10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(6):578-595. doi:10.7150/ijbs.5.578 This issue Cite
Review
Fungal Bioconversion of Lignocellulosic Residues; Opportunities & Perspectives
1. Biorefining Research Initiative, Lakehead University, 955 Oliver Rd, Thunder Bay, Ontario, Canada, P7B 5E1
2. Department of Biology, Lakehead University, 955 Oliver Rd, Thunder Bay, Ontario, Canada, P7B 5E1
Received 2009-6-29; Accepted 2009-8-2; Published 2009-9-4
Abstract
The development of alternative energy technology is critically important because of the rising prices of crude oil, security issues regarding the oil supply, and environmental issues such as global warming and air pollution. Bioconversion of biomass has significant advantages over other alternative energy strategies because biomass is the most abundant and also the most renewable biomaterial on our planet. Bioconversion of lignocellulosic residues is initiated primarily by microorganisms such as fungi and bacteria which are capable of degrading lignocellulolytic materials. Fungi such as Trichoderma reesei and Aspergillus niger produce large amounts of extracellular cellulolytic enzymes, whereas bacterial and a few anaerobic fungal strains mostly produce cellulolytic enzymes in a complex called cellulosome, which is associated with the cell wall. In filamentous fungi, cellulolytic enzymes including endoglucanases, cellobiohydrolases (exoglucanases) and β-glucosidases work efficiently on cellulolytic residues in a synergistic manner. In addition to cellulolytic/hemicellulolytic activities, higher fungi such as basidiomycetes (e.g. Phanerochaete chrysosporium) have unique oxidative systems which together with ligninolytic enzymes are responsible for lignocellulose degradation. This review gives an overview of different fungal lignocellulolytic enzymatic systems including extracellular and cellulosome-associated in aerobic and anaerobic fungi, respectively. In addition, oxidative lignocellulose-degradation mechanisms of higher fungi are discussed. Moreover, this paper reviews the current status of the technology for bioconversion of biomass by fungi, with focus on mutagenesis, co-culturing and heterologous gene expression attempts to improve fungal lignocellulolytic activities to create robust fungal strains.
Keywords: Biomass, Lignocellulose, Bioconversion, Fungi, Cellulases, Cellulosome
1. Introduction
Millions of years ago, atmospheric carbon was captured by plants in a process called photosynthesis, and over time was manifested into crude oil and coal. However, since the industrial revolution, we have used much of these energy sources, causing the excessive release of carbon back into the atmosphere. Thus, over the past 150 years atmospheric CO2 levels have increased from ~280 to ~380 ppm [1,2]. In return, this is potentially causing warmer temperatures worldwide and leading to global climate changes [1,3,4].
Rising energy consumption, depletion of fossil fuels and increased environmental concerns have shifted the focus of energy generation towards biofuel use. Global crude oil production is predicted to decline five times below its current level by 2050. Based on World Energy Council (WEC) calculations, the world-wide primary energy consumption is approximately 12 billion tonnes coal equivalent per year. United Nations calculations have shown that the world's population will increase to about 10 billion people by 2050 which will in turn increase energy demands to at least 24 billion tonnes coal equivalent per year (twice of what we consume today) depending on economic, social and political developments [5,6].
Lignocellulose is a renewable organic material and is the major structural component of all plants. Lignocellulose consists of three major components: cellulose, hemicellulose and lignin. In addition, small amounts of other materials such as ash, proteins and pectin can be found in lignocellulosic residues, in different degrees based on the source [7]. Cellulose, the major constituent of all plant material and the most abundant organic molecule on the Earth, is a linear biopolymer of anhydroglucopyranose-molecules, connected by β-1,4-glycosidic bonds. Coupling of adjacent cellulose chains by hydrogen bonds, hydrophobic interactions and Van der Waal's forces leads to a parallel alignment of crystalline structures known as microfibril [8]. Hemicelluloses, the second most abundant component of lignocellulosic biomass, are heterogeneous polymers of pentoses (including xylose and arabinose), hexoses (mainly mannose, less glucose and galactose) and sugar acids. Composition of hemicelluloses is very variable in nature and depends on the plant source [9,10]. Lignin, the third main heterogeneous polymer in lignocellulosic residues, generally contains three aromatic alcohols including coniferyl alcohol, sinapyl and p-coumaryl. Lignin acts as a barrier for any solutions or enzymes by linking to both hemicelluloses and cellulose and prevents penetration of lignocellulolytic enzymes to the interior lignocellulosic structure. Not surprisingly, lignin is the most recalcitrant component of lignocellulosic material to degrade [7,11].
Lignocellulosic wastes are produced in large amounts by different industries including forestry, pulp and paper, agriculture, and food, in addition to different wastes from municipal solid waste (MSW), and animal wastes (Table 1) [12-17]. These potentially valuable materials were treated as waste in many countries in the past, and still are today in some developing counties, which raises many environmental concerns [18,19]. Significant efforts, many of which have been successful, have been made to convert these lignocellulosic residues to valuable products such as biofuels, chemicals and animal feed [20]. Interestingly, in 2008 approximately 90% of the global ethanol fuel production (15,472.2 out of 17,335.2 Million of Gallons) was concentrated in two countries, Brazil (6,472.2), and The United States of America (9,000) [21]. In Brazil, ethanol is usually produced from cane juice, whereas in the USA, starch-crops such as corn are usually used for ethanol production [7]. Using sugars or corn as the main source for ethanol production caused a great deal of controversy due to its effect on food production and costs, which has made it difficult for ethanol to become cost competitive with fossil fuels. These concerns became a driving force in the generation of new biofuel research using lignocellulosic wastes produced by many different industries. The Iogen Corporation in Canada (http://www.iogen.ca/) is the world's leading operating plant for bioethanol production from lignocellulosic residues, and uses up to 30 tonnes per day of wheat, oat and barley straw to produce up to 0.52 million gallons of ethanol per year [22].
Some of the lignocellulosic residues produced by different industries and potential for ethanol production
| Lignocellulosic Wastes | Annual production | Potential contribution to ethanol production (billion litre/year) | References |
|---|---|---|---|
| World Agricultural Wastes1 | Trillion grams/year (Tg/y) | ||
| Corn stover | 203.62 | 58.6 | [12] |
| Barley straw | 58.45 | 18.1 | [12] |
| Oat straw | 10.62 | 2.78 | [12] |
| Rice straw | 731.34 | 204.6 | [12] |
| Wheat straw | 354.35 | 103.8 | [12] |
| Sorghum straw | 10.32 | 2.79 | [12] |
| Bagasse | 180.73 | 51.3 | [12] |
| Subtotal | 1549.42 | 442.0 | |
| Municipal Solid Waste (MSW) | Million metric tons (million MT) | ||
| USA (2001) | 208 | 13.7 2 | [13] |
| China (1998) | 127 | 8.3 3 | [14] |
| Canada (2002) | 30.5 | 2 4 | [15] |
| Animal Wastes5 | |||
| In Canada (2001) | 177.5 | [16] | |
| In USA (1995) | 160 | [17] |
1 Average values from 1997 to 2001 have been used to calculate world agricultural waste production [12]. 2-4Potential contribution of MSW in USA, China and Canada in 2001, 1998 and 2002 respectively, assuming a conservative yield of 66 L of ethanol/MT of MSW [13-15]. 5The fiber content (including cellulose and hemicellulose) of cattle manure, for example, is 52.6% (dry biomass basis). These sugars can be hydrolyzed and fermented to produce ethanol but the utilization of animal manures is more complicated due to its high protein content [16,17].
In nature, degradation of cellulosic biomass is performed by mixtures of hydrolytic enzymes collectively known as cellulases. The cellulases include endo-acting (endoglucanases) and exo-acting (cellobiohydrolases) enzymes, which act in a synergistic manner in biomass-degrading microbes. Many microorganisms including fungi and bacteria had been found to degrade cellulose and other plant cell wall fibres. By 1976, over 14,000 fungal species capable of degrading cellulose had been isolated, but only a few of them were subjected to in-depth studies [23]. Obviously, fungi contribute significantly to the decay of lignocellulosic residues in nature by producing many different lignocellulolytic enzymes. Most fungal strains produce various enzymes in large amounts which are released in the environment and act in a synergistic manner. The breakdown of lignocellulosic biomass involves the formation of long-chain polysaccharides, mainly cellulose and hemicellulose, and the subsequent hydrolysis of these polysaccharides into their component 5- and 6-carbon chain sugars. In biofuel production, these sugars can be converted to bioethanol through fermentation processes [24].
The primary challenge in biomass conversion to bioethanol is achieving yields that make it cost-competitive with the current fossil-based fuels. Cellulose in the plant cell wall is not readily available to enzymatic hydrolysis (cellulases) due primarily to; (1) low accessibility of (micro-) crystalline cellulose fibers, which prevents cellulases from working efficiently, and (2) the presence of lignin (mainly) and hemicellulose on the surface of cellulose, which prevents cellulases from accessing the substrate efficiently [25]. Thus, pretreatment of lignocellulosic residues before hydrolysis is a prerequisite and this can be performed by different methods (discussed in section 3.1.). High temperature and acid have been used initially for chemical cellulose degradation and they are still involved in pretreatment of lignocellulosic residues at industrial scales. However, this approach is expensive, slow and inefficient [26]. In addition, the overall yield of the fermentation process will be decreased because this pretreatment releases inhibitors such as weak acids, furan and phenolic compounds [27]. Some of these problems could be overcome by applying microorganisms such as fungi. For example, thermophilic fungal species such as Sporotrichum thermophile [28], Thermoascus aurantiacus [29] and Thielavia terrestris [30] have been proposed as good candidates for bioconversion of lignocellulosic residues to sugars and offer the great potential to be used at industrial scales. Applying thermophilic fungal species at industrial scales also allows energy savings because the costly cooling after steam pre-treatment is avoided and saccharification rates are improved. These fungi have been shown to produce cellulases and to degrade native cellulose; however, the enzyme activity in thermophilic organisms (e.g. S. thermophile) is usually low compared to mesophilic fungi such as T. reesei [28].
The initial conversion of biomass into sugars is a key bottleneck in the process of biofuel production and new biotechnological solutions are needed to improve their efficiency, which would lower the overall cost of bioethanol production. Despite the fact that some fungal strains have the advantages of being thermostable and producing cellulases, most of these fungal strains do not produce sufficient amounts of one or more lignocellulolytic enzymes required for efficient bioconversion of lignocellulosic residues to fermentable sugars. Wild-type T. reesei and its best extracellular cellulase producer mutants (e.g. RUT-C30) for example, produce small amounts of β-glucosidase which inhibit further cellulose hydrolysis due to accumulation of the end product inhibitor (cellobiose). In addition, plant cell walls are naturally resistant to microbial and enzymatic (fungal and bacterial) deconstruction, collectively known as “biomass recalcitrance” [11]. These rate-limiting steps in the bioconversion of lignocellulosic residues to ethanol remain one of the most significant hurdles to producing economically feasible cellulosic ethanol. Improving fungal hydrolytic activity and finding stable enzymes capable of tolerating extreme conditions has become a priority in many recent studies.
This review focuses on lignocellulosic bioconversion by the application of different lignocellulolytic enzyme-producing fungi. In addition, this review addresses recent efforts to create robust fungal strains using mutagenesis, co-culturing and heterologous gene expression techniques and how these robust organisms can help overcome some of the critical issues in biofuel production.
2. Lignocellulolytic enzyme-producing fungi
Lignocellulolytic enzymes-producing fungi are widespread, and include species from the ascomycetes (e.g. T. reesei), basidiomycetes including white-rot fungi (e.g. P. chrysosporium), brown-rot fungi (e.g. Fomitopsis palustris) and finally a few anaerobic species (e.g. Orpinomyces sp.) which degrade cellulose in gastrointestinal tracts of ruminant animals [31,32]. Biomass degradation by these fungi is performed by complex mixtures of cellulases [33], hemicellulases [31] and ligninases [7,34], reflecting the complexity of the materials. Cellulases and most hemicellulases belong to a group of enzymes known as glycoside hydrolases (GH). Currently more than 2500 GH have been identified and classified into 115 families (for more information please visit the CAZy web page; www.cazy.org) [35]. Interestingly, the same enzyme family may contain members from bacteria, fungi and plants with several different activities and substrate specifications. However, fungal cellulases (hydrolysis of β-1,4-glycosidic bonds) have been mostly found within a few GH families including 5, 6, 7, 8, 9, 12, 44, 45, 48, 61 and 74 [35,36]. Table 2 summarizes a few different fungi producing different lignocellulolytic enzymes.
2.1. Fungal extracellular cellulases
Hydrolysis of the β-1,4-glycosidic bonds in cellulose can be achieved by many different enzymes known as cellulases which use two different catalytic mechanisms, the retaining and the inverting mechanisms. All GH 12 cellulases, for example, hydrolyze glycosidic bonds by the retaining mechanism whereas family 6 cellulases use the inverting mechanism [33,36]. In both mechanisms, two catalytic carboxylate residues are involved and catalyze the reaction by acid-base catalysis. Many different fungal species have the ability to degrade cellulose by producing extracellular fungal cellulose-degrading enzymes including endo-cleaving (endoglucanases) and exo-cleaving (cellobiohydrolases). Endoglucanases can hydrolyze glycosidic bonds internally in cellulose chains whereas cellobiohydrolases act preferentially on chain ends. The products of the enzymatic reaction are mostly a disaccharide known as cellobiose and, to a lesser extent, cello-oligosaccharides, which will be further hydrolyzed by the third group of enzymes called β-glucosidases [56]. Cellulases mostly have a small independently folded carbohydrate binding module (CBM) which is connected to the catalytic domain by a flexible linker. The CBMs are responsible for binding the enzyme to the crystalline cellulose and thus enhance the enzyme activity [33]. Currently many CBMs have been identified and classified into 54 families, however only 20 families (1, 13, 14, 18, 19, 20, 21, 24, 29, 32, 35, 38, 39, 40, 42, 43, 47, 48, 50 and 52) have been found in fungi. Different fungal cellulolytic enzymes and their main features are summarized in Table 3.
Examples of different fungi producing different lignocellulolytic enzymes and their substrates.
| Group | Fungal strain | Enzymes | Substrate | References | |
|---|---|---|---|---|---|
| Aerobic fungi (Extracellular lignocellulolytic enzymes) | Ascomycetes | T. reesei | Cellulases (CMCase, CBH, BGL), Hemicellulase (xylanase) | Wheat straw | [37,38] |
| T. harzianum | Cellulases (CMCase, CBH), β-1,3-glucanases | Wheat bran, wheat straw | [39,40] | ||
| A. niger | Cellulases, Xylanases | Sugar cane bagasse | [41] | ||
| Pestalotiopsis sp. | Cellulases (CMCase, CBH), Laccase | Forest litter of Quercus variabilis | [42,43] | ||
| Basidiomycetes | P. chrysosporium | Cellulases (CMCase, CBH, BGL), CDH, LiP, MnP, Hemicellulase (xylanases) | Red oak, grape seeds, barley bran, woodchips | [7,44,45] | |
| F. palustris | Cellulases (CMCase, CBH, BGL) | Microcrystalline cellulose | [32,46] | ||
| Anaerobic rumen fungi (Chytridiomycetes) (Cell-wall associated lignocellulolytic enzymes, “cellulosome”) | Anaeromyces | Anaeromyces mucronatus 543 | Cellulase (CMCase), Hemicellulase (xylanase) | Orchard grass hay | [47,48] |
| Caecomyces | Caecomyces communis | Cellulases, Hemicellulases (xylanase, β-D-xylosidase) | Maize stem | [48-50] | |
| Cyllamyces | Cyllamyces aberensis | Cellulases, Xylanases | Grass silage | [48,51] | |
| Neocallimastix | Neocallimastix frontalis | Cellulases, Hemicellulase (xylanase, β-galactosidase) | Cotton fiber, wheat straw | [48,52,53] | |
| Orpinomyces | Orpinomyces sp. | Cellulase (CMCase, CBH, β-glucosidase), Hemicellulases (xylanase, mannanases) | Wheat straw | [31,48,53,54] | |
| Piromyces | Piromyces sp. | Cellulases (CMCase, CBH, β-glucosidase) Hemicellulases (xylanase, mannanases) | Maize stem | [31,48,49,55] |
CMCase: Carboxymethylcellulases (endoglucanase), CBH: Cellobiohydrolases, BGL: β-glucosidases, CDH: Cellobiose dehydrogenase, MnP: Manganese peroxidises, LiP: Lignin peroxidises.
2.1.1. Endo-1,4- β-glucanases (EC 3.2.1.4, endocellulase)
Endoglucanases (EG) are also referred to as carboxymethylcellulases (CMCase), named after the artificial substrate used to measure the enzyme activity. EG initiate cellulose breakdown by attacking the amorphous regions of the cellulose, making it more accessible for cellobiohydrolases by providing new free chain ends. This has been shown by the effect of the enzyme on carboxymethylcellulose and amorphous cellulose [8]. Fungal EGs are generally monomers with no or low glycosylation and have an open binding cleft. They mostly have pH optima between 4.0 and 5.0 and temperature optima from 50 to 70 °C (Table. 3). Studies have shown that many fungi produce multiple EGs. For example, T. reesei produces at least 5 EGs (EGI/Cel7B, EGII/Cel5A, EGIII/Cel12A, EGIV/Cel61A and EGV/Cel45A) whereas three EGs were isolated from white-rot fungus P. chrysosporium (EG28, EG34 and EG44) [44,57]. In addition, some EGs lack a CBM while some other EGs with CBM have been described. For example, four of five EGs in T. reesei including EGI, EGII, EGIV and EGV have CBM whereas EGIII does not have a CBM [36].
2.1.2. Cellobiohydrolases (EC 3.3.1.91, exocellulase)
Cellobiohydrolases (CBH) preferentially hydrolyze β-1,4-glycosidic bonds from chain ends, producing cellobiose as the main product. CBHs have been shown to create a substrate-binding tunnel with their extended loops which surround the cellulose [58,59]. Similar to EGs, CBHs are monomers with no or low glycosylation with pH optima mostly between 4.0 and 5.0, but the temperature optima are wider, from 37 to 60 °C (Table. 3). Studies have shown that some CBHs can act from the non-reducing ends and others from the reducing ends of the cellulosic chains, which increases the synergy between opposite-acting enzymes. For example, T. reesei has been shown to have two CBHs acting from non-reducing (CBHII/Cel6A) and reducing (CBHI/Cel7A) ends, which results in a more efficient cellulolytic degrader. Moreover, both CBHI and CBHII of T. reesei have CBM at the carboxy-terminus or at the amino-terminus of the catalytic module respectively. Cellobiose, the end product of CBHs, acts as a competitive inhibitor, which can limit the ability of the enzymes to degrade all of cellulose molecules in a system [36,44,60].
Overview of the three groups of fungal cellulolytic enzymes and their main features.
| Optimum Substrate | Molecular mass (kDa) | GH family: corresponding structural fold | Optimum temperature (°C) | pH optimum | Glyco-sylation | References | |
|---|---|---|---|---|---|---|---|
| Endo-1,4- β-glucanases (EG) | Cellulose (amorphous regions) | Monomeric (22-45) | 5: (β/α)8 | 50-70 | Mostly 4-5 | None or very low | [33,35,44] |
| 6: Distorted (β/α) | |||||||
| 7, 12: β-jelly roll | |||||||
| 9, 48: (α/α)6 | |||||||
| 45: β barrel | |||||||
| 61: - | |||||||
| 74: 7-fold β-propeller | |||||||
| Cellobiohydrolases (CBH) | Cellulose (crystalline regions) | Monomeric (50-65) | 6: Distorted (β/α) | 37-60 | Mostly 4-5 | None or very low | [33,35,44] |
| 7: β-jelly roll | |||||||
| 9, 48: (α/α)6 | |||||||
| β-glucosidases (BGL) | Cellobiose, cellodextrins | Monomeric, dimeric, trimeric (35-450) | 1: (β/α)8 | 45-75 | Vary1 | Usually very high | [33,35,44,61-63] |
| 3: - |
1pH optima of BGLs vary based on the enzyme localization.
2.1.3. β-glucosidases (EC 3.2.1.21)
β-glucosidases (BGL) have been isolated from many different fungal species including ascomycetes such as T. reesei, and basidiomycetes such as white-rot and brown-rot fungi. β-glucosidases hydrolyze soluble cellobiose and cellodextrins to glucose, and are thus competitively inhibited by glucose. BGLs have been placed in families 1 and 3 of glycoside hydrolases based on their amino acid sequences [64]. Family 3 includes β-glucosidases from fungi, bacteria, and plants whereas family 1 includes β-glucosidases of bacterial, plant and mammalian origins which have galactosidase activity in addition to β-glucosidase activity. BGLs from both families hydrolyze β-1,4-glycosidic bonds using the retaining mechanism [65]. BGLs show the most variability among the cellulolytic enzymes due to their structure and localization. While some BGLs have a simple monomeric structure with around 35 kDa molecular mass (e.g. from Pleurotus ostreatus) [61] some others have dimeric (e.g. from Sporobolomyces singularis with 146 kDa) [62] or even trimeric structures with over 450 kDa (e.g. from Pisolithus tinctorius) [63]. In addition, most of BGLs are glycosylated and in some cases, such as the 300 kDa monomeric BGL from Trametes versicolor, the glycosylation degree is up to 90% [66]. Regarding localization, BGLs can be grouped into three different types including intracellular, cell wall-associated and extracellular [67]. Not surprisingly, pH optima for the enzymes vary based on enzyme localization, however, the temperature optima range from 45 to 75 °C (Table 3). In T. reesei, for example, two β-glucosidases (BGLI/Cel3A & BGLII/Cel1A) have been isolated from culture supernatant, but the enzymes were found to be primarily bound to the cell wall [68]. Moreover, BGL production in T. reesei is very low compared to other cellulolytic fungi such as A. niger. Attempts with some success have been made to improve BGL activity in T. reesei by transformation of the bgl gene from the thermophilic fungus Talaromyces emersonii (cel3a) [69]. More recently, the production of T. reesei β-glucosidase I was enhanced by homologous recombination using xylanase (xyn3) and cellulase (egl3) promoters which improved β-glucosidase activity to 4.0 and 7.5 fold compared to the parent, respectively [70].
2.2. Fungal hemicellulases
Several different enzymes are needed to hydrolyze hemicelluloses, due to their heterogeneity [10]. Xylan is the most abundant component of hemicellulose contributing over 70% of its structure. Xylanases are able to hydrolyze β-1,4 linkages in xylan and produce oligomers which can be further hydrolyzed into xylose by β-xylosidase. Not surprisingly, additional enzymes such as β-mannanases, arabinofuranosidases or α-L-arabinanases are needed depending on the hemicellulose composition which can be mannan-based or arabinofuranosyl-containing [71]. Similar to cellulases, hemicellulases are usually modular proteins and have other functional modules, such as CBM, in addition to their catalytic domains. Also similarly to cellulases, most of the hemicellulases are glycoside hydrolases (GHs), although some hemicellulases belong to carbohydrate esterases (CEs) which hydrolyze ester linkages of acetate or ferulic acid side groups [71,72]. Hemicellulases belong to 20 different GH families (1, 2, 3, 4, 5, 8, 10, 11, 26, 27, 36, 39, 43, 51, 52, 53, 54, 57, 62 and 67) and all of them except for 4 (families 4, 8, 52 and 57) have been found in fungi. All but 1 (family 7) of the 7 different CE families (1, 2, 3, 4, 5, 6 and 7) reported for hemicellulases have been found in fungi [35]. Similarly to cellulases, aerobic fungi such as Trichoderma and Aspergillus secrete a wide variety of hemicellulases in high concentrations (8 and 12 hemicellulases, respectively) and these work in a synergistic manner [71].
2.3. Fungal ligninases
Lignin, the most abundant renewable aromatic polymer on the Earth, is composed of non-phenolic (80-90%) and phenolic structures [73]. It has been shown that fungi degrade lignin by secreting enzymes collectively termed “ligninases”. These include two ligninolytic families; i) phenol oxidase (laccase) and ii) peroxidases [lignin peroxidase (LiP) and manganese peroxidase (MnP)] [74]. White-rot basidiomycetes such as Coriolus versicolor [73], P. chrysosporium and T. versicolor [75] have been found to be the most efficient lignin-degrading microorganisms studied. Interestingly, LiP is able to oxidize the non-phenolic part of lignin, but it was not detected in many lignin degrading fungi. In addition, it has been widely accepted that the oxidative ligninolytic enzymes are not able to penetrate the cell walls due to their size. Thus, it has been suggested that prior to the enzymatic attack, low-molecular weight diffusible reactive oxidative compounds have to initiate changes to the lignin structure (as discussed below) [76,77].
2.4. Oxidative (Non-lignocellulolytic) lignocellulose-degradation mechanisms in higher fungi
A few decades ago, non-enzymatic degradation mechanisms for plant cell-wall polysaccharide degradation were also considered and over the time more evidence for these was found. The non-enzymatic degradation mechanism is mostly assisted by oxidation through production of free hydroxyl radicals (•OH). In fact, many white and brown-rot fungi have been shown to produce hydrogen peroxide (H2O2) which enters the Fenton reaction and results in release of •OH [78,79]. These free radicals attack polysaccharides as well as lignin in plant cell walls in a nonspecific manner providing some cleavages which make it easier for the lignocellulolytic enzymes to penetrate [80,81]. Three different pathways have been found for the generation of free radicals (discussed below) including cellobiose dehydrogenase (CDH) catalyzed reactions, low molecular weight peptides/quinone redox cycling and glycopeptide-catalyzed Fenton reactions (Table 4) [44].
CDH, an extracellular monomeric protein with some glycosylation has been identified in a number of wood- and cellulose-degrading fungi including basidiomycetes (mostly white-rot fungi) and ascomycetes growing on cellulose. The enzyme is able to oxidize cellobiose, higher cellodextrins and other disaccharides or oligosaccharides with β-1,4 linkages. In addition, CDH with (in ascomycetes) or without CBM (in basidiomycetes) have been identified however even in the absence of CBM they are able to bind to cellulose through hydrophobic interactions [82]. It has been shown in some fungi that under cellulolytic conditions CDH production increases which helps cellulases and hemicellulases [83,84]. It is now widely accepted that CDH are able to degrade and modify all three major components of the lignocellulosic residues (cellulose, hemicelluloses and lignin) by producing free hydroxyl radicals in a Fenton-type reaction (for detailed information please refers to the review by Baldrin and Valaskova, 2008 [44]).
It has been shown that white and brown-rot fungi produce low molecular weight chelators which are able to penetrate into the cell wall. For example Gloeophyllum trabeum produces a low molecular weight peptide (known as short fiber generating factor, SFGF) which can degrade cellulose into short fibers by an oxidative reaction [81,85]. It has also been reported that some of these low molecular weight compounds are quinones which have to be converted to hydroquinones by some fungal enzymes (Table 4) and then through Fenton reaction, free hydroxyl radicals will be produced [73].
Different glycopeptides with different molecular weight (ranging from 1.5 to 12 kDa) have been found in many brown-rot fungi such as G. trabeum [86] and white-rot fungi such as P. chrysosporium [77,87]. Similar to the other mechanisms, glycopeptides are able to catalyze redox reactions and thus produce free hydroxyl radicals.
Different mechanisms involved in production of •OH in different fungi
| Fungi | Mechanisms | Other enzymes involved/their function | References |
|---|---|---|---|
| White-rot fungi (e.g. Dichomitus squalens) | CDH catalyzed reaction | Oxalate decarboxylase/regulation of oxalate concentration | [88,89] |
| Brown and white-rot fungi (e.g. Coniophora puteana, P. chrysosporium) | Quinone redox cycling | Benzoquinone reductases, CDH, sugar dehydrogenases/convert quinones to hydroquinones | [83,90] |
| Brown and white-rot fungi (e.g. F. palustris, P. chrysosporium) | Glycopeptides-catalyzed Fenton reaction | Cell wall-associated reductase/reduction of glycopeptides | [91] |
2.5. “Cellulosome”: non-free cellulases in anaerobic fungi
Anaerobic fungi represent a special group of microorganisms inhabiting the gastro-intestinal tract of ruminants and most non-ruminant herbivores. These fungi, along with some anaerobic bacteria (mainly from the class Clostridia e.g. Clostridium thermocellum [92]), produce a range of cellulolytic and hemicellulolytic enzymes in a multienzyme complex known as cellulosome. The first anaerobic gut fungi able to break down ingested lignocellulosic residues were identified in 1975 by Orpin [93] and since then 6 genera and 18 species have been identified some of which are shown in Table 2. The cellulosome, however, was initially discovered in anaerobic bacteria (Clostridium thermocellum) in 1983 [94], and then first described in anaerobic fungi in 1992 (Neocallimastix frontalis) [95]. In anaerobic bacteria, the cellulosome is usually comprised of 20 or more different cellulolytic/hemicellulolytic enzymes. However, in anaerobic fungi such as N. frontalis and Piromyces, cellulosomes include at least six or ten polypeptides, respectively (cellulosome-type complex) [55,95,96]. All hydrolytic enzymes in the cellulosome are bound together by noncatalytic scaffolding proteins. In addition to catalytic subunits, all enzymes have noncatalytic subunits known as “fungal dockerin domains” (FDD), which allow binding to cohesin modules of the scaffolding proteins. Interestingly, 50 fungal FDDs have been identified so far which present different amino acid sequences than those found in bacterial dockerins [31,97].
Anaerobic fungi efficiently hydrolyze cellulose and hemicellulose by producing many lignocellulolytic enzymes. Most of the enzymes are associated with the cellulosome; however, some free enzymes also have been identified. In Piromyces sp. PC2, a cellulosome-producing anaerobic fungus for example, 17 lignocellulolytic enzyme encoding genes have been isolated including ten cellulases, one β-glucosidases, five hemicellulases and one enzyme facilitating protein folding. Interestingly, FDD has been reported only for 11 of the genes, which indicates that these cellulases are cellulosome-associated. Moreover, CBM has been identified only in three of those 17 genes, including two cellulases and one hemicellulase [31]. Interestingly, the major products of cellulose digestion by fungal cellulosomes are glucose which eliminate the costly addition of β-glucosidase, whereas in the case of bacterial cellulosomes, cellobiose is the major product [98]. Despite many advantages of cellulosomes such as synergistic activity between the components and efficient hydrolytic activity on both cellulose and hemicellulose, fungal cellulosomes are much less well characterized compared to bacterial cellulosomes.
3. Bioconversion and biotechnological aspects of lignocellulose degradation by microorganisms
The bioconversion of lignocellulosic residues to valuable materials such as ethanol is more complicated than the bioconversion of starch based residues and thus requires four steps of processing, of which the first three are bio-related processes and the fourth is primarily a chemical engineering process that will not be discussed in great detail in this review; i) pretreatment ii) de-polymerization (saccharification) of cellulose and hemicelluloses to soluble monomer sugars (hexoses and pentoses) by a process known as hydrolysis, iii) conversion of these monomeric sugars to valuable products such as ethanol in a fermentation process and iv) separation and purification of the products (Figure 1). In order to improve the yield, each step in the bioconversion process has to be optimized. In addition, process integration has to be considered in order to minimize process energy demand [22].
3.1. Pretreatment
Pretreatment of the lignocellulosic residues is necessary because hydrolysis of non-pretreated materials is slow, and results in low product yield. Some pretreatment methods increase the pore size and reduce the crystallinity of cellulose (Figure 1). Pretreatment also makes cellulose more accessible to the cellulolytic enzymes, which in return reduces enzyme requirements and thus the cost [99]. Many different pretreatment methods have been used, but they can be categorized into three broad groups: chemical (e.g. acid or alkali), physical/ physicochemical (e.g. physical ball milling or physicochemical steam explosion) and biological pretreatment by microorganisms [100]. In the chemical pretreatment method using acid for example, hemicelluloses will be targeted whereas in alkali-catalyzed pretreatment mainly lignin is removed [22]. It has been suggested that, there will probably not be a general pretreatment procedure and that different raw materials will require different pretreatments [22]. Since many fungal cellulolytic enzymes (fungal-derived cellulases and β-glucosidases) work at lower pH (usually 4-5) acidic pretreatment seems a preferred option when fungal enzymes are chosen for the hydrolysis [22].
Biological pretreatment uses microorganisms and their enzymes selectively for delignification of lignocellulosic residues and has the advantages of a low-energy demand, minimal waste production and a lack of environmental effects [101]. White-rot basidiomycetes possess the capabilities to attack lignin. P. chrysosporium, for example, has been shown to non-selectively attack lignin and carbohydrate [102]. P. chrysosporium was successfully used for biological pretreatment of cotton stalks by solid state cultivation (SSC) and results have shown that the fungus facilitates the conversion into ethanol [101]. Other basidiomycetes such as Phlebia radiata, P. floridensis and Daedalea flavida, selectively degrade lignin in wheat straw and are good choices for delignification of lignocellulosic residues [103]. Ceriporiopsis subvermispora, however, lacks cellulases (cellobiohydrolase activity) but produces manganese peroxide and laccase, and selectively delignifies several different wood species [104].
3.2. Hydrolysis
After pretreatment, cellulose and hemicelluloses are hydrolyzed to soluble monomeric sugars (hexoses and pentoses) using cellulases and hemicellulases, respectively (Figure 1). As mentioned earlier, many fungal species such as Trichoderma, Penicillium, Aspergillus and T. emersonii are able to produce large amounts of extracellular cellulases and hemicellulases. High temperature and low pH tolerant enzymes are preferred for the hydrolysis due to the fact that most current pretreatment strategies rely on acid and heat [105]. In addition, thermostable enzymes have several advantages including higher specific activity and higher stability which improve the overall hydrolytic performance [106]. Ultimately, improvement in catalytic efficiencies of enzymes will reduce the cost of hydrolysis by enabling lower enzyme dosages. Some fungal strains such as T. emersonii [107], T. aurantiacus [29], T. terrestris [30], S. thermophile [28], Chaetomium thermophilum [108] and Corynascus thermophilus [109] can produce thermostable enzymes which are stable and active at elevated temperatures (˃60°C) well above their optimum growth temperature (30 to ~55) [110]. Due to the promising thermostability and acidic tolerance of thermophilic fungal enzymes, they have good potential to be used for hydrolysis of lignocellulosic residues at industrial scales.
Schematic picture for the conversion of lignocellulosic biomass to ethanol, including the major steps. Hydrolysis and fermentation can be performed separately (SHF, indicated by broken arrows) or as simultaneous saccharification and fermentation (SSF). In consolidated bioprocessing (CBP) however, all bioconversion steps are minimized to one step in a single reactor using one or more microorganisms. Different techniques such as mutagenesis, co-culturing and heterologous gene expression have been used to improve sugars utilization of the microbial biocatalyst as well as activity and/or stability of hydrolytic fungal-derived enzymes in order to improve the overall yields. For reduction of production cost, ethanol production can be integrated with a combined heat and power plant using lignin.
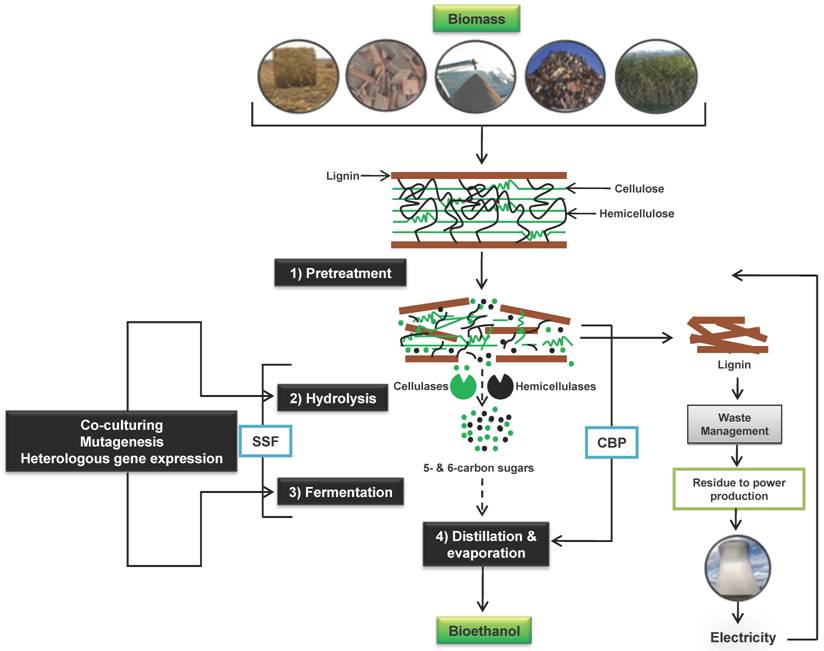
3.3. Fermentation
In the fermentation process, the hydrolytic products including monomeric hexoses (glucose, mannose and galactose) and pentoses (xylose and arabinose) will be fermented to valuable products such as ethanol (Figure 1). Among these hydrolytic products, glucose is normally the most abundant, followed by xylose or mannose and other lower concentration sugars. Saccharomyces cerevisiae is the most frequently and traditionally used microorganism for fermenting ethanol from starch-based residues at industrial scales [22]. S. cerevisiae has a few advantages such as its wide public acceptance, high fermentation rate and high ethanol tolerance that make it a good candidate for fermentation processes. However, S. cerevisiae is unable to efficiently utilize xylose as the sole carbon source or ferment it to ethanol [111]. To make industrial lignocellulosic bioconversion more economically feasible, it is necessary to choose microorganisms capable of fermenting both glucose and xylose. Therefore, many successful attempts have been made to improve xylose fermentation in S. cerevisiae since the first discovery of pentose-fermenting yeasts in 1981 by a Canadian group [112]. These efforts can be classified within two major groups: recombinant (e.g. metabolic engineering of S. cerevisiae with genes from other xylose-fermenting yeasts) and non-recombinant (e.g. adaptation) techniques. These improvements have reached the point where the deficient xylose-fermenting S. cerevisiae can now convert xylose to ethanol at an efficiency close to its theoretical value of 0.51 g g-1 (for extensive review please read Chu and Lee, 2007 [111]).
In addition to xylose, S. cerevisiae is also unable to ferment arabinose, unless supplemented with rich media [113]. Therefore, recombinant S. cerevisiae harbouring xylose-fermenting genes have been engineered with arabinose-metabolizing genes from other microorganisms. The latest recombinant S. cerevisiae (TMB 3400) has been shown to successfully ferment both xylose and arabinose in addition to glucose [114].
During fermentation of lignocellulosic-based biomass, S. cerevisiae faces yet another challenge: the presence of inhibiting compounds including low molecular weight organic acids, furan derivatives, phenolics and inorganic compounds. These compounds are released and formed during pretreatment and/or hydrolysis of the lignocellulosic residues [115]. Thus, it is necessary to detoxify hydrolytic products before the fermentation which increases process cost in addition to sugar loss [116]. Interestingly, S. cerevisiae is one of the least sensitive microorganisms to the inhibitory effect of lignocellulolytic hydrolysate inhibitors. In a recent study for example, glucose and xylose, the hydrolytic products of steam-pretreated corn stover were efficiently co-fermented to ethanol without detoxification using the recombinant S. cerevisiae strain TMB 3400 [117]. It is also possible to adopt recombinant xylose-fermenting S. cerevisiae to the hydrolysate inhibitors by continuous cultivation in the presence of the inhibitors [118].
Attempts have been taken to reduce by-product inhibition. In a recent study for example, wheat straw pellets were subjected to wet explosion pretreatment using three different oxidizing agents, H2O2, O2 and air [119]. Interestingly, the pretreatment with O2 has been shown to be the most efficient in enhancing conversion of the raw material to sugars. Using the method also has been minimized the production of furfural as a by-product which improved enzymatic hydrolysis and minimized the enzyme loading to 10 FPU/g with conversion rate of 70 and 68% for cellulose and hemicellulose respectively [119]. Ammonia fiber explosion (AFEX) pretreatment also has been shown to be a good candidate since it does not produce some inhibitory by-products such as furans. However, the disadvantage of the method is that some of the phenolic compounds in lignin may remain on the pretreated material, which then needs to be washed. This creates wastewater, which causes the process to become environmentally unfriendly [100].
The last two steps of bioconversion of pretreated lignocellulolytic residues to ethanol (hydrolysis and fermentation) can be performed separately (SHF) or simultaneously (SSF) (Figure 1). In the separate hydrolysis and fermentation (SHF), the hydrolysate products will be fermented to ethanol in a separate process. The advantage of this method is that both processes can be optimized individually (e.g. optimal temperature is 45-50 °C for hydrolysis, whereas it is 30 °C for fermentation). However, its main drawback is the accumulation of enzyme-inhibiting end-products (cellobiose and glucose) during the hydrolysis. This makes the process inefficient, and the costly addition of β-glucosidase is needed to overcome end-product inhibition [120]. In simultaneous saccharification and fermentation (SSF), however, the end-products will be directly converted to ethanol by the microorganism. Therefore, addition of high amounts of β-glucosidase is not necessary and this reduces the ethanol production costs [121]. However, the main drawback of SSF is the need to compromise processing conditions such that temperature and pH are suboptimal for each individual step. However, the development of recombinant yeast strains (i.e. improved thermotolerance) is expected to enhance the performance of SSF [1]. Further process integration can be achieved by a process known as consolidated bioprocessing (CBP) which aims to minimize all bioconversion steps into one step in a single reactor using one or more microorganisms. CBP operation featuring cellulase production, cellulose/hemicellulose hydrolysis and fermentation of 5- and 6- carbon sugars in one step have shown the potential to provide the lowest cost for biological conversion of cellulosic biomass to fuels, when processes relying on hydrolysis by enzymes and/or microorganisms are used (Figure 1) [122].
4. Methods used to improve fungal enzyme production, activity and/or stability
In order to increase ethanol yield in the bioconversion process, both cellulose and hemicellulose have to be completely hydrolyzed with minimum sugar degradation. Moreover, all monomeric sugars produced during hydrolysis have to be efficiently fermented to ethanol. Technologies required for bioconversion of lignocelluloses to ethanol and other valuable products are currently available but need to be developed further in order to make biofuels cost competitive compared to other available energy resources such as fossil fuels. Many attempts have been made to improve the overall process yield and cost with a main focus on enzyme production and activity. Not surprisingly, the application of different strains and processes which are selected on the basis of the biomass residues used make comparisons difficult, if not impossible. Nevertheless, the most recent and important improvements in production/activity of fungal enzymes using different techniques such as mutagenesis, co-culturing and heterologous gene expression of cellulases are discussed below and summarized in Table 5.
4.1. Mutagenesis
Many fungal strains have been subjected to extensive mutagenesis studies due to their ability to secrete large amounts of cellulose-degrading enzymes. It has been four decades since Mandels and Weber (late 1960s) screened over 100 wild-type strains of Trichoderma species to isolate the best cellulolytic strain and came up with T. reesei (initially called T. viride QM6a) [123]. Cellulolytic activity of T. reesei QM6a has been improved by using different mutagenesis techniques including UV-light and chemicals at the US Army Natick Laboratory, resulting in the mutant QM 9414 with higher filter paper activity (FPA) [124]. Other studies in different laboratories have also made significant contributions to strain improvements using mutagenesis techniques, leading to development of the mutant strains M7, NG14 [125] and RUT-C30 [126]. T. reesei RUT-C30 is one of the best known mutants, producing 4-5 times more cellulase than the wild-type strain (QM 6a). A recent study by Kovács and et al. (2008) has shown that wild-type Trichoderma atroviride (F-1505) produces the most cellulase among 150 wild-type Trichoderma. Moreover, T. atroviride mutants were created by mutagenesis using N-methyl-N′-nitro-N-nitrosoguanidine (NTG) as well as UV-light. These T. atroviride mutants (e.g. T. atroviride TUB F-1724) produce high levels of extracellular cellulases as well as β-glucosidase when they are grown on pretreated willow [127].
Cellulase and xylanase activities in Penicillium verruculosum 28K mutants were improved about 3-fold using four cycles of UV mutagenesis. The enzyme production was further improved by 2- to 3-fold in a two-stage fermentation process using wheat bran, yeast extract medium and microcrystalline cellulose as the inducer [128]. However, caution has to be taken during strain improvement by mutagenesis. Studies have shown that the best T. reesei mutant (RUT-C30) lacks an 85 Kb genomic fragment and is consequently missing 29 genes which include transcription factors, metabolic enzymes and transport proteins. In fact, these genotype changes are correlated with phenotypic changes such as poor growth on α-linked oligo- and polyglucosides or disturbance of osmotic homeostasis [129].
On the other hand, site-directed mutagenesis (SDM) has played a central role in the characterization and improvement of cellulases including their putative catalytic and binding residues. Different site-directed mutagenesis methods such as saturation mutagenesis, error-prone PCR and DNA shuffling have been used to improve specific enzyme properties. For example, by the application of SDM it was found that Glu 116 and 200 are the catalytic nucleophile and acid-base residues in Hypocrea jecorina (anamorph T. reesei) Cel12A, respectively. In the study, mutant enzymes were produced where Glu was replaced by Asp or Gln at each position (E116D/Q and E200D/Q). The specific activity of these mutants was reduced by more than 98%, suggesting the critical role of these two residues in the catalytic function of the enzyme [130].
In another study, the thermostable endo-1,4-β-xylanase (XynII) mutants from T. reesei were further mutated to resist inactivation at high pH by using SDM. All mutants were resistant to thermal inactivation at alkaline pH. For example, thermotolerance for one mutant (P9) at pH 9 was increased approximately 4-5 °C, resulting in better activity in sulphate pulp bleaching compared to the reference [131]. Also, the catalytic efficiency and optimum pH of T. reesei endo-β-1,4-glucanase II were improved by saturation mutagenesis followed by random mutagenesis and two rounds of DNA shuffling. The pH optimum of the variant (Q139R/L218H/W276R/N342T) was shifted from 4.8 to 6.2, while the enzyme activity was improved more than 4.5-fold [132]. Moreover, the stability of T. reesei endo-1,4-β-xylanases II (XynII) was increased by engineering a disulfide bridge at its N-terminal region. In fact, two amino acids (Thr-2 and Thr-28) in the enzyme were substituted by cysteine (T2C:T28C mutant) resulting in a 15 °C increase in thermostability [133].
4.2. Co-culturing
Fungal co-culturing offers a means to improve hydrolysis of lignocellulosic residues, and also enhances product utilization which minimizes the need for additional enzymes in the bioconversion process. In the case of cellulose degradation, for example, all three enzymatic components (EG, CBH and β-glucosidase) have to be present in large amounts. However, none of the fungal strains, including the best mutants, are able to produce high levels of the enzymes at the same time. T. reesei for example produces CBH and EG in high quantities whereas its β-glucosidase activity is low [134]. A. niger however, produces large amounts of β-glucosidase, but has limited EG components [56]. In addition, hemicellulose hydrolysis must also be considered when lignocellulosic residues are subjected to biomass conversion. However, this will be determined by the pretreatment methods. Specifically in an alkali pretreatment method, a part of lignin will be removed and thus hemicellulose has to be degraded by the use of hemicellulases, whereas in acid-catalyzed pretreatment, the hemicellulose layer will be hydrolyzed [22]. Again, some fungal strains have been shown to work more efficiently on cellulosic residues whereas others produce more hemicellulolytic enzymes and efficiently hydrolyze hemicellulosic portions [20,135]. Conversion of both cellulosic and hemicellulosic hydrolytic products in a single process can be achieved by co-culturing two or more compatible microorganisms with the ability to utilize the materials. In fact, in nature, lignocellulosic residues are degraded by multiple co-existing lignocellulolytic microorganisms. Co-culturing of two or more fungal strains in mixed culture fermentation is widely used in many biological processes including the production of antibiotics, enzymes and fermented food [136]. Mixed fungal cultures have many advantages compared to their monocultures, including improving productivity, adaptability and substrate utilization. Improving fungal cellulolytic activity of T. reesei and A. niger by co-culturing was the subject of extensive research including studies done by Maheshwari [137], Ahmad [138] and Juhász [139]. Moreover, other fungal strains have been co-cultured to obtain better cellulolytic activity such as co-culturing of T. reesei RUT-C30 and A. phoenicis [140] or A. ellipticus and A. fumigatus (Table 5) [141]. There are a few examples of co-culturing fungal strains for the purpose of combining cellulose and hemicellulose hydrolysis such as co-culturing T. reesei D1-6 and A. wentii Pt 2804 in a mixed submerged culture [142] or co-culturing T. reesei LM-UC4 and A. phoenicis QM329 using ammonia-treated bagasse [143]. In the both cases, enzyme activity for cellulases and hemicellulases was significantly increased. The main drawback of co-culturing however is the complexity of growing multiple microorganisms in the same culture [144].
Alternatively to co-culture, microorganisms can be metabolically engineered, which enable one microorganism to complete an entire task from beginning to end. This can be done by altering metabolic flux by blocking undesirable pathway(s) and/or enhancement of desirable pathway(s). For example by application of homologous recombination, the production of T. reesei β-glucosidase I was enhanced using xylanase (xyn3) and cellulase (egl3) promoters which improved β-glucosidase activity to 4.0 and 7.5 fold compared to the parent, respectively. This will permit one fungal strain such as T. reesei to be more efficient on hydrolysis of cellulose to glucose which improve the yield and therefore lower the cost [70].
4.3. Heterologous expression of cellulases
Heterologous expression is a powerful technique to improve production yield of enzymes, as well as activity. In order to make a robust lignocellulolytic fungal strain, many different fungal cellulases with higher and/or specific activity based on the need for a functional cellulase system in the organism have been cloned and expressed. For example, thermostable β-glucosidase (cel3a) from thermophilic fungus T. emersonii was expressed in T. reesei RUT-C30 using a strong T. reesei cbh1 promoter. The expressed enzyme has been shown to be highly thermostable (optimum temperature at 71.5 °C) with high specific activity [69]. In the study for the improvement of biofinishing of cotton, T. reesei cellobiohydrolase (I & II) were overexpressed using additional copy(s) of the genes cloned under T. reesei cbh1 promoter. The results have shown that the expression of CBHI was increased to 1.3- and 1.5-fold with one or two additional copies of the gene, respectively. In the case of CBHII, however, the expression was increased to 3- to 4-fold using just one additional copy of the gene [145]. In addition, chimeric proteins with specific applications have been designed using recombinant DNA technology. For example, an endoglucanase from Acidothermus cellulolyticus was fused to T. reesei cellobiohydrolase and expressed in T. reesei. This bi-functional endo- & exo-acting cellulolytic enzyme has been shown to improve saccharification yields [146]. Moreover, the structural and biochemical information obtained from family GH 12 homologues was used to create a wide range of H. jecorina Cel12 A variants which were heterologously expressed as secreted proteins in A. niger displaying temperature stability changes ranging from none to an increase of 3.9 °C (the most stable variant, P201C) (Table 5) [36].
Some methods which have been used to improve fungal lignocellulolytic activity or stability.
| Methods | Fungal strain | Enzyme | Altered feature | Technique | Reference |
|---|---|---|---|---|---|
| Mutagenesis | T. reesei RUT-C30 | Cellulases | Activity | UV treatment followed by 2 rounds of NTG treatment | [126,129] |
| T. atroviride TUB F-1724 | β-glucosidase | Activity | 2 rounds of NTG treatment followed by UV treatment | [127] | |
| P. verruculosum 28K mutants | Cellulases and xylanases | Activity | Four cycles of UV mutagenesis followed by two-stage fermentation process | [128] | |
| T. reesei P9 | Thermophilic endo-1,4-β-xylanase (XynII) | pH stability (alkalinity), Thermostability | SDM (using PCR and synthetic oligonucleotide primers) (N97R+F93W+H144K) | [131] | |
| T. reesei (Variants L218H, Q139R/N342T) | Endo-β-1,4-glucanase II | Catalytic efficiency, pH optimum | Saturation mutagenesis followed by random mutagenesis and two rounds of DNA shuffling | [132] | |
| T. reesei (T2C:T28C mutant) | Endo-1,4-β-xylanases II (XynII) | Thermostability | PCR and synthetic oligonucleotide primers (Engineering a disulfide bridge at N-terminal region) | [133] | |
| Co-culturing | T. reesei RUT-C30 and A. niger LMA | β-glucosidase | Activity | Fed-batch fermentor on a Cellulose-Yeast extract medium | [138] |
| T. reesei RUT-C30 and A. phoenicis | β-glucosidase | Activity | Shake flask culture | [140] | |
| A. ellipticus and A. fumigatus | β-glucosidase | Activity | Solid state fermentation using pretreated sugarcane bagasse | [141] | |
| T. reesei D1-6 and A. wentii Pt 2804 | Cellulases, xylanases | Activity | Mixed culture medium (M3) supplemented with trace metal & vitamin solutions | [142] | |
| T. reesei LM-UC4 and A. phoenicis QM329 | Cellulases, hemicellulases | Activity | Solid state fermentation using ammonia-treated bagasse | [143] | |
| Heterologous gene expression | T. reesei RUT-C30 | Thermostable β-glucosidase (cel3a) from thermophilic fungus T. emersonii | Activity | Heterologous gene expression using T. reesei cbh1 promoter | [69] |
| T. reesei | Cellobiohydrolase (I & II) | Activity | Overexpression using T. reesei cbh1 promoter | [145] | |
| Acidothermus cellulolyticus and T. reesei | Endoglucanase & cellobiohydrolase | Bi-functional endo- & exo-acting cellulase | Chimeric protein, expressed in T. reesei | [146] | |
| H. jecorina (P201C) | Cel12 A | Thermostability | Mutation followed by heterologous expression in A. niger | [36] |
SDM: site-directed mutagenesis
5. Concluding remarks
Lignocellulolytic microorganisms, especially fungi, have attracted a great deal of interest as biomass degraders for large-scale applications due to their ability to produce large amounts of extracellular lignocellulolytic enzymes. Many successful attempts have been made to improve fungal lignocellulolytic activity including recombinant and non-recombinant techniques. Process integration has also been considered for the purpose of decreasing the production cost, which was partly achieved by performing hydrolysis and fermentation in a single reactor (SSF) using one or more microorganisms (co-culturing). Moreover, recombinant S. cerevisiae with efficient fermenting activity for both 5- and 6-carbon sugars in the presence of inhibitors contributed to process integration. These laboratory improvements should now be verified in pilot and demonstration plants, such as the projects completed at the Iogen pilot plant (Canada). Scaling up the production of lignocellulosic ethanol, however, requires further reduction of the production cost. Overall, in order to improve the technology and reduce the production cost, two major issues have to be addressed: i) improving technologies to overcome the recalcitrance of cellulosic biomass conversion (pretreatment, hydrolysis and fermentation) and ii) sustainable production of biomass in very large amounts. In the case of large scale biomass production, in addition to forest and crop residues, energy crops such as switchgrass and Miscanthus can be grown to meet the needs. On the other hand, biotechnological approaches including systems biology and computational tools are likely good candidates to overcome these issues.
Acknowledgements
We thank Miranda M. Maki and Bruce Rosa for critically reading the manuscript, and for their helpful comments and improvement of the text.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Galbe M, Zacchi G. A review of the production of ethanol from softwood. Appl Microbiol Biotechnol. 2002;59:618-628
2. Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K, Knowlton N, Eakin CM, Iglesias-Prieto R, Muthiga N, Bradbury RH, Dubi A, Hatziolos ME. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737-1742
3. Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci U S A. 2006;103:11206-11210
4. Alonso A, Perez P, Morcuende R, Martinez-Carrasco R. Future CO2 concentrations, though not warmer temperatures, enhance wheat photosynthesis temperature responses. Physiol Plant. 2008;132:102-112
5. United Nations Department of Economic, Social Affairs PD. World Population Prospects, The 2006 Revision, Highlights; Working Paper ESA/P/WP-202. USA: UN. 2007
6. Schiffer H-W. WEC energy policy scenarios to 2050. Energy Policy. 2008;36:2464-2470
7. Sanchez C. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol Adv. 2009;27:185-194
8. Percival Zhang YH, Himmel ME, Mielenz JR. Outlook for cellulase improvement: screening and selection strategies. Biotechnol Adv. 2006;24:452-481
9. Saha BC. Alpha-L-arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol Adv. 2000;18:403-423
10. Saha BC. Hemicellulose bioconversion. J Ind Microbiol Biotechnol. 2003;30:279-291
11. Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804-807
12. Kim S, Dale BE. Global potential bioethanol production from wasted crops and crop residues. Biomass and Bioenergy. 2004;26:361-375
13. Kalogo Y, Habibi S, MacLean HL, Joshi SV. Environmental implications of municipal solid waste-derived ethanol. Environ Sci Technol. 2007;41:35-41
14. Pokhrel D, Viraraghavan T. Municipal solid waste management in Nepal: practices and challenges. Waste Manag. 2005;25:555-562
15. Statistics Canada. Human Activity and the Environment, Annual Statistics 2005, Solid Waste in Canada; Catalogue 16-201-XIE November 2005;pp 8. Canada: Statistics Canada. 2005
16. Champagne P. Feasibility of producing bio-ethanol from waste residues: A Canadian perspective Feasibility of producing bio-ethanol from waste residues in Canada. Resources, Conservation and Recycling. 2007;50:211-230
17. Wen Z, Liao W, Chen S. Hydrolysis of animal manure lignocellulosics for reducing sugar production. Bioresour Technol. 2004;91:31-39
18. Palacios-Orueta A, Chuvieco E, Parra A, Carmona-Moreno C. Biomass burning emissions: a review of models using remote-sensing data. Environ Monit Assess. 2005;104:189-209
19. Levine JS. Biomass burning and global change. Levine JS (eds) (vol. 1) Remote sensing and inventory development and biomass burning in Africa The MIT Press, Cambridge, Massachusetts, USA 1996: pp 35
20. Howard RL, Abotsi E, Jansen van Rensburg EL, Howard S. Lignocellulose biotechnology: issues of bioconversion and enzyme production. African Journal of Biotechnology. 2003;2:602-619
21. Annual World Ethanol Production by Country Estimates, 2008. Renewable Fuels Association (RFA), Licht FO. http://www.ethanolrfa.org/industry/statistics/
22. Hahn-Hagerdal B, Galbe M, Gorwa-Grauslund MF, Liden G, Zacchi G. Bio-ethanol--the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006;24:549-556
23. Mandels M, Sternberg D. Recent advances in cellulase technology. Ferment. Technol. 1976;54:267-286
24. Zhou S, Ingram LO. Synergistic hydrolysis of carboxymethyl cellulose and acid-swollen cellulose by two endoglucanases (CelZ and CelY) from Erwinia chrysanthemi. J Bacteriol. 2000;182:5676-5682
25. Zhang YH, Ding SY, Mielenz JR, Cui JB, Elander RT, Laser M, Himmel ME, McMillan JR, Lynd LR. Fractionating recalcitrant lignocellulose at modest reaction conditions. Biotechnol Bioeng. 2007;97:214-223
26. Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841-845
27. Palmqvist E, Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates. I: inhibition and detoxification. Bioresource Technology. 2000;74:17-24
28. Bhat KM, Maheshwari R. Sporotrichum thermophile Growth, Cellulose Degradation, and Cellulase Activity. Appl Environ Microbiol. 1987;53:2175-2182
29. Gomes I, Gomes J, Gomes DJ, Steiner W. Simultaneous production of high activities of thermostable endoglucanase and beta-glucosidase by the wild thermophilic fungus Thermoascus aurantiacus. Appl Microbiol Biotechnol. 2000;53:461-468
30. Gilbert M, Yaguchi M, Watson DC, Wong KK, Breuil C, Saddler JN. A comparison of two xylanases from the thermophilic fungi Thielavia terrestris and Thermoascus crustaceus. Appl Microbiol Biotechnol. 1993;40:508-514
31. Ljungdahl LG. The cellulase/hemicellulase system of the anaerobic fungus Orpinomyces PC-2 and aspects of its applied use. Ann N Y Acad Sci. 2008;1125:308-321
32. Yoon JJ, Cha CJ, Kim YS, Son DW, Kim YK. The brown-rot basidiomycete Fomitopsis palustris has the endo-glucanases capable of degrading microcrystalline cellulose. J Microbiol Biotechnol. 2007;17:800-805
33. Bayer EA, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol. 1998;8:548-557
34. Weng JK, Li X, Bonawitz ND, Chapple C. Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr Opin Biotechnol. 2008;19:166-172
35. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233-238
36. Sandgren M, Stahlberg J, Mitchinson C. Structural and biochemical studies of GH family 12 cellulases: improved thermal stability, and ligand complexes. Prog Biophys Mol Biol. 2005;89:246-291
37. Chahal DS. Solid-State Fermentation with Trichoderma reesei for Cellulase Production. Appl Environ Microbiol. 1985;49:205-210
38. Kurzatkowski W, Torronen A, Filipek J, Mach RL, Herzog P, Sowka S, Kubicek CP. Glucose-induced secretion of Trichoderma reesei xylanases. Appl Environ Microbiol. 1996;62:2859-2865
39. Sivan A, Elad Y, Chet I. Biological control effects of a new isolate of Trichoderma harzianum on Pythium aphanidermatum. Phytopathology. 1984;74:498-501
40. Khan MH, Ali S, Fakhru'l-Razi A, Alam Z. Use of fungi for the bioconversion of rice straw into cellulase enzyme. J Environ Sci Health B. 2007;42:381-386
41. Park YS, Kang SW, Lee JS, Hong SI, Kim SW. Xylanase production in solid state fermentation by Aspergillus niger mutant using statistical experimental designs. Appl Microbiol Biotechnol. 2002;58:761-766
42. Hao J, Song F, Huang F, Yang C, Zhang Z, Zheng Y, Tian X. Production of laccase by a newly isolated deuteromycete fungus Pestalotiopsis sp. and its decolorization of azo dye. J Ind Microbiol Biotechnol. 2007;34:233-240
43. Hao JJ, Tian XJ, Song FQ, He XB, Zhang ZJ, Zhang P. Involvement of lignocellulolytic enzymes in the decomposition of leaf litter in a subtropical forest. J Eukaryot Microbiol. 2006;53:193-198
44. Baldrian P, Valaskova V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev. 2008;32:501-521
45. Abbas A, Koc H, Liu F, Tien M. Fungal degradation of wood: initial proteomic analysis of extracellular proteins of Phanerochaete chrysosporium grown on oak substrate. Curr Genet. 2005;47:49-56
46. Song BC, Kim KY, Yoon JJ, Sim SH, Lee K, Kim YS, Kim YK, Cha CJ. Functional analysis of a gene encoding endoglucanase that belongs to glycosyl hydrolase family 12 from the brown-rot basidiomycete Fomitopsis palustris. J Microbiol Biotechnol. 2008;18:404-409
47. Lee SS, Ha JK, Cheng KJ. The effects of sequential inoculation of mixed rumen protozoa on the degradation of orchard grass cell walls by anaerobic fungus Anaeromyces mucronatus 543. Can J Microbiol. 2001;47:754-760
48. Doi RH. Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Ann N Y Acad Sci. 2008;1125:267-279
49. Roger V, Grenet E, Jamot J, Bernalier A, Fonty G, Gouet P. Degradation of maize stem by two rumen fungal species, Piromyces communis and Caecomyces communis, in pure cultures or in association with cellulolytic bacteria. Reprod Nutr Dev. 1992;32:321-329
50. Gerbi C, Bata J, Breton A, Prensier G. Glycoside and polysaccharide hydrolase activity of the rumen anaerobic fungus Caecomyces communis (Sphaeromonas communis SENSU ORPIN) at early and final stages of the developmental cycle. Curr Microbiol. 1996;32:256-259
51. Ozkose E, Thomas BJ, Davies DR, Griffith GW, Theodorou MK. Cyllamyces aberensis gen.nov. sp.nov, a new anaerobic gut fungus with branched sporangiophores isolated from cattle. Can. J. Bot. 2001;79:666-673
52. Li XL, Calza RE. Fractionation of cellulases from the ruminal fungus Neocallimastix frontalis EB188. Appl Environ Microbiol. 1991;57:3331-3336
53. Griffith GW, Ozkose E, Theodoroua MK, Davies DR. Diversity of anaerobic fungal populations in cattle revealed by selective enrichment culture using different carbon sources. Fungal Ecology. 2009;2:87-97
54. Chen H, Li XL, Blum DL, Ljungdahl LG. Two genes of the anaerobic fungus Orpinomyces sp. strain PC-2 encoding cellulases with endoglucanase activities may have arisen by gene duplication. FEMS Microbiol Lett. 1998;159:63-68
55. Ali BR, Zhou L, Graves FM, Freedman RB, Black GW, Gilbert HJ, Hazelwood GP. Cellulases and hemicellulases of the anaerobic fungus Piromyces constitute a multiprotein cellulose-binding complex and are encoded by multigene families. FEMS Microbiol Lett. 1995;125:15-21
56. Kumar R, Singh S, Singh OV. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J Ind Microbiol Biotechnol. 2008;35:377-391
57. Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, Goedegebuur F, Houfek TD, England GJ, Kelley AS, Meerman HJ, Mitchell T, Mitchinson C, Olivares HA, Teunissen PJ, Yao J, Ward M. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem. 2003;278:31988-31997
58. Divne C, Stahlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JK, Teeri TT, Jones TA. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994;265:524-528
59. Rouvinen J, Bergfors T, Teeri T, Knowles JK, Jones TA. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990;249:380-386
60. Divne C, Stahlberg J, Teeri TT, Jones TA. High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei. J Mol Biol. 1998;275:309-325
61. Morais H, Ramos C, Matos N, Forgacs E, Cserhati T, Almeida V, Oliveira J, Darwish Y, Iles Z. Liquid chromatographic and electrophoretic characterisation of extracellular beta-glucosidase of Pleurotus ostreatus grown in organic waste. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;770:111-119
62. Ishikawa E, Sakai T, Ikemura H, Matsumoto K, Abe H. Identification, cloning, and characterization of a Sporobolomyces singularis beta-galactosidase-like enzyme involved in galacto-oligosaccharide production. J Biosci Bioeng. 2005;99:331-339
63. Cao WG, Crawford DL. Purification and some properties of beta-glucosidase from the ectomycorrhizal fungus Pisolithus tinctorius Strain Smf. Can J Microbiol. 1993;39:125-129
64. Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280( Pt 2):309-316
65. Dan S, Marton I, Dekel M, Bravdo BA, He S, Withers SG, Shoseyov O. Cloning, expression, characterization, and nucleophile identification of family 3, Aspergillus niger beta-glucosidase. J Biol Chem. 2000;275:4973-4980
66. Evans CS. Properties of the beta-D-glucosidase (cellobiase) from the wood-rotting fungus, Coriolus versicolor. Appl Microbiol Biotechnol. 1985;22:128-131
67. Cai YJ, Chapman SJ, Buswell JA, Chang ST. Production and distribution of endoglucanase, cellobiohydrolase, and beta-glucosidase components of the cellulolytic system of Volvariella volvacea, the edible straw mushroom. Appl Environ Microbiol. 1999;65:553-559
68. Kubicek CP. Release of carboxymethyl-cellulase and β-glucosidase from cell walls of Trichoderma reesei. Eur J Appl Microbiol Biotechnol. 1981;13:226-231
69. Murray P, Aro N, Collins C, Grassick A, Penttila M, Saloheimo M, Tuohy M. Expression in Trichoderma reesei and characterisation of a thermostable family 3 beta-glucosidase from the moderately thermophilic fungus Talaromyces emersonii. Protein Expr Purif. 2004;38:248-257
70. Rahman Z, Shida Y, Furukawa T, Suzuki Y, Okada H, Ogasawara W, Morikawa Y. Application of Trichoderma reesei cellulase and xylanase promoters through homologous recombination for enhanced production of extracellular beta-glucosidase I. Biosci Biotechnol Biochem. 2009;73:1083-1089
71. Shallom D, Shoham Y. Microbial hemicellulases. Curr Opin Microbiol. 2003;6:219-228
72. Bourne Y, Henrissat B. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Opin Struct Biol. 2001;11:593-600
73. Wang L, Yan W, Chen J, Huang F, Gao P. Function of the iron-binding chelator produced by Coriolus versicolor in lignin biodegradation. Sci China C Life Sci. 2008;51:214-221
74. Martinez AT, Speranza M, Ruiz-Duenas FJ, Ferreira P, Camarero S, Guillen F, Martinez MJ, Gutierrez A, del Rio JC. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol. 2005;8:195-204
75. Moredo N, Lorenzo M, Domínguez A, Moldes D, Cameselle C, Sanroman A. Enhanced ligninolytic enzyme production and degrading capability of Phanerochaete chrysosporium and Trametes versicolor. World J Microb Biotechnol. 2003;19:665-669
76. Srebotnik E, Messner K, Foisner R. Penetrability of White Rot-Degraded Pine Wood by the Lignin Peroxidase of Phanerochaete chrysosporium. Appl Environ Microbiol. 1988;54:2608-2614
77. Tanaka H, Itakura S, Enoki A. Hydroxyl radical generation by an extracellular low-molecular-weight substance and phenol oxidase activity during wood degradation by the white-rot basidiomycete Phanerochaete chrysosporium. Holzforschung. 1999;53:21-28
78. Suzuki MR, Hunt CG, Houtman CJ, Dalebroux ZD, Hammel KE. Fungal hydroquinones contribute to brown rot of wood. Environ Microbiol. 2006;8:2214-2223
79. Guillen F, Martinez AT, Martinez MJ. Substrate specificity and properties of the aryl-alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem. 1992;209:603-611
80. Call HP, Mücke I. History, overview and applications of mediated lignolytic systems, especially laccase-mediator-systems (Lignozyme®-process). J Biotechnol. 1997;53:163-202
81. Wang W, Huang F, Mei Lu X, Ji Gao P. Lignin degradation by a novel peptide, Gt factor, from brown rot fungus Gloeophyllum trabeum. Biotechnol J. 2006;1:447-453
82. Renganathan V, Usha SN, Lindenburg F. Cellobiose oxidizing enzymes from the lignocellulose-degrading basidiomycete Phanerochaete chrysosporium - interaction with microcrystalline cellulose. Appl Microbiol Biotechnol. 1990;32:609-613
83. Henriksson G, Johansson G, Pettersson G. A critical review of cellobiose dehydrogenases. J Biotechnol. 2000;78:93-113
84. Baminger U, Subramaniam SS, Renganathan V, Haltrich D. Purification and characterization of cellobiose dehydrogenase from the plant pathogen Sclerotium (Athelia) rolfsii. Appl Environ Microbiol. 2001;67:1766-1774
85. Yang W, Liu J, Wang W, Zhang Y, Gao P. Function of a low molecular peptide generated by cellulolytic fungi for the degradation of native cellulose. Biotechnol Lett. 2004;26:1799-1802
86. Enoki A, Hirano T, Tanaka H. Extracellular substance from the brown rot basidiomycete Gloeophyllum trabeum that produces and reduces hydrogen peroxide. Mater Organism. 1992;27:247-261
87. Tanaka H, Yoshida G, Baba Y, Matsumura K, Wasada H, Murata J, Agawa M, Itakura S, Enoki A. Characterization of a hydroxyl-radical-producing glycoprotein and its presumptive genes from the white-rot basidiomycete Phanerochaete chrysosporium. J Biotechnol. 2007;128:500-511
88. Kurek B, Gaudard F. Oxidation of spruce wood sawdust by MnO(2) plus oxalate: a biochemical investigation. J Agric Food Chem. 2000;48:3058-3062
89. Makela MR, Hilden K, Hatakka A, Lundell TK. Oxalate decarboxylase of the white rot fungus Dichomitus squalens demonstrates a novel enzyme primary structure and non-induced expression on wood and in liquid cultures. Microbiology. 2009
90. Brock BJ, Rieble S, Gold MH. Purification and Characterization of a 1,4-Benzoquinone Reductase from the Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1995;61:3076-3081
91. Enoki A, Tanaka H, Itakura S. Physical and chemical characteristics of glycopeptide from wood decay fungi. Wood Deterioration and Preservation (Goodell B, Nicholas DD & Schultz TP, eds). Washington, DC: Oxford University Press. 2003:140-153
92. Raman B, Pan C, Hurst GB, Rodriguez M Jr, McKeown CK, Lankford PK, Samatova NF, Mielenz JR. Impact of pretreated Switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS ONE. 2009;4:e5271
93. Orpin CG. Studies on the rumen flagellate Neocallimastix frontalis. J Gen Microbiol. 1975;91:249-262
94. Lamed R, Setter E, Bayer EA. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983;156:828-836
95. Wilson CA, Wood TM. The anaerobic fungus Neocallimastix frontalis: isolation and properties of a cellulosome-type enzyme fraction with the capacity to solubilize hydrogen-bond-ordered cellulose. Appl. Microbiol. Biotechnol. 1992;37:125-129
96. Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521-554
97. Nagy T, Tunnicliffe RB, Higgins LD, Walters C, Gilbert HJ, Williamson MP. Characterization of a double dockerin from the cellulosome of the anaerobic fungus Piromyces equi. J Mol Biol. 2007;373:612-622
98. Dijkerman R, Op den Camp HJ, Van der Drift C, Vogels GD. The role of the cellulolytic high molecular mass (HMM) complex of the anaerobic fungus Piromyces sp. strain E2 in the hydrolysis of microcrystalline cellulose. Arch Microbiol. 1997;167:137-142
99. Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol. 2005;96:673-686
100. Taherzadeh MJ, Karimi K. Pretreatment of lignocellulosic wastes to improve ethanol and biogas production: a review. Int J Mol Sci. 2008;9:1621-1651
101. Shi J, Chinn MS, Sharma-Shivappa RR. Microbial pretreatment of cotton stalks by solid state cultivation of Phanerochaete chrysosporium. Bioresour Technol. 2008;99:6556-6564
102. Anderson WF, Akin DE. Structural and chemical properties of grass lignocelluloses related to conversion for biofuels. J Ind Microbiol Biotechnol. 2008;35:355-366
103. Arora DS, Chander MKGP. Involvement of lignin peroxidase, manganese peroxidase and laccase in degradation and selective ligninolysis of wheat straw. Int. Bioterior. Biodegrad. 2002;50:115-120
104. A. Ferraz, Ana M. Córdova, A. Machuca: Wood biodegradation and enzyme production by Ceriporiopsis subvermispora during solid-state fermentation of Eucalyptus grandis. Enzyme and Microbial Technology. 2003;32:59-65
105. Turner P, Mamo G, Karlsson EN. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb Cell Fact. 2007;6:9
106. Viikari L, Alapuranen M, Puranen T, Vehmaanpera J, Siika-Aho M. Thermostable enzymes in lignocellulose hydrolysis. Adv Biochem Eng Biotechnol. 2007;108:121-145
107. Grassick A, Murray PG, Thompson R, Collins CM, Byrnes L, Birrane G, Higgins TM, Tuohy MG. Three-dimensional structure of a thermostable native cellobiohydrolase, CBH IB, and molecular characterization of the cel7 gene from the filamentous fungus, Talaromyces emersonii. Eur J Biochem. 2004;271:4495-4506
108. Li YL, Li DC, Teng FC. [Purification and characterization of a cellobiohydrolase from the thermophilic fungus Chaetomium thermophilus CT2]. Wei Sheng Wu Xue Bao. 2006;46:143-146
109. Rosgaard L, Pedersen S, Cherry JR, Harris P, Meyer AS. Efficiency of new fungal cellulase systems in boosting enzymatic degradation of barley straw lignocellulose. Biotechnol Prog. 2006;22:493-498
110. Maheshwari R, Bharadwaj G, Bhat MK. Thermophilic fungi: their physiology and enzymes. Microbiol Mol Biol Rev. 2000;64:461-488
111. Chu BC, Lee H. Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol Adv. 2007;25:425-441
112. Schneider H, Wang PY, Chan YK, Maleszka R. Conversion of D-xylose into ethanol by the yeast Pachysolen tannophilus. Biotechnol Lett. 1981;3:89-92
113. McMillan JD, Boynton BL. Arbinose utilization by xylose-fermenting yeasts and fungi. Appl Biochem Biotechnol. 1994;45:569-584
114. Karhumaa K, Wiedemann B, Hahn-Hagerdal B, Boles E, Gorwa-Grauslund MF. Co-utilization of L-arabinose and D-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microb Cell Fact. 2006;5:18
115. Larsson S, Quintana-Sainz A, Reimann A, Nilvebrant NO, Jonsson LJ. Influence of lignocellulose-derived aromatic compounds on oxygen-limited growth and ethanolic fermentation by Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2000;84:617-632
116. Nilvebrant NO, Persson P, Reimann A, De Sousa F, Gorton L, Jonsson LJ. Limits for alkaline detoxification of dilute-acid lignocellulose hydrolysates. Appl Biochem Biotechnol. 2003;105:615-628
117. Ohgren K, Bengtsson O, Gorwa-Grauslund MF, Galbe M, Hahn-Hagerdal B, Zacchi G. Simultaneous saccharification and co-fermentation of glucose and xylose in steam-pretreated corn stover at high fiber content with Saccharomyces cerevisiae TMB3400. J Biotechnol. 2006;126:488-498
118. Martin C, Marcet M, Almazan O, Jonsson LJ. Adaptation of a recombinant xylose-utilizing Saccharomyces cerevisiae strain to a sugarcane bagasse hydrolysate with high content of fermentation inhibitors. Bioresour Technol. 2007;98:1767-1773
119. Georgieva TI, Hou X, Hilstrom T, Ahring BK. Enzymatic hydrolysis and ethanol fermentation of high dry matter wet-exploded wheat straw at low enzyme loading. Appl Biochem Biotechnol. 2008;148:35-44
120. Philippidis GP, Smith TK, Wyman CE. Study of the enzymatic hydrolysis of cellulose for production of fuel ethanol by the simultaneous saccharification and fermentation process. Biotechnol Bioeng. 1993;41:846-853
121. Stenberg K, Bollok M, Reczey K, Galbe M, Zacchi G. Effect of substrate and cellulase concentration on simultaneous saccharification and fermentation of steam-pretreated softwood for ethanol production. Biotechnol Bioeng. 2000;68:204-210
122. Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577-583
123. Mandels M, Weber J. The production of cellulases. In Cellulases and their applications. Advan. Chem. Ser. 1969;95:391-414
124. Mandels M, Weber J, Parizek R. Enhanced cellulase production by a mutant of Trichoderma viride. Appl Microbiol. 1971;21:152-154
125. Montenecourt BS, Eveleigh DE. Preparation of mutants of Trichoderma reesei with enhanced cellulase production. Appl Environ Microbiol. 1977;34:777-782
126. Montenecourt BS, Eveleigh DE. Selective screening methods for the isolation of high yielding cellulase mutants of Trichoderma reesei. Adv. Chem. Ser. 1979;181:289-301
127. Kov́acs K, Megyeri L, Szakacsa G, Kubicekc CP, Galbeb M, Zacchi G. Trichoderma atroviride mutants with enhanced production of cellulase and β-glucosidase on pretreated willow. Enzyme and Microbial Technology. 2008;43:48-55
128. Solov'eva IV, Okunev ON, Vel'kov VV, Koshelev AV, Bubnova TV, Kondrat'eva EG, Skomarovskii AA, Sinitsyn AP. The selection and properties of Penicillium verruculosum mutants with enhanced production of cellulases and xylanases. Mikrobiologiia. 2005;74:172-178
129. Seidl V, Gamauf C, Druzhinina IS, Seiboth B, Hartl L, Kubicek CP. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics. 2008;9:327
130. Okada H, Mori K, Tada K, Nogawa M, Morikawa Y. Identification of active site carboxylic residues in Trichoderma reesei endoglucanase Cel12A by site-directed mutagenesis. J Mol Catal. 2000:249-255
131. Fenel F, Zitting AJ, Kantelinen A. Increased alkali stability in Trichoderma reesei endo-1, 4-beta-xylanase II by site directed mutagenesis. J Biotechnol. 2006;121:102-107
132. Qin Y, Wei X, Song X, Qu Y. Engineering endoglucanase II from Trichoderma reesei to improve the catalytic efficiency at a higher pH optimum. Journal of Biotechnology. 2008;135:190-195
133. Fenel F, Leisola M, Janis J, Turunen O. A de novo designed N-terminal disulphide bridge stabilizes the Trichoderma reesei endo-1,4-beta-xylanase II. J Biotechnol. 2004;108:137-143
134. Stockton BC, Mitchell DJ, Grohmann K, Himmel ME. Optimum β-D-glucosidase supplementation of cellulase for efficient conversion of cellulose to glucose. Biotechnol Lett. 1991;13:57-62
135. Johnson KG. Exocellular β-mannanases from hemicellulolytic fungi. World Journal of Microbiology and Biotechnology. 1990;6:209-217
136. Gutierrez-Correa M, Tengerdy RP. Cellulolytic enzyme production by fungal mixed culture solid substrate fermentation. Biotechnol. Lett. 1998;20:45-47
137. Maheshwari DK, Gohade S, Paul J, Verma A. A paper mill sludge as a potential source for cellulase production by Trichoderma reesei QM9123 and Aspergillus niger using mixed cultivation. Carbohydr Polym. 1994;23:161-163
138. Ahamed A, Vermette P. Enhanced enzyme production from mixed cultures of Trichoderma reesei RUT-C30 and Aspergillus niger LMA grown as fed batch in a stirred tank bioreactor. Biochemical Engineering Journal. 2008;42:41-46
139. Juh́asz T, Kozma K, Szengyel Z, Ŕeczey K. Production of β-glucosidase in mixed culture of Aspergillus niger BKMF 1305 and Trichoderma reesei RUT C30, Food. Technol. Biotechnol. 2003;41:49-53
140. Duff SJB. Cellulase and beta-glucosidase production by mixed culture of Trichoderma reesei Rut C30 and Aspergillus phoenicis. Biotechnol. Lett. 1985;7:185-190
141. Gupte A, Madamwar D. Solid state fermentation of lignocellulosic waste for cellulose and β-glucosidase production by co-cultivation by Aspergillus ellipticus and Aspergillus fumigatus. Biotechnol Prog. 1997;13:166-169
142. Panda T, Bisaria VS, Ghose TK. Studies on mixed fungal culture for cellulase and hemicellulase production. Part 1. Optimization of medium for the mixed culture of Trichoderma reesei Dl-6 and Aspergillus wentii Pt 2804. Biotechnology Letters. 1983;5:767-772
143. Dueñas R, Tengerdy RP, Gutierrez-Correa M. Cellulase production by mixed fungi in solid-substrate fermentation of bagasse. World Journal of Microbiology & Botechnology. 1995;11:333-337
144. Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506-577
145. Miettinen-Oinonen A, Paloheimo M, Lantto R, Suominen P. Enhanced production of cellobiohydrolases in Trichoderma reesei and evaluation of the new preparations in biofinishing of cotton. J Biotechnol. 2005;116:305-317
146. Bower B, Larenas E, Mitchinson C. Exo-endo cellulase fusion protein. Patent WO2005093073. 2005
Author contact
![]() Correspondence to: Wensheng Qin, Email: wqinca; Phone: (807) 343-8840; Fax: (807) 346-7796
Correspondence to: Wensheng Qin, Email: wqinca; Phone: (807) 343-8840; Fax: (807) 346-7796

 Global reach, higher impact
Global reach, higher impact