody was omitted (data not shown).
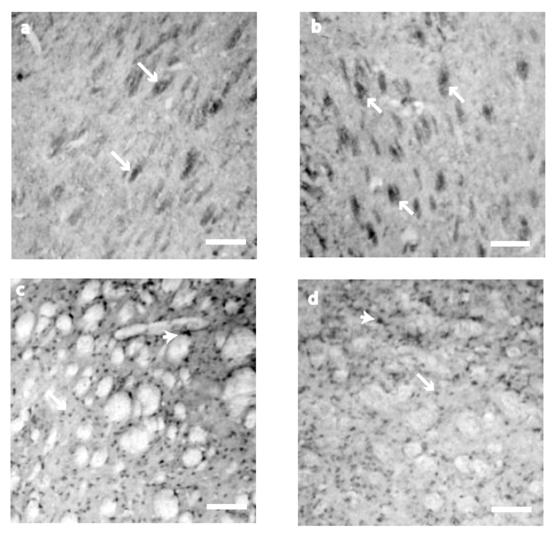
NT-3 expression was somewhat different from the other two neurotrophins evaluated. At P10 and P21, cell staining was circumscribed to neuronal bundles (Fig. 3 a, and b). Somatic expression was observed by P42 (121.28 ± 18) and P80 (89.6 ± 17.5), without significant differences (t 55= 1.211, P = 0.231) in the NT-3 positive cells (Fig. 3 c and d). No cells were detected in control sections for IHC where primary antibody was avoided (data not shown).
BDNF immunostaining detection at the neostriatum. (a) At P10, BDNF expression was higher. The staining was restricted to the cytoplasm of small cells (~15μm). (b) Although the staining was considerably reduced by P21, it was restricted to cell somas. (c) The figure shows BDNF staining by P42. (d) At P80, there was a reduction in BDNF expression. Scale bar 160 μm.
NT-4/5 immunostaining detection at the neostriatum. (a) At P10 striatal tissue did not show NT-4/5 immunostaining. (b) On P21, NT-4/5 showed its highest staining in cell bodies of ~15μm (arrows) although they can be seen in cell somas of about ~25μm as well (arrowheads). At P42 (c) and P80 (d) NT-4/5 did not change its expression between those two postnatal days, but when NT-4/5 positive cells were compared between P42 and P21, the staining was significantly different (see the text for details). Scale bar 160μm.
NT-3 immunostaining detection at the neostriatum. At P10 and P21 (a, b), NT-3 immunostaining was restricted to the neural bundles (arrows). At P42 (c) and P80 (d) the staining was found in the cell somas of ~15 μm (arrows) and big cells of ~25μm of diameter (arrowheads). Scale bar 160μm.
A summary of neurotrophins expression in all developmental times evaluated is illustrated in Figure 4.
Number of immunostained cells for BDNF, NT-3 y NT-4/5 in the postnatal neostriatum. The bars show the number of stained cells for BDNF (white bar), NT-3 (dense bar) and NT-4/5 (sparse bar). BDNF, NT-3 and NT-4/5 staining displayed significant differences (F3=7.907, P<0.001; t 55= 1.211, P = 0.231; and F2=4.860, P=0.011, respectively) in their postnatal expression. p=0.01*, p<0.001**
Neurotrophins relative mRNA level expression
It is known that neurotrophins expression is low and varies along the postnatal development; therefore to determine whether IHC staining of neurotrophins was correlated with gene expression, RT-PCR technique was carried. We examined relative mRNA levels of NGF, BDNF, NT-3 y NT-4/5, at P10, P21, and P42. β-actin was used as a housekeeping gene and mRNA level of each neurotrophin was normalized with β-actin endogenous expression. Neurotrophins mRNA levels were present in all postnatal ages evaluated (Fig. 5), although theirs mRNA levels varies in each age. mRNA levels of NGF did not change over the evaluated period (F2=5.119, P=0.060). mRNABDNF levels exhibited a significant reduction (F2=29.712, P<0.001) at P21 and P42 in comparison with P10 (P10 vs. P21, t=5.135, P<0.006; P10 vs. P42, t=7.547, P<0.001; Bonferroni´s t-test). mRNANT-3 and mRNANT-4/5 levels did not display any significant difference (F2= 3.213, P=0.113; F2=2.289, P=0.183 respectively) at the evaluated days.
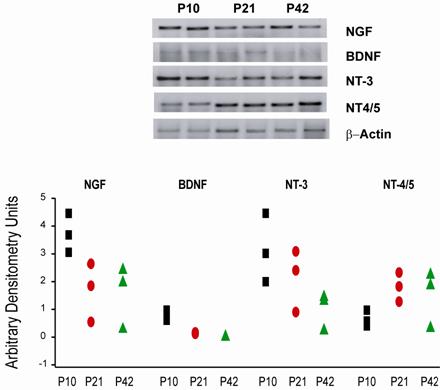
Relative RT-PCR of neurotrophins mRNA in striatal tissue from mice at P10, P21 and P42. The upper panel shows representative PCR products of neurotrophins from 2 different mice at P10, P21, P42 stained with sybr green on a 2 % agarose electrophoresis gel. The lower panel displays data for neurotrophins mRNA from mice striatum at P10 (squares, n=3), P21 (ellipses, n=3) and P42 (triangles, n=3). Data were analyzed by measuring the area and density of the electrophoresis bands and are expressed as arbitrary densitometry units.
Discussion
Neurotrophins are trophic factors whose actions are crucial for differentiation, survival and the organization of neuronal connections during postnatal development. Previous studies have revealed low levels of neurotrophin mRNAs in the striatum [1-9]. These earlier findings prompted us to elucidate neurotrophin protein temporal expression in normal striatal tissue, in attempts to better understand striatal abnormalities in which the lack of trophic factors during development may underlie degenerative diseases like Parkinson or Huntington diseases. In the present study we show that in mice dorsal neostriatum, temporal neurotrophin IHC expression at postnatal days 10th, 21st, 42nd and 80th is differential. We observed that when BDNF was already expressed in the neostriatum, NT-3 was expressed in the corticostriatal bundles and NT-4/5 staining was absent. With neurotrophin relative RT-PCR analysis, we observed that temporal expression of BDNF was different in comparison with NGF, NT-3 and NT-4/5.
Temporal and spatial IHC expression of neurotrophins at the postnatal striatum
Under our experimental conditions, BDNF was the only neurotrophin IHC expressed at P10 and declined with increasing postnatal age. BDNF is highly expressed during embryonic period and its expression changes during brain development [3]. Our results for BDNF protein expression correlate well with the presence of its neurotrophin receptor mRNATrkB in the neostriatum, which is highly expressed during embryonic development and declines at postnatal ages [20]. It has been demonstrated that neurons as well as glial cells in the striatum exhibit BDNF immunoreactivity [21] therefore, BDNF positive cells in our IHC experiments can be either type of cell.
NT-3 displayed expression in striatal bundles at P10 and P21 and in striatal cells at P42, sustaining its level of ICH expression up to P80. This is the first report in mice striatal tissue documenting differences in spatial expression of NT-3 at P10 and P21, where it was circumscribed to neuronal bundles. It has been recognized that, NT-3 is anterogradely transported from the cortex similar to BDNF, consequently, NT-3 expression in the neostriatal bundles at P10 and P21 may represent anterograde transport of NT-3 from the cortex as it has been suggested to occur [22]. In an earlier report, it was demonstrated that NT-3 mRNA is highly expressed in developing neurons in the cingulate cortex. That expression was transient during early stages of development and later the expression was seen in the hippocampus, supporting the notion that NT-3 promotes the development of cells that are anatomically connected [23]. NT-3 influence on striatal function may be related to different neuronal populations than those affected by BDNF. Its temporal expression also suggests that, NT-3 is not necessary as a differentiation or survival factor for striatal cells. In addition NT-3 activates a different type of neurotrophin receptors, the TrkC receptors [24]. Alternatively, NT-3 may influence the same population BDNF affects during another developmental time as a result of different developmental strategies and developmental speed.
NT-4/5 is expressed in the neostriatum, when BDNF is declining (Fig. 2 and 4). NT- 4/5 should arrive to the neostriatum between postnatal day 10th and 21st, however, it is not known where the NT-4/5 is coming from, or if it is produced inside of the nucleus by neostriatal cells. Nevertheless, differences in temporal expression between BNDF and NT-4/5 are important to note, because both neurotrophins activate the same TrkB receptor [24]. It is possible that the actions of both neurotrophins are not the same, or NT-4/5 actions may substitute for BDNF actions on striatal cells. Alternatively, each neurotrophin may affect distinct neuronal populations at different developmental stages. Other studies have documented that NT-4/5 is not necessary for early postnatal development of the neostriatum, still this neurotrophin is found all over the brain [25].
The developmental differences in neurotrophins temporal expression in the mice striatum may reflect the requirements of diverse group of cells for specific trophic factors at different postnatal stages, or it may suggest that every cellular group depends upon the presence of different neurotrophins at several stages of the postnatal development.
Neurotrophins mRNA expression in postnatal striatum
To establish whether the expression of neurotrophins detected by IHC procedure was a consequence of gene expression, the mRNA levels of NGF, BDNF, NT-3, and NT-4/5, were evaluated at P10, P21 and P42. P80 was omitted from this analysis because there were no differences in neurotrophins expression between P42 and p80 in IHC experiments. Although β-actin was used as internal standard; we included NGF as a positive control because it is recognized that NGF mRNA levels are high between P2 and P30 [4].
mRNANGF was high and remained high at all ages evaluated as reported previously [4].This result was important to show that RNA was not degraded and PCR was working correctly.
BDNF presence in the neostriatum has been controversial due to the lack of specific antibodies; nevertheless it has been detected by the ELISA technique [8]. In our experiments, mRNABDNF levels were evaluated at P10, P21, and P45. mRNABDNF expression was low and reduced with time in agreement with our IHC results. In another study mRNA BDNF showed its maximal level by the third postnatal week (~P21) when the PCR was carried out with the RNAse protection technique [6, 7], hence discrepancies in the expression level obtained in the present study may due to differences in the experimental procedure, still our data are in consonance with those reporting that BDNF presence in the striatal tissue declines with time [3].
NT-3 expression level of its mRNA is regulated differentially in comparison to BDNF, and has been reported that mRNANT-3 levels are constant during postnatal development [7]. In our experimental conditions after normalizing to the housekeeping gene, mRNANT-3 expression at P10, P21, and P42 did not exhibit any statistical change, which agrees with previous experimental reports [6, 7]. In light of our IHC experiments, we believe that part of the mRNANT-3 expression comes from mRNA of corticostriatal fibers, although we can not ruled out changes due to sample processing or biological changes (see below).
We did not observe significant changes in the mRNANT-4/5 level expression in the postnatal days evaluated. Although with IHC procedure there was no expression of NT- 4/5 at P10. It has been reported that NT-4/5 is expressed in low amounts by postnatal development [25] and increases in adult rodents [9, 12]; therefore, differences between mRNA and IHC expression of NT-4/5 may be due to insufficient levels of mRNA for protein translation (see below).
Comparing our mRNA results vs. our IHC data, mRNABDNF levels followed the same expression pattern than those obtained with IHC procedures. However, NT-3 showed high expression levels at P10 and P21 with the RT-PCR technique but not with IHC. mRNANT-4/5 expression level was also observed at P10 but not in the tissue processed with the IHC protocol. It is known that expression levels of mRNA and protein may vary in the cells, this could be due to multiple factors, e.g. low levels of protein expression, technological sensitivities, sample processing, anatomical origin of tissue and biological differences between transcript and protein abundance. A previous study showed that protein expression does not always correlate with expression levels of mRNA [26]. There are several possible explanations for these discrepancies like post-transcriptional and post-translational modifications; in addition, proteins may have different half-lives [27]. Gene expression analysis is more sensitive than immunocytochemical procedures; therefore it is possible that genes expressed in our study are at levels not high enough for translated protein expression detected by IHC. Satisfactory immunostaining requires preservation of protein tertiary structure in the region of the relevant epitopes [28] and it may be that some of neurotrophins antibodies are less immunoreactive than others.
It is hoped that continued improvements in commercially available antibodies, could further elucidate the relationship between mRNA levels and corresponding protein expression. In addition to the comparison of IHC and mRNA levels by RT-PCR, there will be the need for further evaluation utilizing techniques such as in situ hybridization, western blotting, microarray and proteomic analysis, to have a better idea about neurotrophins expression through all development.
Functional implications of neurotrophins in the postnatal striatum
Changes in neurotrophic factors during postnatal development may reveal differential nourishment requirements for cell populations in the neostriatum. BDNF is transported from the cortex to the neostriatum along the corticostriatal pathway where it has a trophic effect on medium spiny neurons (MSN) [6, 29], besides its function in cell survival, BDNF induces neuronal differentiation in neurons positive to calbindin [30] and DARPP-32 [31], specific markers for MSN. Actually, there is a reduction in calbindin and DARPP-32 positive neurons in BDNF knock out mice [32, 33]. In addition, there is an increase in apoptotic death during development in animals that do not express TrkB receptors [32] which are those targeted by BDNF. However, its expression in postnatal and adult tissue may be linked to other functions that require low amounts of BDNF such as modulation of synaptic transmission.
The presence of NT-3 in striatal tissue is not surprising, because this neurotrophin exhibits neurotrophic effects on striatal, cortical, and nigral neurons and increases the number of GABAergic and calbindin positive neurons similar to BDNF [30]. This neurotrophin also promotes survival and differentiation of DARPP-32 positive neurons, increases their neurites and somatic area [33]. Notably, NT-3 can be found in glial cells [34], as well as in white matter [35]. In our studies, NT-3 was expressed in the neostriatal bundles at P10 and P21; we do not know if NT-3 is synthesized in glial cells, nonetheless, the presence of this neurotrophin in corticostriatal bundles suggest that, NT-3 may arrive to the striatum by anterograde transport from cortex and perhaps there is an interaction between neuronal fibers and glial cells at the neostriatum [22].
NT-4/5 temporal expression at P21, P42 and P80, suggests a possible use of this neurotrophin in therapeutic manipulations because, it has been demonstrated that NT-4/5 increases the survival of GABA, calbindin and calretinin positive neurons in the neostriatum, and shares several of the BDNF neurotrophic effects because, it also acts on TrkB receptors [24, 30]. Therefore, if BDNF exhibits a synthesis failure, which has been demonstrated in some degenerative illness [14], TrkB receptors might be activated with NT-4/5 to protect medium spiny neurons from cell death. Nevertheless this stimulation should be given in early phases of cellular harm, before TrkB receptors reduce their expression due to neuronal damage [36]. Perhaps early stimulation with NT-4/5 will stop TrkB receptor decrement.
BDNF and NT-3, but not NT-4/5, regulates soma morphology in medium spiny neurons without changing GAD immunostaining or mRNAGAD expression [13]. It has been demonstrated that BDNF and NT-4/5 control different functions in the same cell type, for example BDNF but not NT-4/5, increases the somatic area in neurons [37-39]. According to our results (Fig. 4), BDNF and NT-4/5 may act on different neuronal populations; however, both neurotrophins may affect the same cell population at different times due to changes in their expression time course. Although both neurotrophins activate TrkB receptors, their temporal expression is different, and NT-3 activates TrkB receptors in a lower proportion and activates TrkC receptors as well [40].
Conclusions
As neurotrophins have important actions on neuron's survival, differentiation and maintenance, any failure in spatial and temporal neurotrophic supply may harm neostriatal cells; hence, the knowledge about their postnatal expression shall be crucial to understanding those pathologies afflicting the neostriatum early in development. In the present study, the most important result was to visualize differences in neurotrophin´s temporal expression in the neostriatum. In particular, our data supporting NT-3 protein expression in fiber bundles opens the possibility of revising its function in the neostriatum as well NT-4/5 function, even though it is unclear which cells NT-4/5 is synthesized.
Acknowledgements
Initial experiments and image acquisition of IHC procedures were performed at the laboratory of Dr. Gutiérrez, (IIB, UNAM). We thank A. Vilches for his technical help in PCR experiments and to Dr. R. Villalobos (UBIMED, FES-I, UNAM) and Dr. MK Lobo (Mount Sinai University, NY) for their comments to the manuscript. This work was supported by CONACyT Grant No. 42598. V.Z. received a CONACyT fellowship.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Huang E.J, Reichardt L.F. Neurotrophins: roles in neuronal development and function. Annu. Rev. Neurosci. 2001;24:677-736
2. Birling M, Price J. Influence of growth factors on neuronal differentiation. Curr. Opin. Cell Biol. 1995;6:878-884
3. Katoh-Semba R, Semba R, Takeuchi I.K, Kato K. Age-related changes in levels of brain-derived neurotrophic factor in selected brain regions of rats, normal mice and senescence-acelerated mice: a comparison to those of nerve growth factor and neurotrophin-3. Neurosci. Res. 1989;31:227-234
4. Mobley W.C, Woo J.E, Edwwards R.H, Riopelle R.J, Longo F.M, Weskamp G, Otten U, Valletta J.S, Johnston M.V. Development regulation of nerve growth factor in its receptor in the rat caudate-putamen. Neuron. 1989;3:655-664
5. Hofer M, Pagliusi S.R, Hohn A, Leibrock J, Barde Y.A. Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J. 1990;9:2459-2464
6. Canals J.M, Marco S, Checa N, Michels A, Pérez-Navarro E, Arenas E, Alberch J. Differential regulation of the expresión of NGF BDNF and NT-3 after excitotoxicity in a rat model of Huntington's disease. Neurobiol. Dis. 1998;5:357-364
7. Checa N, Canals J.M, Alberch J. Developmental regulation of BDNF and NT-3 expression by quinolinic acid in the striatum and its main connections. Exp. Neurol. 2000;165:118-124
8. Yurek D, Fletcher-Turner A. Differential expression of GDNF BDNF, and NT-3 in the aging nigrostriatal system following a neurotoxic lesion. Brain Res. 2001;891:228-23
9. Timmusk T, Bellaurdo N, Metsis M, Persson H. Widespread and developmentally regulated expression of neurotrophin-4 mRNA in rat brain and peripheral tissues. Eur. J. Neurosci. 1993;5:605-613
10. Ip N.Y, Ibañez C.F Nye S.H, McClain J Jones P.F, Gies D.R Belluscio L, Le Beau M.M Espinosa R, Squinto S.P. Mammalian neurotrophin-4 structure, chromosomal localization, tissue distribution, and receptor specificity. Proc. Natl. Acad. Sci. USA. 1992;89:3060-3064
11. Mobley W.C, Rutkowski J.L, Tennekoon G.I, Buchanan K, Johnston M.V. Choline acetyl-transferase activity in striatum of neonatal rats increased by nerve growth factor. Science. 1985;229:284-287
12. Berkemeier L.R, Winslow J.W, Kaplan D.R, Nikolics K, Goeddel D.V, Rosenthal A. Neurotrophin-5: a novel neurotrophic factor that activates trkA and trkB. Neuron. 1991;7:857-866
13. Pérez-Navarro E, Alberch J, Neveu I, Arenas E. Brain-derived neurotrophic factor, neurotrophin-3 and neurotrophin-4/5 differentially regulate the phenotype and prevent degenative changes in striatal projection neurons after excitotoxicity in vivo. Neuroscience. 1999;91:1257-1264
14. Zucato C, Ciammola A, Rigamonti D, Leavit B.R, Goffredo D, Conti L, MacDonald M.E, Friedlander R.M, Silani V, Hayden M.R, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493-498
15. Di Figlia M. Excitotoxic injury of the neostriatum: a model for Huntington's disease. Trends Neurosci. 1990;13:286-289
16. Beal M.F. Aging, energy, and oxidative stress in neurodegenerative diseases. Ann Neurol. 1995;38:357-366
17. Shen H, Cheng J.M, Cheng K. Expression of neurotrophin mRNAs in the dorsal root ganglion after spinal nerve injury. Mol, Brain Res. 1999;64:186-192
18. Bonini P, Pierucci D, Cicconi1 S, Porzio O, Lauro R, Lionel N.J.L, Marlier L, Borbón P. Neurotrophins and Neurotrophin Receptors mRNAs Expression in Pancreatic Islets and Insulinoma Cell Lines. J Pancreas. 2001;2:105-111
19. Kawakami T, Wakabayashi Y, Isono T, Aimi Y, Okada Y. Expression of neurotrophin messenger RNAs during rat urinary bladder development. Neurosci. Lett. 2002;329:77-80
20. Jung A.B, Bennett J.P.J. Development of striatal dopaminergic function. III: Pre- and postnatal development of striatal and cortical mRNAs for the neurotrophin receptors trkBTK and trkC and their regulation by synaptic dopamine. Dev Brain Res. 1996;94:133-143
21. Chen L.W, Hu H.J, Liu H.L, Yung K.K.L, Chan Y.S. Identification of Brain -Derived Neurotrophic factor in nestin expressing astroglial cells in the neostriatum of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-treated mice. Neurosci. 2004;126:941-953
22. Altar C.A, DiStefano P.S. Neurotrophin trafficking by anterograde transport. Trends Neurosci. 1998;21:433-437
23. Friedman W.J, Ernfors P, Persson H. Transient and persistent expression of NT-3/HDNF in the rat brain during postnatal development. J. Neurosci. 1991;11:1577-1584
24. Checa N, Canals J.M, Gratacos E, Alberch J. TrkB and TRC are differentially regulated by excitotoxicity during development of the basal ganglia. Exp. Neurol. 2001;172:282-292
25. Ibañez C.F. Neurotrophin-4: the odd one out in the neurotrophin family. Neurochem. Res. 1996;21:787-793
26. Gygi S.P, Rochon Y, Franza B.R, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720-1730
27. Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117
28. Pollard K, Lunny D, Holgate C.S, Jackson P, Bird C.C. Fixation, processing, and immunochemical reagent effects on preservation of T-lymphocyte surface membrane antigens in paraffin-embedded tissue. J. Histochem. Cytochem. 1987;35:1329-1338
29. Altar C, Cai N, Blivien T, Juhasz M, Conner J, Acheson A, Lindsay R, Wiegand S. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997;389:856-860
30. Ventimiglia R, Mather P.E, Jones B.E, Lindsay R.M. The neurotrophins BDNF, NT-3 and NT-4/5 promote survival and morphological and biochemical differentiation of striatal neurons. Eur. J. Neurosc. 1995;7:213-222
31. Nakao N, Kokaia Z, Odin P, Lindvall O. Protective effects of BDNF and NT-3 but not PDGF against hypoglycemic injury to cultured striatal neurons. Exp. Neurol. 1995;131:1-10
32. Jones K, Fariñas I, Backus C, Reichardt L. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994;79:989-999
33. Ivkovic S, Polonskaia O, Fariñas I, Ehrlich E. Brain-derived neurotrophic factor regulates maturation of the DARPP-32 phenotype in striatal medium spiny neurons: studies in vivo and in vitro. Neuroscience. 1997;79:509-516
34. Rudge J.S, Alderson R.F, Pasnikowski E, Mc Clain J, Ip N.Y, Lindsay R.M. Expression of ciliary neurotrophic factor and the neurotrophins -nerve growth factor, brain-derived neurotrophic factor, and neurotrophins 3 in cultured hippocampal astrocytes. Eur J. Neurosci. 1992;4:459-471
35. Zhou X.F, Rush R.A. Localization of neurotrophins-3-like immunereactivity in the rat central nervous system. Brain Res. 1994;643:162-172
36. Ginés S, Bosh M, Marco S, Gavaldà N, Díaz-Hernández M, Lucas JJ, Canals JM, Alberch J. Reduced expression of the TrKB receptor in Huntington´s disease mouse models and in human brain. Eur J Neurosci. 2006;23:649-658
37. Ardelt A, Flaris N, Roth K. Neurotrophin-4 selectively promotes survival of striatal neurons in organotypic culture. Brain Res. 1994;647:340-344
38. Arenas E, Akerud P, Wong V, Boylan C, Persson H, Lindsay R, Altar C. Effects of BDNF and NT-4/5 on striatonigral neuropeptides or nigral GABA neurons in vivo. Eur. J. Neurosci. 1996;8:1707-1717
39. Hyman C, Juhasz M, Jackson C, Wright P, Ip N, Lindsay R. Overlapping and distinct actions of the neurotrophins BDNF, NT-3 and NT-4/5 on cultured dopaminergic and GABAergic neurons of the ventral mesencephalon. J. Neurosci. 1994;14:335-347
40. Barbacid M. The Trk family of neurotrophin receptors. J. Neurobiol. 1994;25:1386-1403
Author contact
![]() Correspondence to: Dr. Elizabeth Hernández Echeagaray, Laboratorio de Neurofisiología del Desarrollo y la Neurodegeneración. Unidad de Biomedicina FES-I, Universidad Nacional Autónoma de México. Av. De Los Barrios # 1, Los Reyes Iztacala, C.P. 54090, Tlalnepantla, México. elihernandeziztacala.unam.mx; Phone + 52 55 5623 1111 ext 154; FAX + 52 55 5623-1138
Correspondence to: Dr. Elizabeth Hernández Echeagaray, Laboratorio de Neurofisiología del Desarrollo y la Neurodegeneración. Unidad de Biomedicina FES-I, Universidad Nacional Autónoma de México. Av. De Los Barrios # 1, Los Reyes Iztacala, C.P. 54090, Tlalnepantla, México. elihernandeziztacala.unam.mx; Phone + 52 55 5623 1111 ext 154; FAX + 52 55 5623-1138

 Global reach, higher impact
Global reach, higher impact