10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(7):655-664. doi:10.7150/ijbs.6.655 This issue Cite
Research Paper
Nuclear factor-Y (NF-Y) regulates transcription of mouse Dmrt7 gene by binding to tandem CCAAT boxes in its proximal promoter
Department of Genetics, College of Life Sciences, Wuhan University, Wuhan 430072, P. R. China
* These authors contribute equally to this work.
Received 2010-8-20; Accepted 2010-10-23; Published 2010-10-25
Abstract
Dmrt7, a member of the Dmrt family of genes, is required for spermatogenesis. However, promoter functions of the gene Dmrt7 remain unknown. We have cloned and characterized the proximal promoter region of the mouse Dmrt7 gene. Functional analysis of the 5' flanking region by sequential deletion mutations revealed crucial positive elements between -60 and +1, in which two highly conserved and tandem CCAAT boxes: the CCAAT box1 (-48/-44) and the CCAAT box2 (-7/-3) are located. Site-directed mutagenesis studies demonstrated that both CCAAT boxes are indispensable to the promoter activity. Electrophoretic mobility shift assays (EMSAs) and gel-supershift assays indicated that transcription factor NF-Y binds to the promoter. Chromatin immunoprecipitation (ChIP) analysis demonstrated that NF-Y interacts in vivo with the promoter of the Dmrt7 gene in testis. Co-transfection and reporter analysis showed that over-expression of NF-Ys increased transcription of the Dmrt7-luc gene whereas expression of a dominant-negative NF-Ya decreased the transcription. This suggests that NF-Y can activate the Dmrt7 promoter. These results provide evidence of a transcription regulatory mechanism that controls Dmrt7 gene expression in mouse testis.
Keywords: Endocrinology, Spermatogenesis, DM domain, Transcriptional regulation
Introduction
The genes doublesex in Drosophila and mab-3 in Caenorhabditie elegans were first identified as transcription factors containing a conserved zinc finger-like DNA binding domain (DM domain), which plays a key role in sexual development (1-3). Many organisms have multiple Dmrt (doublesex and mab-3 related transcriptional factor) genes, the number of which varies between species. For example Drosophila, Caenorhabditis elegans, fish, mice and human have four, eleven, six, seven and eight copies respectively (4-9).
From arthropods to nematodes to vertebrates, most Dmrt genes are expressed in gonads and play roles in sex determination, which serves as a rare example of conserved mechanisms underlying sex regulation during evolution. Dmrt1 was the first DM domain gene identified in vertebrates (10). Previous research with Dmrt1-deficient mice strongly demonstrated that this gene was required for testis differentiation after determination, but dispensable for ovary development (11). In birds, DMRT1 is sex-linked (12, 13) on chromosome Z and there is higher dosage of the gene in males (ZZ) as compared with females (ZW) (10, 13-15). The expression of Dmrt1 in turtles and alligators was found to be related to sexual differentiation and was higher in developing male gonads than in female ones as well (16). Furthermore, the Dmrt1y/DMY gene, which is the homolog of Dmrt1 in medaka, transposed to chromosome Y and became a master gene in male differentiation (17, 18).
However, evidence is being accumulated that some proteins in the DMRT family are involved in non-gonadal development. A null mutation for the gene Dmrt1, an ancient conserved gene common to sex-determining pathways, resulted in profound abnormalities in the development of anterior neural plate derivatives in Ciona (19). Both Dmrt2a and Dmrt2b are expressed in presomitic mesoderm and developing somites and contribute to somitogenesis and the creation of left-right asymmetry in the lateral-plate mesoderm (20-23). In chicken and mice, Dmrt3 is expressed similarly in the forebrain, spinal cord, and nasal placode (24), while Dmrt3 in zebrafish is expressed in the olfactory placode and the neural tube (25). Xenopus Dmrt4 is expressed in the developing olfactory system and is required for neurogenesis (26). Both Dmrt5 and Dmrt6 are expressed primarily in the brain (9, 27).
In addition to Dmrt1, Dmrt7 expression is restricted to embryonic gonads and adult testis (9). Dmrt7 deficient mice show male infertility with spermatogenic arrest at the pachytene stage and defects in regulation of sex chromatin (28, 29). Kawamata M, et al. reported a possible post transcriptional role of poly-adenylation in Dmrt7 expression (30). However, the transcriptional mechanisms regulating Dmrt7 need to be studied further, which will contribute substantially to our understanding of the pathway of sex differentiation and spermatogenesis. Here we report on the nature of the mouse Dmrt7 promoter. Deletion and site-directed mutagenesis were used to identify regions of the promoter that are required for transcriptional activity. EMSA and ChIP analysis was employed to determine the binding relationship between transcription factor NF-Y and the Dmrt7 promoter. Finally, our study showed that NF-Y up-regulated the expression of Dmrt7 by binding to two tandem CCAAT-boxes in the proximal promoter region of the gene.
Materials and Methods
In silico sequence analysis. Transcription factor binding sites were predicted using MatInspector (www.genomatrix.de) and TFSEARCH (www.cbrc.jp/research/db/TFSEARCH.html).
Plasmid constructions. Six deletion mutants of the mouse Dmrt7 promoter were constructed using PCR cloning from mouse genomic DNA and primers: forward primer, 5'TATACGCGTCTAGAGGTCACACACAAAATCAGAAG3' for construct -948/+116, 5'TGCACGCGTAAAGGCAGGCTAAAAAGCCTGCC3' for construct -407/+116, 5'GGGACGCGTCAGCCTCGCTCTGGCTGAGGT3' for construct -245/+116, 5'TTGACGCGTCGGAGAAGCGGGTAGGCAAGAAA3' for construct -104/+116, 5'CGGACGCGTATTGCAAACCCTATTGGCTGCGC3' for construct -60/+116, 5'CTCACGCGTCCTTGTGTGAAGGAGCGAGCGG3' for construct +1/+116, and a common reverse primer, 5'CACAAGCTTCACAGCCTCGAGCCGAATCACAG3'. PCR products were double-digested with MluI and HindIII and cloned into the MluI - HindIII site of the pGL3-basic vector (Promega, Madison,WI).
Site-directed mutagenesis for two CCAAT boxes was performed using the following primers as follows: Mut1 for CCAAT box1 mutant: 5'TGCAAACCCTCTGATCTGCGCGGCGCCG3' and 5'CGGCGCCGCGCAGATCAGAGGGTTTGCA3'; mut2 for CCAAT box2 mutant, 5'GTGCTTGGAGCTCCTGATTCCTTGTGTG3' and 5'CACACAAGGAATCAGGAGCTCCAAGCAC3'; mut1/2 for both CCAAT box 1 and 2 mutants using mut1 as a template: 5'GTGCTTGGAGCTCCTGATTCCTTGTGTG3' and 5'CACACAAGGAATCAGGAGCTCCAAGCAC3'. All constructs were sequenced.
For the construction of expression plasmids, the full-length cDNAs of mouse wild-type NF-Ya, NF-Yb and NF-Yc were PCR-amplified from mouse testis cDNAs and cloned into EcoRI site of the pCX-EGFP vector by replacing the EGFP fragment to generate expression plasmids (pCX-NFYa, pCX-NFYb and pCX-NFYc) using primers:, 5′CGTGAATTCGCCATGGAGCAGTATA3′ and 5′CTCGAATTCTTAGGAAACTCGGATGA3′ for NF-Ya; 5′ATAGAATTCATCATGACAATGGACGG3′ and 5′ TCCGAATTCTCATGAAAACTGAATTTG3′ for NF-Yb; 5′GCCGAATTCAAAATGTCCACAGAAG3′ and 5′CTCGAATTCTCAGTCTCCAGTCACCT3′ for NF-Yc. The mouse dominant negative plasmid for NF-Ya (NF-YAm29) was a generous gift from Dr. Roberto Mantovani (University of Milano). All constructs were confirmed by DNA sequence analyses.
Cell culture, transient transfection and dual-luciferase reporter assay. GC-1 (a cell line from type B spermatogonia of mouse and show characteristics of a stage between type B spermatogonia and primary spermatocytes) and COS 7 cells (a cell line was obtained by immortalizing a CV-1 cell line derived from kidney cells of the African green monkey) were maintained in high glucose Dulbecco′s modified Eagle′s medium supplemented with 10% fetal bovine serum, plated in 48-well plates and transfected using LipofectamineTM2000 (Invitrogen, Carlsbad, CA, USA). The cells were transfected with 0.4 µg of each deletion construct (-948/+116, -407/+116, -245/+116, -104/+116, -60/+116, or +1/+116) along with 10 ng/well pRT-TK (an internal control). For luciferase assays, 0.2 µg of construct -60/+116 or mutated constructs, and 0.2 µg NF-Y expression vectors (NF-Ya, NF-Yb and NF-Yc) or the dominant negative NF-Y plasmid (NF-YAm29) were co-transfected into the cells. Empty vector pGL3-basic or pCX (pCX-EGFP without the EGFP coding sequence between two EcoRI sites) was added to equalize the final DNA content in each well. Luciferase activity was measured 24 hours after transfection using the dual-luciferase reporter assay system (Promega, Madison, WI, USA) and a Modulus Single Tube Multimode Reader (Turner Biosystems, Sunnyvale, CA, USA) following the manufacturer's protocol. Assays were performed in triplicate and expressed as means ± S.D.
Electrophoretic mobility shift assays (EMSA). Oligonucleotides corresponding to the CCAAT box1 and box2 of the Dmrt7 promoter were synthesized and annealed into double strands. Their sequences are as follows: 5′TGCAAACCCTATTGGCTGCGCGGCGCCG3′ and 5′CGGCGCCGCGCAGCCAATAGGGTTTGCA3′ for oligo1; 5′TGCAAACCCTCTGATCTGCGCGGCGCCG3′ and 5′CGGCGCCGCGCAGATCAGAGGGTTTGCA3′ for oligo1-ccaat mut; 5′GTGCTTGGAGCTCATTGGTCCTTGTGTG3′ and, 5′CACACAAGGACCAATGAGCTCCAAGCAC3′ for oligo2; 5′GTGCTTGGAGCTCCTGATTCCTTGTGTG3′ and 5′CACACAAGGAATCAGGAGCTCCAAGCAC3′ for oligo2-ccaat mut. Radiolabeled probes were generated by incubation of 250 ng annealed oligonucleotides with 20 µCi [γ-32P] dATP in the presence of T4 Polynucleotide Kinase (Promega, Madison, WI, USA) for 1 h at 37°C, and were subsequently separated from free nucleotides for purification using a G-50 column (Amersham Biosciences, Uppsala, Sweden). Mouse testis nuclear extract used for Electrophoretic mobility shift assays was prepared as described previously (31). Then incubated at room temperature for 30 min with a 100,000 dpm radiolabeled probe and 1 µg poly (dI-dC) in a buffer of 10 mmol/l Tris-HCl, pH 7.5, 50 mmol/l NaCl, 1 mmol/l dithiothreitol, 1 mmol/l EDTA, and 5% glycerol. For supershift experiments, binding reactions were subsequently incubated with anti-NF-Ya (Abcam Inc., Cambridge, CA, USA) for 30 min at room temperature. For competition experiments, the unlabeled competitor oligos (in 50-fold molar excess) were added together with probe at the start of the incubation. Samples were resolved in 5% polyacrylamide gels in 0.5% TBE running buffer at 10 V/cm for 2 h. The dried gel was exposed to a phosphorimager cassette and scanned with typhoon 9200 instrument (GE-Healthcare, Amersham bioscience, Uppsala, Sweden).
Chromatin immunoprecipitation (ChIP) and quantitative real-time PCR. Samples of mouse testis and liver were chopped into small pieces with a scalpel in cold phosphate-buffered saline (PBS) and cross-linked in 1% formaldehyde-PBS for 15 min with constant shaking. The tissues were rinsed in cold PBS and homogenized with a Dounce homogenizer in 1 ml cold cell lysis buffer (10 mM Tris-Cl, pH 8.0, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) supplemented with protease inhibitors (Roche Diagnostics Ltd, Mannheim, Germany). The cells were incubated at 4°C for 5 min to allow the release of the nuclei. The nuclei were collected by centrifugation at 13,000 × g for 5 min. After lysis in the buffer (1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 50 mM Tris-Cl, pH 8.1), sonication was performed with a Sonic Dismembrator model 100 sonicator (Fisher Scientific, Inc., Pittsburgh, PA, USA). The supernatant chromatin after centrifugation was immunoprecipitated with no antibody (beads only), preimmuno IgG (IgG) and anti-NF-Ya (Abcam Inc., Cambridge, CA, USA), together with Protein G PLUS-Agarose (Santa Cruz, Biotech, CA, USA) respectively. DNA isolated from the immunoprecipitated complex was amplified by PCR with primers flanking both CCAAT boxes. The primer sequences are: 5′AGAAGCGGGTAGGCAAGA3′ and 5′CAGTGGCGAAGAGGAACG3′. The amplified PCR fragments were analyzed on a 2% agarose gel. A region of 257 bp around 10 kb downstream of the first exon of the Dmrt7 was amplified as a control with primers: 5′AATAAGTTTCCAACCGTGAAGT3′ and 5′TATCCAAGGAGATGGGTAAGTG3′. The PCR products were cloned into T-easy vector (Promega, Madison, WI, USA) and sequenced for confirmation. Precipitated DNA in the ChIP assay was amplified by quantitative real-time PCR using the multichannel RotorGene 3000 (Corbett Research, Australia). The cycling conditions were: 5 min at 94°C; 45 cycles of 94°C, 20 s; 60°C, 20 s and 72°C, 15 s in a 25 µL reaction mix containing Sybr Green I. The P1 region primers were 5′AGAAGCGGGTAGGCAAGA3′; and 5′CAGTGGCGAAGAGGAACG3′. The P2 region primers were 5′AATAAGTTTCCAACCGTGAAGT3′; and 5′TATCCAAGGAGATGGGTAAGTG3′. Each sample was performed in triplicate at least. Data were analyzed by the software Rotor-gene version 4.6 and then plotted in Microsoft Excel.
Western blot analysis
Western blots were performed according to routine protocols. Aliquots of tissue extracts (50 mg) were separated by electrophoresis in 10% SDS-polyacrylamide gel (PAGE) and transferred onto a 0.45 μm membrane (Hybond-P; Amersham Pharmacia Biotech). Dmrt7 was detected by Dmrt7 antibody (1:500, GenWay Biotech, Inc., San Diego, CA) and β-Actin was used as an internal control (1:1000, Santa Cruz Biotech, CA, USA). Membranes were incubated with primary antibody for 1 h at room temperature, washed in TBST and probed with HRP conjugated secondary antibody (1:10000, Santa Cruz Biotech, CA, USA). After several washes in TBST, detection of the secondary antibody was performed using the SuperSignal Chemiluminescent Substrate system (Pierce, Rockford, USA). The chemical illumination signals were exposed onto Fuji medical X-ray film.
Results
Identification of regulatory elements in the mouse Dmrt7 promoter region
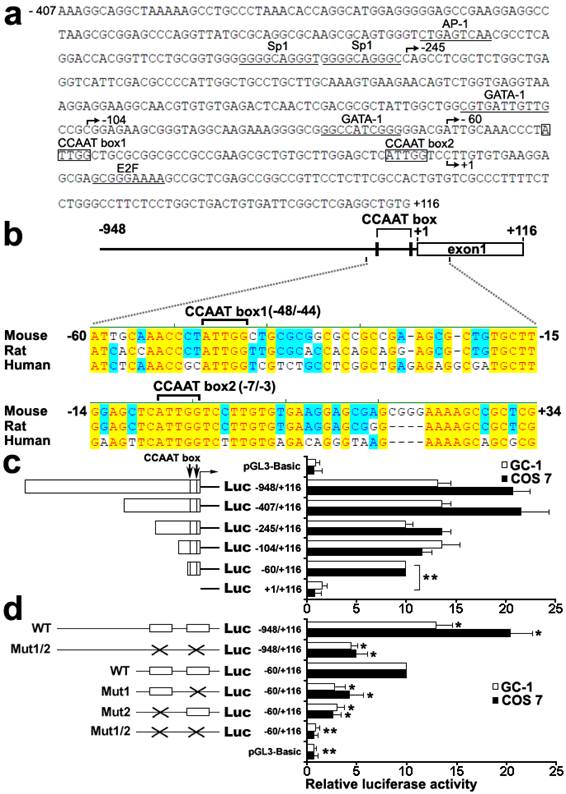
To characterize functional elements in the Dmrt7 promoter, the sequence from -407 to +116 of Dmrt7 was run in MatInspector and TFSEARCH to find putative transcriptional elements. Several possible transcription-factor binding sites, which included AP-1, Sp1, GATA1, E2F and CCAAT box, were identified (Figure 1a). BLAST alignment of the promoter region of mouse, rat, and human Dmrt7 genes revealed two highly conserved CCAAT boxes: the CCAAT box1 (-48/-44) and the CCAAT box2 (-7/-3) (Figure 1b). To determine the functional elements required for promoter activity, six 5'-deletion mutants (-948/+116, -407/+116, -245/+116, -104/+116, -60/+116 and +1/+116) were constructed and analyzed for promoter activity using luciferase reporter in GC-1 and COS 7 cells (Figure 1c). The maximum luciferase activity was observed in the region of the -407/+116. The deletion of the region between -245 and -104 appears to increase promoter activity in GC-1 cells, whereas the opposite result was obtained in COS 7 cells. To explain this discrepancy, we speculate that there could be other GC-1 specific transcription factors that could bind to this region and play a negative role in Dmrt7 promoter activity. Deletion of the -60 to +1 sequence markedly decreased the activity, indicating the region of the -60 to +1 is essential for promoter function (Figure 1c). Two highly conserved CCAAT boxes were located in this region. To confirm the role of both CCAAT boxes, point mutants were created by site-directed mutagenesis (Figure 1d). The mutation of CCAAT box1 in -60/+116 construct resulted in a decrease of approx.60-70% compared to the promoter activity of -60/+116 construct, the CCAAT box2 mutation in -60/+116 construct approx.70-80%. Nearly no activity was observed in the double mutations of both boxes in -60/+116 construct. However, the double mutations of both CCAAT boxes in the -948/+116 region resulted in only a decrease of approx.40-50% compared to the promoter activity of -60/+116 construct. Although there may be other important cis-regulatory sites upstream of the -60/+116, which may compensate the loss of these CCAAT boxes (Figure 1d), the region of the -60/+116 including two highly conserved CCAAT boxes is important for the promoter function. These results showed that both CCAAT box1 and CCAAT box2 are important for transcriptional activation of the mouse Dmrt7 gene.
Transcription factor NF-Y binds to Dmrt7 promoter in vitro and in vivo
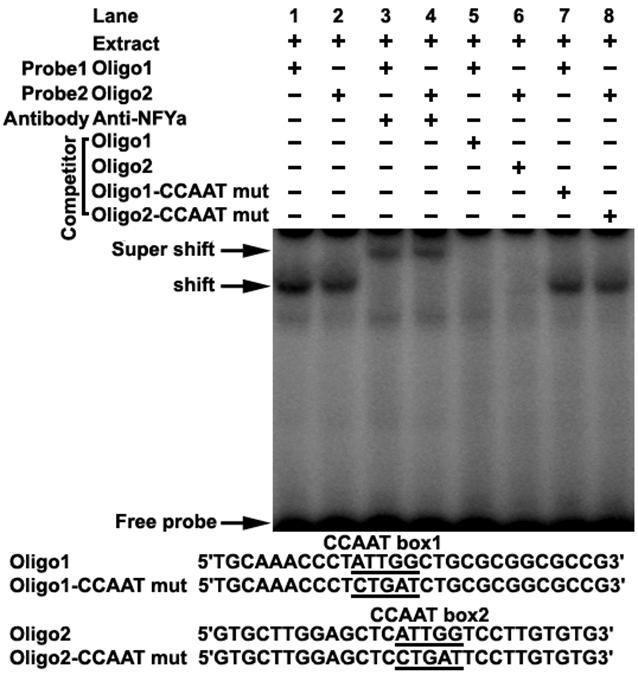
Transcription factor prediction using the MatInspector and TFSEARCH showed that potential transcription factor NF-Y may bind to the CCAAT boxes. To determine if NF-Y binds to the CCAAT boxes, electrophoretic mobility shift assays were carried out using nuclear extracts prepared from mouse testis and double stranded oligonucleotides. Incubation of probe1 spanning from -58 to -31 (containing CCAAT box1) with the extract gave rise to the formation of a DNA-protein complex (Figure 2 lane1), which could be competed by the addition of an excessive amount of unlabelled oligo1 DNA (Figure 2 lane5), but not by the oligo1 DNA with mutated CCAAT box (Figure 2 lane7). Furthermore, addition of NFYa antibody to the binding reaction caused the disappearance of the specific DNA/protein complex and the appearance of the super-shift band (Figure 2 lane3). These results indicate that the CCAAT box1 within the promoter region is capable of binding with transcription factor NF-Ya in vitro. A similar result was also observed in incubation of probe2 spanning from -20 to +8 (containing CCAAT box2) with mouse testis nuclear extract (Figure 2). These results indicate that transcription factor NF-Ya can bind to Dmrt7 promoter in vitro.
Identification and function characterization of the mouse Dmrt7 promoter. a), Nucleotide sequence of the promoter region (from -407 to +116) of mouse Dmrt7. The putative binding sites for transcriptional factors are underlined. The CCAAT boxes are boxed. The numbering of the nucleotides starts at the first nucleotide of the cDNA (+1). b), Sequence alignment of 1 kb upstream of the first exon of Dmrt7 from mouse, rat and human. Two highly conserved CCAAT boxes are indicated. Conserved nucleotides among all three species are denoted by red letters and are shaded in yellow. The positions of the nucleotides in the mouse Dmrt7 gene are indicated. +1 represents the first nucleotide of the cDNA. c), Deletion analysis of the mouse Dmrt7 promoter using the luciferase assay. GC-1 or COS 7 cells were transiently transfected with 0.4 µg of the deletion constructs together with 10 ng Renilla luciferase plasmid, pRT-TK, which served as an inner control for transfection efficiency. The relative activities to the -60/+116 construct of a series of deletion mutants was determined by the luciferase assay. The results are the mean ± S.D. of three independent experiments. *, P < 0.05; **, P < 0.001. d), Mutation analysis in the CCAAT boxes of the Dmrt7 promoter by the luciferase assay. The -948/+116 and -60/+116 constructs were used as a template to generate point mutants. GC-1 or COS 7 cells were transiently transfected with 0.4 µg of these constructs together with 10 ng Renilla luciferase plasmid, pRT-TK. The relative activities of the -60/+116 construct of these mutants was determined by the luciferase assay. The results are the mean ± S.D. of three independent experiments. *, P < 0.05; **, P < 0.001 with compared to the -60/+116 wild-type construct. The box indicates the intact CCAAT box. The fork indicates the corresponding mutations.
Electrophoretic mobility shift assay of NF-Y binding to Dmrt7 promoter. The oligo1 corresponding to -58/-31 and oligo2 corresponding to -20/+8 were γ-32P-ATP labelled and incubated with 5µg of nuclear extract of mouse testis in the absence or presence of 50-fold excess various competitor DNA (mutant or non-labeled oligo) or anti-NFYa antibody, as indicated on the top of the gel image. Samples were resolved on 5% polyacrylamide gels in 0.5% TBE running buffer at 10 V/cm for 2 h. The specific DNA/protein complex and the super-shift bands were indicated by arrows. The shift bands cannot be competed with mutated DNA competitor (lane 7, 8). The sequences of oligo1, oligo2 and corresponding mutants are shown in the lower panel.
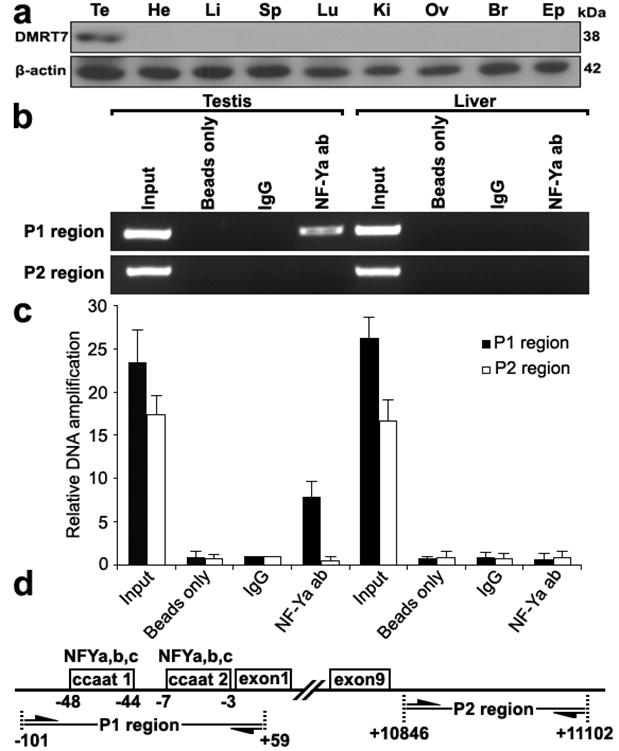
To further determine whether NF-Y binds to the mouse Dmrt7 promoter in vivo, we performed chromatin immunoprecipitation analysis. Because Dmrt7 protein was only detected in testis (Figure 3a), we chose the testis for ChIP analysis. As shown in Figure 3b, a 160 bp of DNA fragment was amplified from the precipitate by NF-Ya antibody from testis, but not from control tissue liver or negative control immunoprecipitations using no antibody (beads only) or normal rabbit IgG. The amplified fragment was confirmed by sequencing. To rule out the possibility of non-specific binding of the NF-Ya antibody, an additional PCR amplification of a distinct genomic region was performed on all of the precipitated chromatin DNAs, which showed no amplification out of the NF-Ya-binding region (Figure 3b,d). A quantitative ChIP PCR was also performed to quantify the binding extent of NF-Ya on the Dmrt7 promoter (Figure 3c). These results indicate that NF-Y specifically binds to the Dmrt7 promoter in vivo.
ChIP assay of NF-Y binding to the Dmrt7 promoter in testis. a), Western blot analysis of the Dmrt7 protein in the adult testis, heart, liver, spleen, lung, kidney, ovary, brain and epididymis. The Dmrt7 expression was only detected in the testis. β-actin was used as an internal control. b), Interaction of NF-Y with the Dmrt7 promoter in vivo was determined by chromatin immunoprecipitation analysis. Samples of mouse testis and liver were chopped into small pieces and cross-linked in 1% formaldehyde to cross-link endogenous proteins and DNA. Samples of sonicated chromatin were immunoprecipitated with anti-NFYA, no antibody (beads only) and preimmuno IgG (control) respectively. DNA isolated from immunoprecipitated material was amplified by PCR with primers to amplify the 160-bp sequence of mouse Dmrt7 promoter corresponding to the -101 to +59 region. Primers for an unrelated region were used as negative control. The amplified PCR fragments were analyzed on 2% agarose gel. c), A q-ChIP PCR was performed to quantify the binding extent of NF-Ya on the Dmrt7 promoter. All values were initially expressed relative to relevant IgG DNA content. d), The relative positions of the primers are shown.
NF-Y activates the mouse Dmrt7 promoter
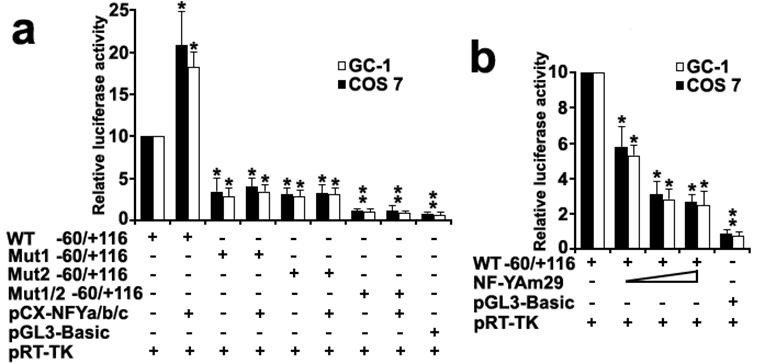
To investigate the role of transcription factor NF-Y in the activation of the Dmrt7 promoter, both GC-1 and COS 7 cells were co-transfected with luciferase reporter driving by wild-type Dmrt7 promoter (-60/+116 construct) or its mutants for CCAAT sites, and NF-Ya/b/c expression plasmids. The luciferase activity increased obviously when a wild-type promoter construct (-60/+116) was co-transfected with NF-Ya/b/c (Figure 4a). However, the luciferase activity was not upregulated obviously when co-transfected with mutants for CCAAT box1 or box2 and was not upregulated absolutely when co-transfected with double mutants for CCAAT box1 and box2 (Figure 4a). This result indicated tandem arrangement of the CCAAT box1 and box2 may ensure maximum promoter activity of the Dmrt7. In addition, replacing the NF-Ya/b/c expression plasmid with the dominant-negative construct (NF-YAm29) resulted in a marked reduction of luciferase activity in a dosage-dependent manner (Figure 4b). NF-YAm29 has three amino acid substitutions at the C-terminal region of the NF-Ya, which impairs its DNA binding and acts as a dominant-negative mutant by sequestering the NF-Yb/Yc subunits into a defective complex (32). These results indicated that NF-Y can activate the Dmrt7 promoter.
Co-transfection analysis of the mouse Dmrt7 promoter. a), Effects of expression of NF-Y on the transcriptional activity of the Dmrt7 5'-flanking sequence (-60/+116). Cells were transfected with 0.2µg -60/+116 construct or mutants and 0.2µg NF-Y expression plasmids (pCX-NFYa, pCX-NFYb, pCX-NFYc). b), Effects of expression of the dominant negative mutant of NF-Y on the transcriptional activity of the Dmrt7 5'-flanking sequence (-60/+116). The dominant negative NF-Ya expression plasmid NF-YAm29 (0.1µg, 0.2µg, 0.3µg) was cotransfected with 0.2µg -60/+116 construct, which inhibits Dmrt7-luciferase activity. pGL3-basic was employed as a negative control. pRT-TK served as an inner control for transfection efficiency. The relative luciferase activities to the -60/+116 construct were determined by luciferase assay. Cells were harvested 24 h post-transfection, and luciferase activity was measured and normalized to Renilla luciferase activity. All transfection experiments were repeated at least three times and the activities are showed relative to that of the -60/+116 construct, which was set to 10. The results are the mean ± S.D. of three independent experiments. *, P < 0.05; **, P < 0.001 with compared to the -60/+116 wild-type construct.
Discussion
Dmrt7 is a member of the DM domain family of genes, which encodes proteins with a conserved DNA binding DM domain. In addition to Dmrt1, some DM domain proteins such as Dmrt3, Dmrt4 and Dmrt7 may be involved in sexual development. Dmrt7 mRNA could be detected in testis and fetal ovary but not in other tissues (9). The expression of Dmrt7 protein was detected in testis (29), but not in the fetal ovary. Dmrt7 mutants were infertile with spermatogenic arrest at the pachytene stage, while the mutant females showed normal fertility (28, 29). Possible post transcriptional role of poly-adenylation in Dmrt7 expression regulation was previously proposed (30). Because transcriptional regulation of the gene Dmrt7 during spermatogenesis is an important process, in this study, we have explored promoter function of the gene and found that nuclear factor-Y (NF-Y) regulates transcription of mouse Dmrt7 gene through binding to two conserved CCAAT boxes in its proximal promoter region. Deletion analysis of the 5' flanking region of the mouse Dmrt7 revealed crucial positive elements between -60 and +1, in which two evolutionarily conserved CCAAT boxes (the CCAAT box1 and box2) were identified. The conservation of transcription factor binding sites between orthologue gene promoters may indicate their highly significant roles in maintaining transcriptional regulation of Dmrt7 gene. Further site-directed mutagenesis and functional analysis were performed to confirm their role in maintenance of promoter function of Dmrt7. The results suggest that binding by NF-Y to these two tandem CCAAT boxes is important for regulation of the basal transcription of the Dmrt7 gene in the mouse testis.
NF-Y protein (also known as CBF) binds with high specificity to the CCAAT sequence and is a ubiquitous heteromeric transcription factor (33). NF-Y is a complex composed of three subunits: NF-Ya (CBF-B, HAP2 in yeast), NF-Yb (CBF-A, HAP3) and NF-Yc (CBF-C, HAP5), which are all required for DNA-binding (34-36). Both NF-Yb and NF-Yc have conserved histone folding motifs resembling H2B-H2A and their heterodimerization is essential for NF-Ya association (33, 37). NF-Ya is a regulatory subunit of the trimeric complex, whose levels vary in different cell types and/or growth conditions (38). It has been suggested that NF-Y can gain access to its genomic locations even in the absence of methyl histone marks and then leads the positioning of methyl histone marks typical of active chromatin independently (39). NF-Y appears to be essential for the activation of transcription (40). A number of mammalian promoters have been shown to contain CCAAT boxes and have been regulated by NF-Y alone or together with other transcriptional factors (41-44). In accordance with these results, the transcription factor NF-Y may activate the mouse Dmrt7 gene and play an important role in the regulation of Dmrt7 expression in testis. This, therefore, suggests that NF-Y regulation of Dmrt7 plays a functional role in spermatogenesis.
In summary, we have found two important CCAAT boxes in the 5' flanking region of the mouse Dmrt7 promoter. NF-Y is a major transcriptional activator of the Dmrt7 gene through binding to these two CCAAT boxes. These results provide evidence of the regulatory mechanisms that control the expression in testis of the mouse Dmrt7 gene and will form the basis for understanding mammalian spermatogenesis.
Acknowledgements
We are grateful to Dr. Roberto Mantovani at Universita di Milano for the dominant negative NF-YA expression plasmid (NF-YAm29). The work was supported by the National Natural Science Foundation of China, the National Key Basic Research project (2006CB102103), the Key Transgenic New Organism Project (2009ZX08009-148B) and Program of Wuhan Subject Chief Scientist.
Conflict of Interest
There are no conflicts of interest with the financing of this project and its subject matter.
References
1. Erdman SE, Burtis KC. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 1993;12(2):527-535
2. Raymond CS, Shamu CE, Shen MM. et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391(6668):691-695
3. Zarkower D. Establishing sexual dimorphism: conservation amidst diversity? Nat Rev Genet. 2001;2(3):175-185
4. Lints R, Emmons SW. Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 2002;16(18):2390-2402
5. Kondo M, Froschauer A, Kitano A. et al. Molecular cloning and characterization of DMRT genes from the medaka Oryzias latipes and the platyfish Xiphophorus maculatus. Gene. 2002;295(2):213-222
6. Volff JN, Zarkower D, Bardwell VJ. et al. Evolutionary dynamics of the DM domain gene family in metazoans. J Mol Evol. 2003;57(Suppl 1):S241-249
7. Brunner B, Hornung U, Shan Z. et al. Genomic organization and expression of the doublesex-related gene cluster in vertebrates and detection of putative regulatory regions for DMRT1. Genomics. 2001;77(1-2):8-17
8. Ottolenghi C, Fellous M, Barbieri M. et al. Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics. 2002;79(3):333-343
9. Kim S, Kettlewell JR, Anderson RC. et al. Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr Patterns. 2003;3(1):77-82
10. Raymond CS, Kettlewell JR, Hirsch B. et al. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999;215(2):208-220
11. Raymond CS, Murphy MW, O'Sullivan MG. et al. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14(20):2587-2595
12. Nanda I, Shan Z, Schartl M. et al. 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat Genet. 1999;21(3):258-259
13. Nanda I, Zend-Ajusch E, Shan Z. et al. Conserved synteny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: a comparative (re)view on avian sex determination. Cytogenet Cell Genet. 2000;89(1-2):67-78
14. Smith CA, McClive PJ, Western PS. et al. Conservation of a sex-determining gene. Nature. 1999;402(6762):601-602
15. Shan Z, Nanda I, Wang Y. et al. Sex-specific expression of an evolutionarily conserved male regulatory gene, DMRT1, in birds. Cytogenet Cell Genet. 2000;89(3-4):252-257
16. Kettlewell JR, Raymond CS, Zarkower D. Temperature-dependent expression of turtle Dmrt1 prior to sexual differentiation. Genesis. 2000;26(3):174-178
17. Matsuda M, Nagahama Y, Shinomiya A. et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417(6888):559-563
18. Nanda I, Kondo M, Hornung U. et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci U S A. 2002;99(18):11778-11783
19. Tresser J, Chiba S, Veeman M. et al. doublesex/mab3 related-1 (dmrt1) is essential for development of anterior neural plate derivatives in Ciona. Development. 2010;137(13):2197-2203
20. Meng A, Moore B, Tang H. et al. A Drosophila doublesex-related gene, terra, is involved in somitogenesis in vertebrates. Development. 1999;126(6):1259-1268
21. Saude L, Lourenco R, Goncalves A. et al. terra is a left-right asymmetry gene required for left-right synchronization of the segmentation clock. Nat Cell Biol. 2005;7(9):918-920
22. Zhou X, Li Q, Lu H. et al. Fish specific duplication of Dmrt2: characterization of zebrafish Dmrt2b. Biochimie. 2008;90(6):878-887
23. Liu S, Li Z, Gui JF. Fish-specific duplicated dmrt2b contributes to a divergent function through Hedgehog pathway and maintains left-right asymmetry establishment function. PLoS One. 2009;4(9):e7261
24. Smith CA, Hurley TM, McClive PJ. et al. Restricted expression of DMRT3 in chicken and mouse embryos. Gene Expr Patterns. 2002;2(1-2):69-72
25. Li Q, Zhou X, Guo Y. et al. Nuclear localization, DNA binding and restricted expression in neural and germ cells of zebrafish Dmrt3. Biol Cell. 2008;100(8):453-463
26. Huang X, Hong CS, O'Donnell M. et al. The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc Natl Acad Sci U S A. 2005;102(32):11349-11354
27. Guo Y, Li Q, Gao S. et al. Molecular cloning, characterization, and expression in brain and gonad of Dmrt5 of zebrafish. Biochem Biophys Res Commun. 2004;324(2):569-575
28. Kawamata M, Nishimori K. Mice deficient in Dmrt7 show infertility with spermatogenic arrest at pachytene stage. FEBS Lett. 2006;580(27):6442-6446
29. Kim S, Namekawa SH, Niswander LM. et al. A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet. 2007;3(4):e62
30. Kawamata M, Inoue H, Nishimori K. Male-specific function of Dmrt7 by sexually dimorphic translation in mouse testis. Sex Dev. 2007;1(5):297-304
31. Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47(5):767-776
32. Mantovani R, Li XY, Pessara U. et al. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269(32):20340-20346
33. Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239(1):15-27
34. Coustry F, Maity SN, de Crombrugghe B. Studies on transcription activation by the multimeric CCAAT-binding factor CBF. J Biol Chem. 1995;270(1):468-475
35. Sinha S, Maity SN, Lu J. et al. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci U S A. 1995;92(5):1624-1628
36. Kim IS, Sinha S, de Crombrugghe B. et al. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol Cell Biol. 1996;16(8):4003-4013
37. Romier C, Cocchiarella F, Mantovani R. et al. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278(2):1336-1345
38. Marziali G, Perrotti E, Ilari R. et al. The activity of the CCAAT-box binding factor NF-Y is modulated through the regulated expression of its A subunit during monocyte to macrophage differentiation: regulation of tissue-specific genes through a ubiquitous transcription factor. Blood. 1999;93(2):519-526
39. Donati G, Gatta R, Dolfini D. et al. An NF-Y-dependent switch of positive and negative histone methyl marks on CCAAT promoters. PLoS One. 2008;3(4):e2066
40. Nicolas M, Noe V, Ciudad CJ. Transcriptional regulation of the human Sp1 gene promoter by the specificity protein (Sp) family members nuclear factor Y (NF-Y) and E2F. Biochem J. 2003;371(Pt 2):265-275
41. Zhu J, Giannola DM, Zhang Y. et al. NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4. Blood. 2003;102(7):2420-2427
42. Huang DY, Kuo YY, Lai JS. et al. GATA-1 and NF-Y cooperate to mediate erythroid-specific transcription of Gfi-1B gene. Nucleic Acids Res. 2004;32(13):3935-3946
43. Koessler H, Kahle J, Bode C. et al. Human replication-dependent histone H3 genes are activated by a tandemly arranged pair of two CCAAT boxes. Biochem J. 2004;384:317-326
44. Kadariya Y, Nakatani K, Nishioka J. et al. Regulation of human methylthioadenosine phosphorylase gene by the CBF (CCAAT binding factor)/NF-Y (nuclear factor-Y). Biochem J. 2005;387:175-183
Author contact
![]() Corresponding author: Professors Rongjia Zhou and Hanhua Cheng, Department of Genetics and Center for Developmental Biology, College of Life Sciences, Wuhan University, Wuhan 430072, P. R. China, Fax: 0086-27-68756253, E-mail: rjzhouedu.cn, hhchengedu.cn
Corresponding author: Professors Rongjia Zhou and Hanhua Cheng, Department of Genetics and Center for Developmental Biology, College of Life Sciences, Wuhan University, Wuhan 430072, P. R. China, Fax: 0086-27-68756253, E-mail: rjzhouedu.cn, hhchengedu.cn

 Global reach, higher impact
Global reach, higher impact