10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(7):745-757. doi:10.7150/ijbs.5.745 This issue Cite
Research Paper
A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus
University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany
Received 2009-10-15; Accepted 2009-11-27; Published 2009-12-3
Abstract
Male moths respond to conspecific female-released pheromones with remarkable sensitivity and specificity, due to highly specialized chemosensory neurons in their antennae. In Antheraea silkmoths, three types of sensory neurons have been described, each responsive to one of three pheromone components. Since also three different pheromone binding proteins (PBPs) have been identified, the antenna of Antheraea seems to provide a unique model system for detailed analyzes of the interplay between the various elements underlying pheromone reception. Efforts to identify pheromone receptors of Antheraea polyphemus have led to the identification of a candidate pheromone receptor (ApolOR1). This receptor was found predominantly expressed in male antennae, specifically in neurons located beneath pheromone-sensitive sensilla trichodea. The ApolOR1-expressing cells were found to be surrounded by supporting cells co-expressing all three ApolPBPs. The response spectrum of ApolOR1 was assessed by means of calcium imaging using HEK293-cells stably expressing the receptor. It was found that at nanomolar concentrations ApolOR1-cells responded to all three pheromones when the compounds were solubilized by DMSO and also when DMSO was substituted by one of the three PBPs. However, at picomolar concentrations, cells responded only in the presence of the subtype ApolPBP2 and the pheromone (E,Z)-6,11-hexadecadienal. These results are indicative of a specific interplay of a distinct pheromone component with an appropriate binding protein and its related receptor subtype, which may be considered as basis for the remarkable sensitivity and specificity of the pheromone detection system.
Keywords: Insect, olfaction, pheromone detection, receptor, pheromone binding protein
Introduction
Volatile chemical signals from the environment which indicate host plants, oviposition places, predators or mating partners are essential for many aspects of insect life. Hence, many insects have developed intricate olfactory systems for detection and discrimination of relevant compounds even at extremely low concentrations [1-3]. In moths, the finding of mating partners is highly dependent on the sensitive registration of female-released pheromones by specialized detection units (sensilla trichodea) on the male antenna [4, 5]. These hair-like structures house 1-3 pheromone-responsive neurons, which extend their sensory dendrites into the fluid-filled sensillum shaft [6, 7]. In Antheraea moth species the male-specific trichoid sensilla are particularly large and thus easily accessible for experimental approaches. This, together with early knowledge on the composition of the female-released sex pheromone blend [8, 9], has made Antheraea an attractive model in olfactory research for almost five decades and has motivated substantial electrophysiological, biochemical and molecular biological studies [10-18]. Electrophysiological recordings from sensilla trichodea of Antheraea polyphemus have classified three sensory neuron types, each tuned to the detection of one of the Antheraea sex-pheromone components (E,Z)-6,11-hexadecadienyl acetate (AC1), (E,Z)-6,11-hexadecadienal (AL) and (E,Z)-4,9-tetradecadienyl acetate (AC2) [6]. Interestingly, in accordance with the three pheromone-responsive neuron types, also three pheromone binding protein subtypes (ApolPBP1, ApolPBP2, ApolPBP3) of A. polyphemus have been identified [13, 19]. Subsequent binding studies and structural analyzes have shown that the three PBP subtypes differentially interact with the three pheromonal compounds of Antheraea [11, 20, 21], suggesting that a distinct PBP type may contribute to the detection of a certain pheromone component. This notion was supported by comparative studies on the sibling species Antheraea pernyi, where the male antenna also exhibits AC1-, AC2- and AL-specific neurons [6] and three PBP types [19], each with binding preference for one of the three pheromone components [11]. Recent studies in other moths [22, 23] and in the fruitfly Drosophila melanogaster [24] further substantiate the conception of specific roles of different PBPs in pheromone detection. Functional studies have shown that both, a distinct binding protein and a distinct receptor, contribute to the selective and sensitive response to a distinct pheromone component [22, 23, 25]. The existence of three neuron types on the antenna of Antheraea implies that each of these neurons may express a distinct receptor type specifically tuned to one pheromone component. Therefore, in this study attempts were made to identify candidate pheromone receptors of Antheraea, to assess their functional properties and their possible interplay with pheromone binding proteins.
Materials and Methods
Animals and tissue preparation
Antheraea polyphemus cocoons were obtained from Bill Oehlke (Montague, Prince Edward Island, Canada). Animals were allowed to develop to adults at 25°C. After hatching, males and females were separated. Antennae were dissected from cold-anaesthetized animals. Antennae for RNA isolation were immediately frozen using liquid nitrogen and stored at -70°C.
Pheromone components
(E,Z)-6,11-hexadecadienyl acetate (AC1), (E,Z)-6,11-hexadecadienal (AL) and (E,Z)-4,9-tetradecadienyl acetate (AC2) were synthesized by Chemtech B.V. (Amsterdam, The Netherlands).
Identification of receptor sequences
To identify genes encoding putative pheromone receptors in Antheraea pernyi and Antheraea polyphemus probes based on verified and candidate pheromone receptors of Heliothis virescens [26] and Bombyx mori [27, 28] were employed to screen cDNA libraries made from antennae of male A. polyphemus or A. pernyi. Digoxigenin (DIG-)-labeled probes for library screening were obtained by standard PCR using specific sense and antisense primers amplifying receptor coding regions, the PCR DIG labeling mix (Roche, Mannheim, Germany) and plasmids carrying the corresponding cDNAs. PCR conditions were 1 min 40 s at 94°C, then 21 cycles with 94°C for 30 s, 55°C for 40 s and 72°C for 1 min, with a decrease of the annealing temperature by 0.5°C per cycle. Subsequently, 19 further cycles at the condition of the last cycling step were performed, followed by incubation for 7 min at 72°C. PCR products were gel purified using the Geneclean II Kit (Q-BIOgene, Irvine, CA) and diluted in hybridization solution (30% formamide, 5x SSC, 0.1% lauroylsarcosine, 0.02% SDS, 2% blocking reagent [Roche], 100 µg/ml denatured herring sperm DNA).
For cDNA library screening, phage DNA was transferred to and immobilized on Hybond-N+ nylon transfer membranes (Amersham Biosciences, Freiburg, Germany). Hybridization to DIG-labeled probes was performed at 30°C as described [29]. After hybridization the membranes were washed twice for 5 min in 2x SSC, 0.1% SDS at room temperature, followed by three washes in 2x SSC, 0.1% SDS for 20 min each at 30°C. Hybridized probes were detected using anti-DIG AP-conjugated antibodies (Roche) and CSPD (Applied Biosystems, Foster City, CA) as substrate. cDNA inserts from positive phage were isolated, subcloned into the pBluescript II SK+ vector and sequenced.
This approach led to the identification of a cDNA clone in A. pernyi carrying a long open reading frame displaying high sequence identity to the candidate pheromone receptor sequences used for screening. Using this clone (AperOR1) as probe to screen the A. polyphemus cDNA library, a highly related cDNA could be identified in A. polyphemus (ApolOR1). As both identified clones were truncated at the 5' end, RACE-PCR was performed using the GeneRacer Kit (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. Analysis of the RACE products allowed the completion of the ApolOR1 and AperOR1 coding regions.
Sequencing and sequence analysis
Sequencing was performed on an ABI310 sequencing system using vector and cDNA derived primers and the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems).
Sequence analyzes were made using HUSAR (Heidelberg unix sequence analysis resources; http://genius.embnet.dkfz-heidelberg.de/). The unrooted neighbor joining tree was calculated with MEGA 4.0 [30] based on a ClustAL alignment [31] including protein sequences indicated in the figure legend.
Reverse transcription (RT-)PCR
Total RNAs from antennae of male and female A. polyphemus were isolated using TRIzol reagent (Invitrogen). Poly (A)+ RNA was isolated from total RNA with oligo (dT)25 magnetic dynabeads (Dynal, Oslo, Norway), transcribed into cDNA as previously described [29] and used in RT-PCR experiments. For specific amplification of ApolOR1 the primer pair 5'-GATTACGCTATGAAGACACA-3' and 5'-CCTTTACTCTCTTCCACCGA-3' was used. To test the integrity of the prepared cDNA, degenerate primers (5'-AAYTGGGAYGAYATGGARAA-3', 5'-GCCATYTCYTGYTCRAARTC-3') directed against conserved regions of insect actins were used [32]. PCR conditions were as described above for preparation of DIG-labeled screening probes. PCR products were analyzed on agarose gels and visualized by ethidium bromide staining. Based on the primer design the expected sizes for RT-PCR products were 412 bp for ApolOR1 and 450 bp for actin.
In situ hybridization
Antennae of 1-2 days old male or female A. polyphemus moths were embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek Europe, Zoeterwoude, The Netherlands) and frozen at -22ºC. Cryosections (12 µm) of antennae were thaw mounted on SuperFrost Plus slides (Menzel-Gläser, Braunschweig, Germany) and air-dried at room temperature for at least 30 min. In situ hybridization with DIG- and/or biotin-labeled antisense RNA probes and visualization of hybridization was performed as reported previously [29]. Briefly, hybridized DIG-labeled probes were detected by an anti-DIG AP-conjugated antibody in combination with HNPP/Fast Red (HNPP Fluorescent Detection Set, Roche). Biotin-labeled probes were visualized using the Tyramide Signal Amplification System (Perkin Elmer, Boston, MA), including an anti-biotin streptavidin HRP-conjugate and FITC-tyramides as substrate. Sections were mounted with PBS/glycerol (1:3). Biotin- or DIG-labeled antisense RNA probes for ApolOR1, ApolPBP1 [13], ApolPBP2 and ApolPBP3 [19] were generated using a T3/T7 RNA transcription system (Roche) following recommended protocols. Briefly, linearized recombinant pBluescript II SK+ plasmids were mixed with transcription buffer, DIG- or biotin-labeling mix and T3 or T7 RNA polymerase and incubated for 3 h at 37°C. Plasmids contained the cDNAs for ApolOR1 (Acc. No. FN556591), ApolPBP1 (Acc. No. X17559), ApolPBP2 (Acc. No. AJ277266) or ApolPBP3 (Acc. No. AJ277267). After DNAse treatment for 30 min at 37°C, RNA was ethanol precipitated and dissolved in water. ApolOR1-antisense RNA was shortened by alkaline hydrolysis to an average length of ~800 bp according to [33]. Shortened ApolOR1-antisense RNA and non-treated PBP-antisense RNAs were finally diluted in hybridization buffer.
Hybridized sections were analyzed on a Zeiss LSM510 Meta laser scanning microscope (Zeiss, Oberkochen, Germany). Figures were arranged in Powerpoint (Microsoft) and further processed using Photoshop (Adobe). Images were not altered except for adjustment of brightness or contrast for uniform tone within a single figure.
Expression of ApolOR1 in Flp-In T-REx293 cells
For functional analysis of ApolOR1 a stable ApolOR1-expressing cell line was generated using the Flp-In-System (Invitrogen, Karlsruhe, Germany) in combination with a modified HEK293 cell line stably carrying a mouse Gα15 gene (Flp-In T-REx293/Gα15; kindly provided by E. Tareilus and R. Gouka, Unilever R+D, Vlardingen, The Netherlands).
For generation of the stable ApolOR1 cell line the coding region of ApolOR1 was first PCR-amplified using a specific sense primer adding a NotI restriction site and an antisense primer including a XhoI site. The resulting product was gel purified, digested with the appropriate restriction enzymes and ligated into the NotI/XhoI site of a modified pcDNA5-FRT/TO vector (Invitrogen) carrying a FLAG-sequence [22], thus bringing ApolOR1 into frame with the FLAG-tag. Correct amplification and integration into the vector was verified by sequencing. To generate the stable cell line, Flp-In T-REx293/Gα15 cells were transfected with the ApolOR1/pcDNA5-FRT/TO construct together with the plasmid pOG44 (Invitrogen) in a 1:10 ratio using Lipofectamine 2000 (Invitrogen) according to the supplier's protocol. 48 h after transfection, cells were selected for receptor integration into the genome using media supplemented with 100 mg/L hygromycin instead of zeocin. Integration of the ApolOR1 sequence into the genome was further verified by PCR using receptor specific primers and genomic DNA isolated from hygromycin-resistant cells. Flp-In T-REx293/Gα15 cells positive for ApolOR1 were maintained and used for calcium imaging experiments.
Calcium imaging of ApolOR1-expressing cells
For calcium imaging ApolOR1/Flp-In T-REx293/Gα15 cells (1 x 105) were seeded onto cover slips (Ø 15 mm) coated with poly-L-lysine and incubated for 24 h in DMEM with 5 µg/ml tetracycline to induce receptor expression. After a wash in Ringer solution (138 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1.5 mM MgCl2, 10 mM glucose, 10 mM Hepes, pH 7.3) cells were loaded with Fura-2 AM (Invitrogen) by incubating them for 45 min at 37°C with 4 μmol/L of the Ca2+-sensitive dye in Ringer solution. Details of the equipment used for monitoring calcium signals in single cells have been described previously [22, 23]. Briefly, cover slips with Fura-2 loaded cells were placed into a flow-chamber on the stage of an inverted microscope equipped for epifluorescence and rinsed with Ringer solution (warmed to 37°C) at a flow rate of 1 ml/10 sec. Test solutions (400 µl) were applied at the same flow rate via syringes connected to the flow system by three-way valves. To monitor changes in [Ca2+]i concentration in single cells, the intensity of fluorescent light emission at 510 nm was measured over time, using excitation wavelengths of 340 nm and 380 nm. The Metafluor imaging system and Metafluor 4 software (Visitron Systems, Puchheim, Germany) was used for data acquisition and analysis of images. The ratio of fluorescence emission at 340 nm/380 nm excitation was used as index with an increase indicating a rise in [Ca2+]i. Fluorescence intensity ratios for at least 30 individual ApolOR1-expressing cells per measurement were determined before (F0) and after stimulation (F; peak of the response). F/F0 values of individual cells were determined and averaged in a single experiment. Following the application of test substances, the cell viability was tested by application of 10 mM ATP in Ringer solution. Between stimulations, cells were washed with Ringer solution for at least 5 min. In experiments without PBPs, hydrophobic pheromone components were diluted from stock solutions in hexane using Ringer solution with 0.1% DMSO. All dilutions were prepared freshly and used within 3 hours. To analyze the ability of the different PBPs to solubilize and transport specific pheromone components, substances from stock solutions in hexane were added to recombinant protein in Ringer solution and incubated for 30 minutes on ice. Prior to calcium imaging experiments the solution was warmed to 37°C.
Expression and purification of pheromone binding proteins
Bacterial expression of Antheraea polyphemus PBP1, PBP2 and PBP3 and purification of the PBPs from a periplasmic preparation of E. coli BL21 (DE3) were performed as reported in previous studies [11, 20, 34, 35]. To remove possible hydrophobic ligands which have been found to co-purify with PBPs expressed in E. coli, binding proteins were delipidated according to the protocol described in [36]. The delipidated PBPs were finally dissolved in Ringer solution. The protein concentration was determined spectrometrically at 280 nm applying absorption coefficients determined for the proteins by the ProtParam program (ExPASy molecular biology server; Swiss Institute of Bioinformatics).
Results
Identification of candidate pheromone receptor sequences
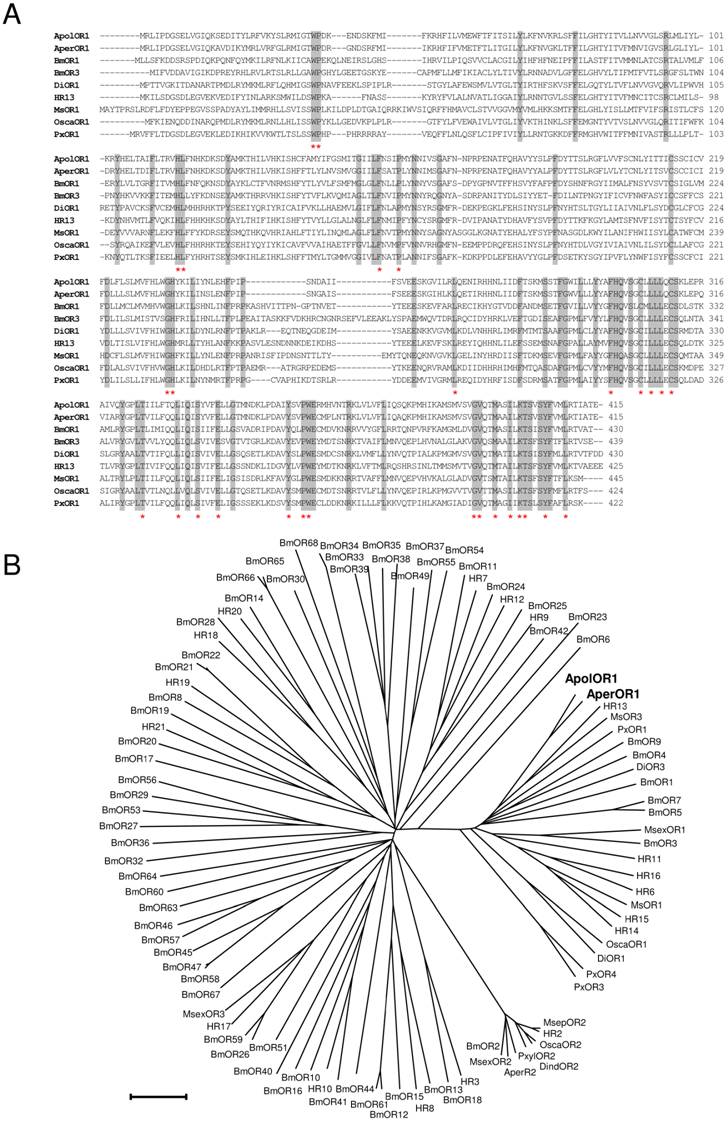
In this study, attempts were made to identify and characterize pheromone receptors in the two sibling species Antheraea pernyi and Antheraea polyphemus. We applied a homology based screening approach using probes based on sequences encoding putative and verified pheromone receptors of the moth species Heliothis virescens and Bombyx mori [26, 28, 37]. This led to the identification of a relevant cDNA from Antheraea pernyi (AperOR1) which was subsequently employed as screening probe to isolate a highly related cDNA from Antheraea polyphemus (ApolOR1). As both cDNAs were truncated at the 5' end, full-length sequences were completed by RACE experiments. The amino acid sequences of ApolOR1 and AperOR1 (Fig. 1A) share a high number of amino acid residues at identical positions with an overall sequence identity of 83%. A comparison of the Antheraea sequences with previously described candidate and verified pheromone receptors of several other moth species revealed highly conserved amino acids present in all sequences with increasing numbers towards the C-terminal end (Fig. 1A, asterisks). In a neighbor joining tree including olfactory receptors from several moth species (Fig. 1B), the novel receptors ApolOR1 and AperOR1 are categorized in the subfamily of candidate and functionally verified pheromone receptors [22, 23, 27, 28, 37-40]. Both ApolOR1 and AperOR1 share a high sequence identity (50%) with HR13, the receptor for the main sex-pheromone component in H. virescens [23] and with the bombykol receptor BmOR1 of Bombyx mori [22, 27] (47% and 49%, respectively). Slightly higher identities of 51% (ApolOR1) and 53% (AperOR1) are observed with the candidate pheromone receptor MsOR3 of Mythimna separata [38].
Sequence comparisons. (A) Alignment of amino acid sequences of ApolOR1, AperOR1 and of selected verified pheromone receptors. Identical amino acids are shaded grey. Amino acids conserved throughout all putative and verified pheromone receptor sequences forming a subfamily with the Antheraea receptors in (B) are indicated by asterisks. (B) Sequence relatedness of moth olfactory receptors. Neighbor joining tree based on the identity between olfactory receptors of different moth species. The distance tree was calculated using the MEGA program and is based on a ClustAL alignment of the sequences indicated. Branch lengths are proportional to percentage sequence difference. Scale bar: 10% difference.
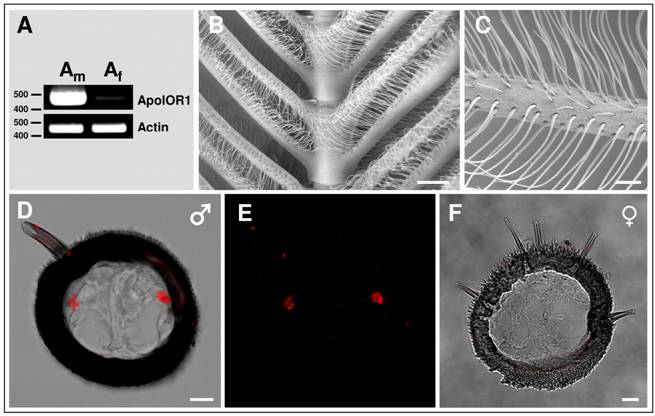
Expression of ApolOR1 in the antenna of A. polyphemus
Sex-pheromone receptors of moths are exclusively or preferentially expressed in the antennae of males [26, 28, 37-40]. To investigate the sex-specific expression of ApolOR1, RT-PCR experiments were performed with cDNA prepared from male antennae or female antennae of A. polyphemus. The results depicted in Fig. 2A indicate a very strong band with cDNA from male antennae; in contrast, only a very faint band was obtained with cDNA from female antennae. In a next step, attempts were made to visualize ApolOR1-expressing cells on the sexually dimorphic antenna. In Antheraea polyphemus, male animals possess large, bipectinate antennae with long side branches. Each segment of the antennal stem carries two opposing pairs of side branches, forming a feather-like overall structure (Fig. 2B). Laterally on the surface of the male antennal side branches, about 60.000 pheromone-sensitive sensilla trichodea per antenna are arranged in several rows (Fig. 2C); low numbers of other sensilla types (e.g. sensilla basiconica) are positioned more medially [6, 41]. Each sensillum trichodeum contains one to three bipolar olfactory receptor neurons; the vast majority houses two neurons [6].
To visualize and characterize the antennal cells expressing ApolOR1, in situ hybridization experiments were performed on cross sections through the side branches of male antennae. The results (Fig. 2D and E) indicate that the ApolOR1-specific antisense RNA probe typically labeled two cells per cross section which were located laterally on opposing sides of the section. This location corresponds to the position of trichoid sensilla on the antennal surface, suggesting that ApolOR1 is expressed in a sensory cell of this pheromone-sensitive sensillum type. In situ experiments performed on cross sections through the short side branches of the female antenna which lack pheromone-sensitive s. trichodea never showed any ApolOR1-positive cells (Fig. 2F).
Expression of ApolOR1 in the antenna of A. polyphemus. (A) Sex-biased expression of ApolOR1. RT-PCR was performed with a specific primer pair for ApolOR1 and cDNAs prepared from male and female antennae. Primers for actin were used to control the integrity of the cDNA. The positions of marker bands (bp) are indicated. (B and C) Electron micrographs of the male antenna. (D-F) Localization of ApolOR1-expressing cells on the antenna. In situ hybridization with DIG-labeled ApolOR1 antisense RNA was carried out on cross sections through antennal side branches. Hybridization signals were visualized by red fluorescence. (D and E) Male antenna. (D) Projection of a stack of optical sections. The red fluorescence channel and the transmitted-light channel have been overlaid. Scale bar: 10 µm. In (E) only the red fluorescence channel is shown. (F) Hybridization of an ApolOR1 probe to an antennal section of a female. Selected image from a stack of optical sections. The transmitted-light and red fluorescence channel have been overlaid. Scale bars: 200 µm in (B), 50 µm in (C), 10 µm in (D) and (F).
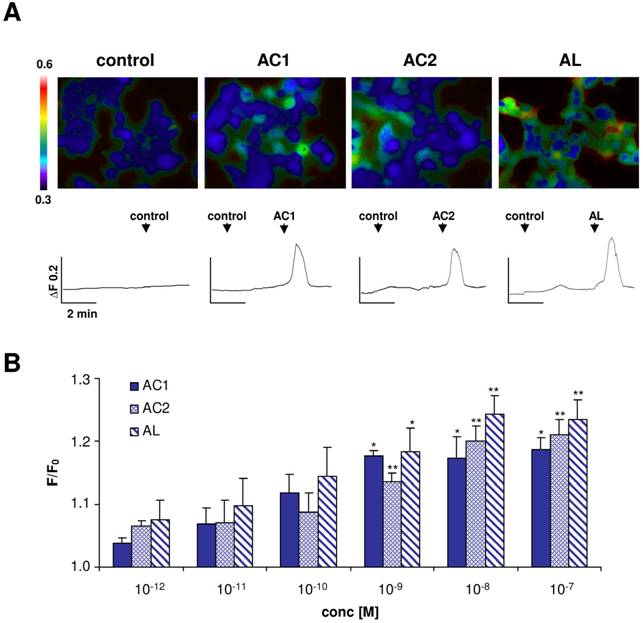
Responsiveness of ApolOR1 to pheromone components
To investigate if one of the three Antheraea sex-pheromone components might activate ApolOR1, we generated a HEK293 cell line which stably expressed this receptor type. The responsiveness of these cells to the pheromonal compounds AC1, AC2 and AL (1 nM), dissolved in the organic solvent DMSO, was analyzed by calcium imaging experiments (Fig. 3A). ApolOR1 cells did not respond to a control stimulus (0.1% DMSO, 0.1% hexane) but a clear increase in intracellular calcium levels was observed upon application of AC1, AC2 or AL. Closer examination of the responses recorded from single cells showed a rapid and transient time course of the calcium signals. Subsequently, the response of these cells to different concentrations of the pheromonal compounds was determined. To quantify the dose-dependent responses, the fluorescence intensity ratios of at least 30 ApolOR1-expressing cells were determined before (F0) and after stimulus application (F; peak of response). Mean F/F0 values from independent experiments were averaged and normalized to experiments with a control stimulus. The results indicate that all three pheromone components led to a dose-dependent response; the response to AL was slightly stronger than to AC1 and AC2 (Fig. 3B). Although explicit responses of single cells were already observed at 10-100 pM, statistically significant responses to all three compounds were first observed at 1 nM. Together, these results indicate that ApolOR1-expressing cells do respond to pheromonal compounds of Antheraea. In fact, under the experimental conditions (compounds solubilized by means of DMSO), all three substances elicited a response of ApolOR1-cells. This observation confirms results of previous studies which demonstrated that heterologously expressed moth pheromone receptors respond to more than one pheromone compound, suggesting that the receptors alone do not account for the specific responses of pheromone-responsive neurons observed in vivo [22, 23, 25]. Moreover, these studies have provided evidence that PBPs contribute to the specificity and sensitivity of the pheromone detection system.
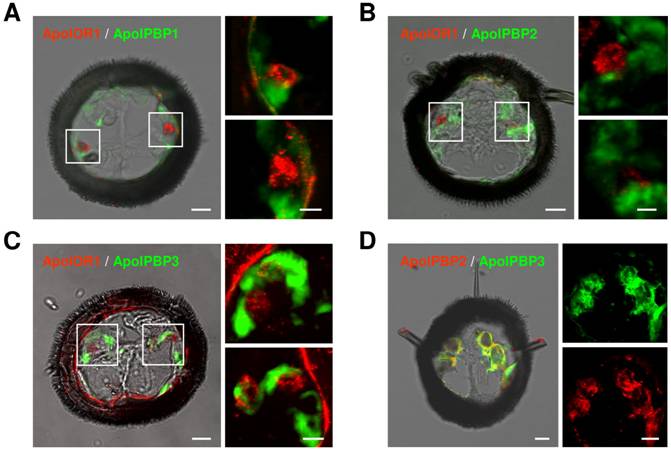
ApolOR1-expressing neurons are surrounded by PBP-expressing cells
In A. polyphemus, three different PBP subtypes have been described [13, 19]. To get a first hint at which of the three PBP subtypes could mediate responses of ApolOR1 to its cognate ligand in vivo, we set out to examine their expression in the antenna. Toward this goal we performed double in situ hybridization experiments on sections through the male antenna employing a DIG-labeled ApolOR1 probe in combination with biotin-labeled probes for the different PBPs (Fig. 4A-C). The combination of the ApolOR1 probe (red) with the ApolPBP1 probe (green) revealed ApolOR1-expressing cells closely associated with ApolPBP1-expressing cells (Fig. 4A). Experiments employing the probe combinations ApolPBP2/ApolOR1 or ApolPBP3/ApolOR1 resulted in similar patterns (Fig. 4B and C). These findings show that ApolOR1-expressing neurons are ensheathed by cells expressing ApolPBP1, ApolPBP2 and ApolPBP3 and imply a co-expression of all three PBP subtypes in supporting cells. To scrutinize this notion we performed double in situ hybridization experiments with combinations of the different PBPs. Fig. 4D exemplarily shows an experiment using a DIG-labeled ApolPBP2 probe and a biotin-labeled ApolPBP3 probe. The result shows staining of the same cells by the two PBP probes indicating co-expression of both PBPs in the same cells. Comparable experiments employing other PBP probe combinations led to similar staining patterns (data not shown). Hence, these results support the notion that all three PBP subtypes are present within the sensillum lymph of trichoid sensilla bathing the dendrites of ApolOR1-expressing neurons. Accordingly, all three PBPs could be implicated in the solubilization and the transfer of hydrophobic pheromonal compounds and in the activation of ApolOR1.
Ability of ApolPBPs to mediate pheromone responses of ApolOR1
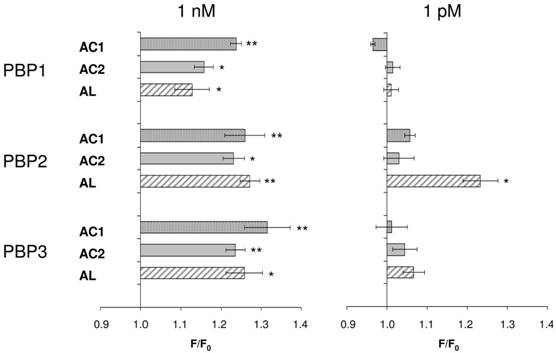
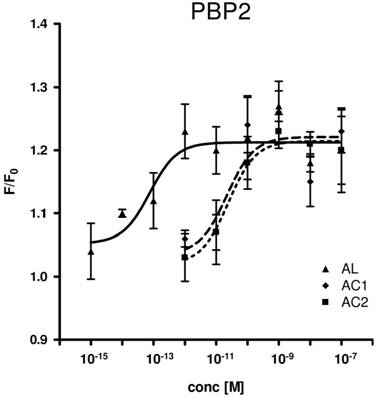
Consequently, we assessed all three PBP types for their capability to replace the organic solvent DMSO in the functional calcium imaging assay of ApolOR1-expressing cells (Fig. 5). In initial imaging experiments a concentration of 1 nM AC1, AC2 or AL was employed. At this ligand concentration, all three PBPs were able to mediate a significant response of ApolOR1-cells to the three compounds. The responses obtained with ApolPBP2 or ApolPBP3 were slightly stronger than with ApolPBP1 (Fig. 5). When the response of ApolOR1-cells in the presence of PBPs was determined at a 1000-fold lower pheromone concentration (1 pM), a different picture emerged. With a concentration of 1 pM pheromone, only PBP2 in combination with AL led to a strong response of the ApolOR1-cells (Fig. 5). Neither ApolPBP1 nor ApolPBP3 in combination with any of the Antheraea pheromonal compounds resulted in an activation of ApolOR1-cells. These results suggest a specific interplay between ApolOR1, ApolPBP2 and AL. To further explore the specificity of PBP2 and AL to activate the ApolOR1-cells, all three pheromonal compounds were analyzed at different concentrations (Fig. 6). The dose-response experiments revealed that a maximal ApolOR1-cell response mediated by ApolPBP2 could be elicited with all three compounds, albeit at very different concentrations. It was found that a combination of ApolPBP2 and AL elicited responses already at a pheromone concentration as low as 10 fM whereas for ApolPBP2/AC1 and ApolPBP2/AC2 first responses were observed at about 10 pM; at this concentration the combination ApolPBP2/AL already elicited a maximal response. Thus, in the presence of ApolPBP2, the sensitivity of the ApolOR1-cells to the pheromone AL is about three orders of magnitude higher than to the two other compounds. At low concentrations, ApolPBP2 mediates a specific response of ApolOR1-cells to AL and confers a very high sensitivity of the system to this pheromone component.
Responses of ApolOR1-expressing Flp-In T-REx293/Gα15 cells to stimulation with different Antheraea pheromonal compounds. (A) Pseudocolour images indicate calcium levels in response to application of a control solution (0.1% DMSO, 0.1% hexane) or to stimulation with AC1, AC2 or AL (1 nM) solubilized with 0.1% DMSO. Traces show responses of representative cells from the measurements shown in the pseudocolour images above. Responses are shown as changes in Fura-2 fluorescence intensity ratios (340/380 nm) over time. (B) Responses of ApolOR1-cells to stimulation with pheromonal compounds solubilized with 0.1% DMSO at different concentrations. Bars represent the mean F/F0 ± SE from 3-7 independent measurements with at least 30 cells each. Response values have been normalized to the response to the control solution. Statistic analysis was done by one-way ANOVA followed by Dunnett's post test. Significant increases are indicated by asterisks, *p < 0.05, **p < 0.01.
Expression of ApolOR1 and ApolPBPs in the A. polyphemus antenna. Double in situ hybridizations were performed with combinations of DIG-labeled and biotin-labeled antisense RNA probes on cross sections through male antennal side branches. Hybridization of probes was visualized by detection systems leading to red (DIG-label) or green (biotin-label) fluorescence. (A-C) Hybridizations using combinations of ApolOR1 and ApolPBP probes. (A) ApolOR1 and ApolPBP1, (B) ApolOR1 and ApolPBP2, (C) ApolOR1 and ApolPBP3. The images to the left represent selected pictures taken from stacks of optical sections and show an overlay of the red and green fluorescence channels and the transmitted-light channel. Scale bars: 10 µm. Higher magnifications of the areas boxed are shown on the right sides, the upper of the two pictures showing the left boxed area and the lower pictures showing the right boxed area. Only the red and green fluorescence channels are shown; images represent projections of stacks of optical sections. Scale bars: 5 µm. (D) Double in situ hybridization with ApolPBP2 (DIG-label) and ApolPBP3 (biotin-label). Depiction as in A-C, details of the left image are displayed on the right (red or green fluorescence channel). Scale bars: 10 µm.
PBP-mediated responses of ApolOR1-expressing cells. The Antheraea pheromone components AC1, AC2 and AL were tested in combination with ApolPBP1, ApolPBP2 or ApolPBP3 (1µM). PBPs were preincubated with 1 nM (left) or 1 pM solutions (right) of pheromonal compounds and used to stimulate ApolOR1-expressing cells. Responses are displayed as mean F/F0 ± SE ratios determined from 3-5 independent measurements with at least 30 cells each. Values have been normalized to responses from stimulations with PBPs only. Significant calcium increases in comparison to controls are indicated by asterisks, *p < 0.05, **p < 0.01 in a one-way ANOVA followed by Dunnett's post test.
Dose-response profile of ApolPBP2-mediated responses of ApolOR1-cells to different concentrations of Antheraea pheromonal compounds. AL, continuous line; AC1, dashed line; AC2, dotted line. ApolPBP2 (1µM) was preincubated with different concentrations of AC1, AC2 or AL and used for stimulation of ApolOR1-expressing cells. Responses are displayed as mean F/F0 ± SE ratios determined from 3-8 independent measurements with at least 30 cells each. Mean responses of cells have been normalized to measurements with ApolPBP2 only.
Discussion
In the present study, we have identified receptors from A. polyphemus (ApolOR1) and A. pernyi (AperOR1) which could clearly be assigned to the relatively conserved group of moth pheromone receptors (Fig. 1B). The predominant expression of ApolOR1 in male antennae, more precisely in cells located beneath long pheromone-sensitive sensilla trichodea (Fig. 2) further supported the notion that they may represent receptors for pheromones. This view was substantiated by analyzes of HEK293 cells which stably expressed the ApolOR1 receptor. When pheromones dissolved in DMSO were applied, ApolOR1-expressing cells responded to AC1, AC2 and AL in a similar manner (Fig. 3). This rather broad response profile of ApolOR1 to pheromonal compounds was in contrast to the specific reaction profile of pheromone-responsive neurons in the male antenna [6] and suggested that the response profile of ApolOR1 in isolation does not account for the observed in vivo specificity. Previous studies have provided strong indications that PBPs may contribute to the specificity of pheromone induced responses [22, 23, 25, 42]. The first hint at an involvement of PBPs in pheromone detection was the discovery of the first moth PBP in A. polyphemus antennal preparations as apparently single protein band which binds a tritium-labeled pheromone component [16]. Subsequent studies identified three cDNAs encoding distinct PBP subtypes of similar molecular weight [13, 19] suggesting that the originally reported single band may in fact have been comprised of three proteins. In accordance with this, we could show expression of all three PBP subtypes in the antenna. Moreover, our in situ hybridization studies indicate co-localization in the same sensillum, implying that each PBP might play a specific role in presenting one of the three pheromone components to its respective receptor. The three ApolPBP subtypes have recently been shown to interact differentially with the pheromone components AC1, AC2 and AL [20, 21]; ApolPBP1 preferentially binds to AC1, ApolPBP2 to AL and ApolPBP3 to AC2 [11, 19, 43]. While the ApolPBPs can also bind molecules other than their preferred pheromone ligand [21, 34], only the appropriate pheromonal compound seems to induce a specific change in ApolPBP conformation [20]. This observation was recently confirmed by analyzes of the Drosophila binding protein LUSH which can also bind a variety of chemicals [24, 44, 45] but only binding of the pheromone 11-cis vaccenyl acetate (cVA) leads to a specific conformational change [24].
The effort to explore which combination of binding protein subtype and pheromone component may be most suitable for activating the ApolOR1 receptor revealed that at low pheromone concentration only the combination of ApolPBP2 and the pheromone AL activated the ApolOR1-cells (Fig. 5). The observation that at high pheromone concentrations all three ApolPBPs mediated a response suggests that under these conditions PBPs act as rather unspecific solubilizers for hydrophobic compounds, similar to solvents like DMSO, whereas at physiologically more relevant concentrations the suitable PBPs mediate a more specific reaction. These findings could be explained by different binding affinities of the PBPs for their respective ligands. At low concentrations only ApolPBP2 might be capable of interacting with AL with high temporal probability whereas ApolPBP1 and ApolPBP3 can only offer similar interactions at higher AL concentrations. While competitive binding assays have indicated very similar affinities of an ApolPBP for the three Antheraea pheromone components [34] other studies could determine significant differences in the binding affinities of moth PBPs for different pheromone components of the respective species [46, 47]. In addition to an increased specificity at low pheromone concentrations, also the sensitivity for the appropriate PBP/pheromone combination was increased by orders of magnitude. These results are in line with earlier studies monitoring the response to pheromones by recording from tip-opened sensilla of Antheraea. It was found that perfusion of appropriate PBPs into the sensillar hairs significantly decreased the threshold for pheromone-induced responses [48, 49]. Early studies have estimated that a sensillum lymph concentration of 1 pM pheromone is sufficient to elicit a response of pheromone-sensitive neurons [50, 51]. Thus, the pheromone receptors would have to be activated at or below 1 pM. In our in vitro system ApolOR1-cells responded robustly to picomolar pheromone concentrations when the right PBP/pheromone combination was applied, indicating responses at very low, physiologically relevant, concentrations. One explanation for the remarkable sensitivity of the system when a pheromone component is applied together with the appropriate PBP could be a 'cooperative' receptor activation, i. e. activation by the pheromone molecule on the one hand and by an 'activated' PBP on the other hand. This view is supported by recent studies analyzing the Drosophila pheromone receptor Or67d, which have demonstrated that the receptor can be activated by high concentrations of cVA alone, but also by a 'constitutively active' mutant LUSH protein [24]. Thus, simultaneous binding of a pheromone molecule and an 'activated' PBP to a pheromone receptor could increase pheromone receptor sensitivity.
Together, the findings of this study indicate that ApolOR1 serves as the receptor for AL, a sex-pheromone component of A. polyphemus and suggest that at physiological pheromone concentrations the highly specific and sensitive response of AL-neurons in sensilla trichodea on the male antenna is based on a molecular interplay of the pheromone receptor ApolOR1 and the pheromone binding protein ApolPBP2. Such an interplay between a PBP and a pheromone receptor could also facilitate the detection of other components of the pheromone blend. In addition to the AL cell, more than 80% of the large trichoid sensilla on the male antenna are equipped with a neuron sensitive to AC1 while smaller trichoid sensilla frequently contain neurons responsive to all three compounds [6]. The presence of all three PBP subtypes in these sensilla suggests that they selectively deliver the respective pheromone components to the respective pheromone receptors, which would account for the differential specificities of the pheromone-responsive neurons. Future studies aimed at the identification and functional characterization of additional Antheraea pheromone receptors will contribute to elucidate whether 'functional receptor/PBP units' also exist for the other Antheraea pheromone components and if such 'units' may represent a general principle in moth pheromone detection.
Acknowledgements
The authors thank Prof. W. Amselgruber and K. Müller, Institute of Anatomy and Physiology of Domestic Animals, University of Hohenheim for making the scanning electron micrographs. Gesa Dreesman, Pablo Pregitzer, and Nelli Dick are acknowledged for excellent technical assistance. We also thank anonymous reviewers for helpful comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (grant KR1786 / 3-3).
Data deposition: The nucleotide sequences reported in this paper have been deposited in the EMBL data base with accession numbers FN556591 (ApolOR1) and FN556592 (AperOR1).
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Benton R. Sensitivity and specificity in Drosophila pheromone perception. Trends Neurosci. 2007;30:512-9
2. de Bruyne M, Baker TC. Odor detection in insects: volatile codes. J. Chem. Ecol. 2008;34:882-97
3. van der Goes van Naters, Carlson JR. Receptors and neurons for fly odors in Drosophila. Curr. Biol. 2007;17:606-12
4. Blomquist GJ, Vogt RG. Insect pheromone biochemistry and molecular biology. The biosynthesis and detection of pheromones and plant volatiles. London: Elsevier Academic Press. 2003
5. Schneider D. 100 Years of Pheromone Research / An Essay on Lepidoptera. Naturwissenschaften. 1992;79:241-50
6. Meng LZ, Wu CH, Wicklein M, Kaissling K-E, Bestmann H-J. Number and sensitivity of three types of pheromone receptor cells in Antheraea pernyi and A. polyphemus. J. Comp. Physiol. [A]. 1989;165:139-46
7. Steinbrecht RA, Gnatzy W. Pheromone receptors in Bombyx mori and Antheraea pernyi. I. Reconstruction of the cellular organization of the sensilla trichodea. Cell Tissue Res. 1984;235:25-34
8. Kochansky J, Tette J, Taschenber EF, Cardé RT, Kaissling K-E, Roelofs WL. Sex pheromone of the moth, Antheraea polyphemus. J. Insect Physiol. 1975;21:1977-83
9. Bestmann H-J, Attygalle AB, Brosche T, Erler J, Platz H, Schwarz J, Vostrowsky O, Cai-Hong W. Identification of three sex pheromone components of the female saturniid moth Antheraea pernyi (Lepidoptera: Saturniidae). Z. Naturforsch. 1987;42c:631-6
10. Schneider D, Lacher V, Kaissling K-E. Die Reaktionsweise und das Reaktionspektrum von Riechzellen bei Antheraea pernyi (Lepidotera, Saturniidae). Zeitschrift für vergleichende Physiologie. 1964;48:632-62
11. Maida R, Ziegelberger G, Kaissling KE. Ligand binding to six recombinant pheromone-binding proteins of Antheraea polyphemus and Antheraea pernyi. J. Comp. Physiol. [B]. 2003;173:565-73
12. Rogers ME, Sun M, Lerner MR, Vogt RG. Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J. Biol. Chem. 1997;272:14792-9
13. Raming K, Krieger J, Breer H. Molecular cloning of an insect pheromone-binding protein. FEBS Lett. 1989;256:215-8
14. Ishida Y, Leal WS. Rapid inactivation of a moth pheromone. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14075-9
15. Steinbrecht RA, Laue M, Ziegelberger G. Immunolocalization of pheromone-binding protein and general odorant-binding protein in olfactory sensilla of the silk moths Antheraea and Bombyx. Cell Tissue Res. 1995;282:203-17
16. Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161-3
17. Damberger FF, Ishida Y, Leal WS, Wuthrich K. Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J. Mol. Biol. 2007;373:811-9
18. Katre UV, Mazumder S, Prusti RK, Mohanty S. Ligand binding turns moth pheromone-binding protein into a pH sensor: Effect on the Antheraea polyphemus PBP1 conformation. J. Biol. Chem. 2009;284(46):32167-77
19. Maida R, Krieger J, Gebauer T, Lange U, Ziegelberger G. Three pheromone-binding proteins in olfactory sensilla of the two silkmoth species Antheraea polyphemus and Antheraea pernyi. Eur. J. Biochem. 2000;267:2899-908
20. Mohl C, Breer H, Krieger J. Species-specific pheromonal compounds induce distinct conformational changes of pheromone binding protein subtypes from Antheraea polyphemus. Invert. Neurosci. 2002;4:165-74
21. Bette S, Breer H, Krieger J. Probing a pheromone binding protein of the silkmoth Antheraea polyphemus by endogenous tryptophan fluorescence. Insect Biochem. Mol. Biol. 2002;32:241-6
22. Grosse-Wilde E, Svatos A, Krieger J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses. 2006;31:547-55
23. Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 2007;25:2364-73
24. Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255-65
25. Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS. Pheromone reception in fruit flies expressing a moth's odorant receptor. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16538-43
26. Krieger J, Grosse-Wilde E, Gohl T, Dewer YME, Raming K, Breer H. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Natl. Acad. Sci. U. S. A. 2004;101:11845-50
27. Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, Yasukochi Y, Touhara K, Nishioka T. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16653-8
28. Krieger J, Grosse-Wilde E, Gohl T, Breer H. Candidate pheromone receptors of the silkmoth Bombyx mori. Eur. J. Neurosci. 2005;21:2167-76
29. Krieger J, Raming K, Dewer YME, Bette S, Conzelmann S, Breer H. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur. J. Neurosci. 2002;16:619-28
30. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596-9
31. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673-80
32. Ishida Y, Leal WS. Cloning of putative odorant-degrading enzyme and integumental esterase cDNAs from the wild silkmoth, Antheraea polyphemus. Insect Biochem. Mol. Biol. 2002;32:1775-80
33. Angerer LM, Angerer RC. In situ hybridization to cellular RNA with radiolabelled RNA probes. In: (ed.) Wilkinson DG. In situ hybridization. Oxford: IRL Press. 1992:15-32
34. Campanacci V, Krieger J, Bette S, Sturgis JN, Lartigue A, Cambillau C, Breer H, Tegoni M. Revisiting the specificity of Mamestra brassicae and Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. J. Biol. Chem. 2001;276:20078-84
35. Oldham NJ, Krieger J, Breer H, Fischedick A, Hoskovec M, Svatos A. Analysis of the silkworm pheromone binding protein-pheromone complex by electrospray-ionization mass spectrometry. Angew. Chemie Int. Ed. 2000;39:4341-3
36. Oldham NJ, Krieger J, Breer H, Svatos A. Detection and removal of an artefact fatty acid from the binding site of recombinant Bombyx mori pheromone-binding protein. Chem. Senses. 2001;26:529-31
37. Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638-42
38. Mitsuno H, Sakurai T, Murai M, Yasuda T, Kugimiya S, Ozawa R, Toyohara H, Takabayashi J, Miyoshi H, Nishioka T. Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 2008;28:893-902
39. Miura N, Nakagawa T, Tatsuki S, Touhara K, Ishikawa Y. A male-specific odorant receptor conserved through the evolution of sex pheromones in Ostrinia moth species. Int. J. Biol. Sci. 2009;5:319-30
40. Patch HM, Velarde RA, Walden KK, Robertson HM. A candidate pheromone receptor and two odorant receptors of the hawkmoth Manduca sexta. Chem. Senses. 2009;34:305-16
41. Keil TA. Reconstruction and morphometry of silkmoth olfactory hairs: a comparative study of sensilla trichodea on the antennae of male Antheraea polyphemus and Antheraea pernyi (Insecta, Lepidoptera). Zoomorphology. 1984;104:147-56
42. Pophof B. Pheromone-binding proteins contribute to the activation of olfactory receptor neurons in the silkmoths Antheraea polyphemus and Bombyx mori. Chem. Senses. 2004;29:117-25
43. Leal WS, Chen AM, Erickson ML. Selective and pH-dependent binding of a moth pheromone to a pheromone-binding protein. J. Chem. Ecol. 2005;31:2493-9
44. Kruse SW, Zhao R, Smith DP, Jones DN. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat. Struct. Biol. 2003;10:694-700
45. Zhou JJ, Zhang GA, Huang W, Birkett MA, Field LM, Pickett JA, Pelosi P. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: evidence for odour recognition and discrimination. FEBS Lett. 2004;558:23-6
46. Du G, Prestwich GD. Protein structure encodes the ligand binding specificity in pheromone binding proteins. Biochemistry. 1995;34:8726-32
47. Plettner E, Lazar J, Prestwich EG, Prestwich GD. Discrimination of pheromone enantiomers by two pheromone binding proteins from the gypsy moth Lymantria dispar. Biochemistry. 2000;39:8953-62
48. Pophof B. Moth pheromone binding proteins contribute to the excitation of olfactory receptor cells. Naturwissenschaften. 2002;89:515-8
49. van den Berg MJ, Ziegelberger G. On the function of the pheromone binding protein in the olfactory hairs of Antheraea polyphemus. J. Insect Physiol. 1991;37:79-85
50. Vogt RG, Riddiford LM, Prestwich GD. Kinetic properties of a sex pheromone-degrading enzyme: the sensillar esterase of Antheraea polyphemus. Proc. Natl. Acad. Sci. U. S. A. 1985;82:8827-31
51. Schneider D, Kasang G, Kaissling K-E. Determination of the olfactory threshold of Bombyx mori with tritium-labelled bombykol. Naturwissenschaften. 1968;55:395
Author contact
![]() Corresponding author: Dr. Jürgen Krieger, University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany. E-mail: kriegerde
Corresponding author: Dr. Jürgen Krieger, University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany. E-mail: kriegerde

 Global reach, higher impact
Global reach, higher impact