10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(4):426-439. doi:10.7150/ijbs.7.426 This issue Cite
Research Paper
Medaka tert produces multiple variants with differential expression during differentiation in vitro and in vivo
1. Department of Biological Sciences, National University of Singapore, 10 Kent Ridge Crescent, Singapore 119260;
2. Division of Bioengineering, School of Chemical and Biomedical Engineering, Nanyang Technological University, 70 Nanyang Drive, Singapore 637457;
3. School of Life Sciences, Huazhong Normal University, Wuhan 430079, China;
4. Yangtze River Fisheries Research Institute, Chinese Academy of Fisheries Sciences, 41 Jianghan Road, Jingzhou City, Hubei 434000, China;
5. National Institute for Basic Biology, Okazaki, Japan
* Present address: The Solomon H. Snyder Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
Received 2011-1-1; Accepted 2011-4-1; Published 2011-4-15
Abstract
Embryonic stem (ES) cells have immortality for self-renewal and pluripotency. Differentiated human cells undergo replicative senescence. In human, the telomerase reverse transcriptase (Tert), namely the catalytic subunit of telomerase, exhibits differential expression to regulate telomerase activity governing cellular immortality or senescence, and telomerase activity or tert expression is a routine marker of pluripotent ES cells. Here we have identified the medaka tert gene and determined its expression and telomerase activity in vivo and in vitro. We found that the medaka tert locus produces five variants called terta to terte encoding isoforms TertA to TertE. The longest TertA consists of 1090 amino acid residues and displays a maximum of 34% identity to the human TERT and all the signature motifs of the Tert family. TertB to TertE are novel isoforms and have considerable truncation due to alternative splicing. The terta RNA is ubiquitous in embryos, adult tissues and cell lines, and accompanies ubiquitous telomerase activity in vivo and in vitro as revealed by TRAP assays. The tertb RNA was restricted to the testis, absent in embryos before gastrulation and barely detectable in various cell lines The tertc transcript was absent in undifferentiated ES cells but became evident upon ES cell differentiation, in vivo it was barely detectable in early embryos and became evident when embryogenesis proceeds. Therefore, ubiquitous terta expression correlates with ubiquitous telomerase activity in medaka, and expression of other tert variants appears to delineate cell differentiation in vitro and in vivo.
Keywords: medaka, pluripotency, senescence, telomerase, tert, TRAP
Introduction
Since the report of Hayflick and Moorehad in 1961 that normal human fibroblasts have only a limited capacity for proliferation in culture [1], many types of normal somatic cell cultures from humans, rodents, birds and other species have been shown to exhibit a finite capacity for cell division. Most normal somatic cells in vivo also cannot divide indefinitely [2]. Hayflick noted that most tumor cells can proliferate without limitation, and proposed that replicative senescence of normal cells might also contribute to organismal ageing [1]. Replicative senescence is the progressive decline in the ability to proliferate, which is an intrinsic property of most normal somatic cells. In vivo, replicative senescence is limited to cells that have the ability to divide, but does not apply to postmitotic cells such as mature muscle and neurons. During replicative senescence, cells sense the number of division they have run, not chronological time [2].
According to the telomere hypothesis for replicative senescence, the telomere length acts as a mitotic clock for sensing the number of division, and telomere shortening leads to senescence and subsequent death [3]. The telomere is a special structure at the chromosome ends to prevent them from end-to-end fusion or being recognized as DNA damage [4]. In vertebrates, telomeres consist of tandem repeats of TTAGGG [5] and telomere-associated proteins [6]. As a result of an inherent inability of DNA polymerase to replicate the 5' ends of linear chromosomes, cell division leads to progressive telomeric shortening to a critical length that is proposed to act as a trigger for cell growth arrest and cellular senescence [3]. Immortal cells such as totipotent stem cells and tumor cells can proliferate without an apparent limitation, because these cells possess telomerase activity that overcomes telomeric attrition by adding telomeric repeats to the chromosome ends and thus prevent cells from replicative senescence [7].
Expression of telomerase activity has been examined in diverse organisms. In unicellular eukaryotes, cell division corresponds to asexual reproduction, and telomerase is constitutively expressed to maintain telomeres from generation to generation. In multicellular organisms, telomerase studies have focused on mammals that grow only in the embryonic and juvenile stages but not in the adult and scenescent stages. In these organisms, the need of distinct cells and tissues are decoupled from the requirement for organismal reproduction. This is most evident in humans: telomerase activity in normal adult somatic tissues is limited to stem cells with high proliferation potential, but is found in the germline and cancer cells [8]. Primary human cells senesced, whereas their transgenic counterparts ectopically expressing telomerase became immortalized and extended their life-span (Bodnar et al., 1998). In fact, forced telomerase expression has since been widely used for cell immortalization [9,10]. In contrast, telomerase activity has been detected ubiquitously in all somatic adult tissues of lower vertebrate such the rainbow trout [11] and invertebrates such as lobster [12], where the growth is indeterminate, and age is proportional to size. In the zebrafish showing functional ageing and very gradual senescence, telomerase activity has been found throughout embryogenesis and in adult muscles [13].
Telomerase is a ribonucleoprotein complex consisting of two subunits: the catalytic subunit telomerase reverse transcriptase (TERT) that adds telomeric repeats to elongate telomere, and the telomerase RNA (TR) subunit that contains a template region complementary to the telomeric sequence for reverse transcription [14]. In human, TERT is the primary regulator for enzyme activity as its expression correlates with telomerase activity [15], whereas TR is ubiquitously expressed in all tissues irrespective of telomerase activity [16]. This notion is supported by the fact that telomerase activity can be restored upon ectopic Tert expression [17]. Therefore, the analysis of tert RNA expression and the direct determination of telomerase activity by telomeric repeat amplification protocol (TRAP) are often used as reliable measure of telomerase expression. Recent studies have revealed that Tert possesses telomerase activity-independent functions to attain cancer stem cell characteristics in glioma cells by inducing EGFR expression [18] and to protect ATM-deficient hematopoietic stem cells from ROS-induced apoptosis [19].
The tert cDNA has been isolated from yeast [20] and tetrapod vertebrates including frog [21], chicken [22], mouse [23], hamster [24], dog [25] and human [26]. Vertebrate Tert proteins share several motifs in three regions. The N-terminal region contains four evolutionarily conserved vertebrate motifs (v1 to v4), followed by a telomerase specific T-motif [21]. The central region contains seven well-conserved reverse transcriptase (RT) motifs (1, 2, A, B, C, D and E) that define the catalytic region and display sequence similarity to other RTs [27]. Specifically, motifs 1, 2 and A correspond to the finger domain of other RTs, whereas motifs B-E corresponds to the core catalytic domain. The C-terminus harbors three conserved motifs (v5 to v7) essential for telomerase activity [28].
Telomerase activity and expression exhibit salient differences in diverse animals. In general, ubiquitous telomerase expression has been documented in lower vertebrates and invertebrates, in contrast to preferential or specific expression in proliferating human cells such as stem cells and cancer cells. The role of telomerase in cell growth, differentiation and embryonic development remains to be elucidated in non-mammalian animals. The teleost fish medaka (Oryzias latipes) is an excellent model for vertebrate development [29]. This fish has ease for maintenance under laboratory conditions, easy embryology, a short generation time of ten weeks, a short life span of three years, many inbred or mutant strains, powerful experimental tools such as embryo microinjection for gene knockdown [30] or chimeric assay [31]. Importantly, medaka has embryonic stem (ES) cells [32,33], adult spermatogonia capable of test-tube sperm production [34] and even haploid ES cells [35]. The medaka genome draft sequence is available (http://dolphin.lab.nig.ac.jp/medaka). These features make medaka a suitable model for telomerase biology in vivo and in vitro.
This study was aimed at the examination of medaka tert expression and roles in ES cells and early embryonic development. We show that the medaka tert locus can produce multiple variants besides the standard terta encoding the longest isoform TertA, the homolog of the mammalian Tert protein. Furthermore, we reveal that terta RNA expression and telomerase activity are ubiquitous in vivo and in vitro, whereas other tert variants exhibit differential expression in vivo and in vitro during ES cell differentiation. Therefore, the multiple tert variants have differential expression during cell differentiation.
Materials and Methods
Fish
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Advisory Committee for Laboratory Animal Research in Singapore and approved by this committee (Permit Number: 27/09). Medaka of orange-red variety, South Korean population (SOK), albino strains i1 and i3 were maintained as described [36]. Embryos were maintained at 26~28°C and staged as described [37]. Unless otherwise indicated, chemicals were purchased from Sigma, enzymes and PCR reagents were from Promega.
Cell culture
Medaka cell cultures were grown at 28°C under ambient air. Three stem cell lines and four new cell cultures were used in this study. MES1 [32] and HX1 [35] are medaka diploid and haploid ES cell lines and were maintained for undifferentiated growth or induced differentiation via embryoid body (EB) formation as described. SG3 is a medaka spermatogonial stem cell line derived from the adult testis and was maintained as described [34]. Besides the three stem cell lines, we established four new cell lines that appear to consist of differentiated cells. These are S125, Sok, Or1 and i1, which were derived from blastula embryos of SOK and the adult testes of SOK, orange-red variety and i1 medaka, respectively. Cell cultures from blastula embryos were derived according to [32] and testicular cultures according to [34]. Cells were subcultured at a 1:3 splitting ratio. All the seven cell lines used in this study had undergone ≥50 passages and thus ≥100 doublings.
Gene identification and isolation
When we started this work, there was no fish tert sequence available. In order to isolate a medaka tert gene by using the homology cloning approach, the chicken Tert was used as a query for a blast search against a medaka genome draft sequence (http://dolphin.lab.nig.ac.jp/medaka/). This led to the identification of scaffold 3719 which predicts partial sequence of a putative Tert. On a sequence alignment of this predicted amino acid (aa) sequence with other known vertebrate Tert proteins generated several highly conserved domains. The genomic sequences encoding these domains were used to design primers TF and TR (Table 1) for isolating a partial cDNA by reverse transcriptase-polymerase chain reaction (RT-PCR). Total RNA was extracted from medaka blastula embryos of by using the TRIzol reagent (Invitrogen, Carlsbad, CA).
First-strand cDNA (5' and 3'-RACE-Ready) was generated from ~ 1-μg of total RNA in a 10-μl volume by using the SMARTTM RACE cDNA amplification Kit (Clontech, Mountain View, CA). RT-PCR was run in 25 μl of 1 X PCR buffer containing 2 mM MgCl2, 0.2 μM of each primer, 200 μM dNTPs each, 50-200 ng cDNA template and 1 U Taq DNA polymerase for 35 cycles (94°C 30 s, 60°C 30 s and 72°C 3 min 10 sec at 94ºC, 10 sec at 53ºC, and 3 min at 72ºC for 35 cycles, followed by 10 min at 72ºC with Taq DNA polymerase (Invitrogen). The PCR product was separated on 1% agarose gels, documented using a UV transilluminator coupled with a CCD camera (Advanced American Biotechnology, Fullerton, CA) and recovered by using UltraCleanTM 15 DNA Purification Kit (MO BIO Laboratories, Carlsbad, CA). After TA-cloning into pGEM-T easy vector (Promega, Madison, WI), plasmid DNA was prepared by using the DNA Purification Kit (iNtRON Biotechnology, Kyunki-Do, Korea), and the inserts were sequenced by using the BigDye Terminator v3.1 Cycle Sequencing Kit on ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Eight clones were found to contain an identical insert of 3,003 nt for a partial but continuous open reading frame.
The partial cDNA was extended to its full length by RACEs. For 5' RACE, a 5' RACE-Ready cDNA and two nested gene specific primers (5TR and 5TRn) were used, while 3'RACE was run by using a 3' RACE-Ready cDNA and two nested gene primers (3TF and TFn). Hot-start PCR was run for 12 cycles at 94ºC for 10 sec, Ta (annealing temperature, decreasing from 74ºC at a rate of 1ºC per cycle) for 10 sec and 72ºC for 1 min, followed by 25 cycles of 10 sec at 94ºC, 10 sec at 68ºC, and 1 min at 72ºC. A fragment of 503 bp and 199 bp was obtained from 5' and 3'RACE reaction, respectively. The PCR products were similarly cloned and sequence. The full-length cDNA was assembled and PCR-cloned from the 5'-RACE-Ready cDNA template by using terminal primers fTF (AAAATGACATCCGGGGATTTGTCGAGCG) and fTR (CCGCTCAGATCACATCTGCATAGCCAGGAAGTCCAG).
Sequence analysis
The PCR products were cloned into pGEM-T vector. The inserts were sequenced in both directions on the Applied Biosystems 3130xl (Applied Biosystems, MA). BLAST searches were run against public databases by using BLASTN for nucleotide sequences and BLASTP for protein sequences. Multiple sequence alignment was conducted by using the Vector NTI suite 8. Phylogenetic trees were constructed by using the DNAMAN package (http://www.lynnon.com/).
RNA isolation and RT-PCR analysis
Total RNA samples were extracted from medaka embryos at different developmental stages, adult tissues (brain, heart, liver, ovary, testis, skin, intestine), and cell lines of ES cells (MES1; Hong et al, 1996), spermatogonial cells (SG3; Hong et al., 2004b), and serial cultures of adult testis-derived, uncharacterized cell types (Liu T & YH, unpublished). The synthesis of cDNA from 1-2 μg of total RNA was primed by oligo dT18 by using the Molony murine leukemia virus reverse transcriptase (Promega, Madison, WI). RT-PCR was run for 30 cycles (β-actin) and 35 cycles (tert) for 10 sec at 94ºC, 10 sec at 60ºC and 50 sec at 72ºC by using cDNA-specific primers listed in Table 1. Specifically, fTF and fTR flanking the full-length cDNA sequence were used for cDNA cloning, and RTF (spanning exon 11 and 12) and RTR (spanning exon 15 and 16) were used for RT-PCR analysis.
Primers used for medaka tert cloning and PCR analysis*
| Gene (accession) | Primer | Sequence | Ta (ºC) |
|---|---|---|---|
| Tert (DQ248968.1) | TF | AAAAGGAGGAGAAATGTTTTGGCC | 55 |
| TR | TCACATCTGCATAGCCAGGAAGTCCAG | ||
| 3TF | AGCGCCGGTTGGAGAGGCTTCTGGGAG | 70 | |
| 3TFn | CTGCTGGACTTCCTGGCTATGCAGATGTGA | ||
| 5TR | CAGGTAGCGGGTGACGTCTGTGCCGAG | 72 | |
| 5TRn | AGCGGAAGTGGTCTGCATCCCGGTCCTCCT | ||
| fTF | TCCTTGAATCACCCATACTGC | 54 | |
| fTR | TTCCATTTTCTTTTCTTTTAATCACTTTAAC | ||
| vTF | cgcccAAGCTTATGACATCCGGGGATTTGTCGAGCG | 60 | |
| vTR | cgcggACTAGTCATCTGCATAGCCAGGAAGTCCAGC | ||
| RTF | CAAAGACTATTCCAGCTATGCAGGCCTCTC | 60 | |
| RTR | CCAACCGGCGCTTCCATTTGTGCAGG | ||
| tertb-RTF | cgagctgtaatcaggtggatttc | 59 | |
| tertb-RTR | cactgacggtacgtcttctttc | ||
| nanog (NP_001153902) | Forward | CTCCACATGTCCCCCCTTATC | 60 |
| Reverse | AGGATAGAATAGTCACATCAC | ||
| oct4 (AY639946) | Forward | GCTTTCTTTGGCGTAAACTCGTC | 60 |
| Reverse | TCATCCTGTCAGGTGACCTACC | ||
| no tail (ENSORLG00000011262) | Forward | ATGAGCGCGTCGAACCCGGAC | 60 |
| Reverse | AGACGGGCGCTTTCATCCAGT | ||
| β-actin (D89627) | Forward | TGGGATGACATGGAGAAGATCTG | 56 |
| Reverse | TGGGAATTCCAATCCAGACAGAGTATT |
* Irrelevant sequences are in small letter, and introduced restriction sites are in bold. Ta, annealing temperature
Telomerase activity assay
Telomerase activity was examined by the sensitive telomeric repeat amplification protocol (TRAP) (Eisenhauer et al., 1997) by using the TRApeze® Telomerase Detection Kit (Chemicon #S7700, Temecula, CA). For this, 20-50 embryos or 40-100 mg tissues were washed with phosphate buffered saline (PBS) and homogenized in 200 μl of ice-cold CHAPS lysis buffer containing 200 units/ml RNasin® ribonuclease inhibitor (Promega, Madison, WI). Crude protein samples were quantified by using the Bio-Rad protein assay reagent (Bio-Rad, Foster City, CA). Aliquots of 500 ng protein (cell or tissue extract) or from an equal number of embryos was assayed for telomerase activity in a 25-μl volume containing 20 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 63 mM KCl, 0.005% Tween-20, 1 mM EGTA, 50 μM of dNTPs each, and 0.1 μg of primer TS (AATCCGTCGAGCAGAGTT). After 30 min incubation at 30ºC, the reaction was stopped by heating at 94ºC for 3 min. Following addition of 0.1 μg of primer RP (CCCTTACCCTTACCCTTACCCTAA) and 1 unit of Taq DNA polymerase, the telomerase reaction products were PCR-amplified for 33 cycles (10 sec at 94ºC, 10 sec at 55ºC, and 30 sec at 72ºC). The PCR products were resolved on 0.75-mm thick, 12.5% denaturing polyacrylamide gels at 200 V for 1.5 h. The gels were stained for 15-20 min in 1 μg/ml SYBR Green I Nucleic Acid Stain (Cambrex, East Rutherford, NJ) and documented on a gel imaging system (Synoptics, Cambridge). Negative controls were similarly run except that protein samples were inactivated by heating for 15 min at 90ºC before addition to the reaction mixture.
Microscopy and Photography
Observation and imaging on Leica MZFIII stereo microscope with Nikon E4500 digital camera (Nikon Corp), Zeiss Axiovertinvert 2 invert microscope and Axiovert 200 upright microscope with a Nikon E4500 digital camera or a Zeiss AxioCam M5Rc digital camera (Zeiss Corp., Germany) were described previously [35].
RESULTS
Medaka tert cDNA and its encoded Tert protein
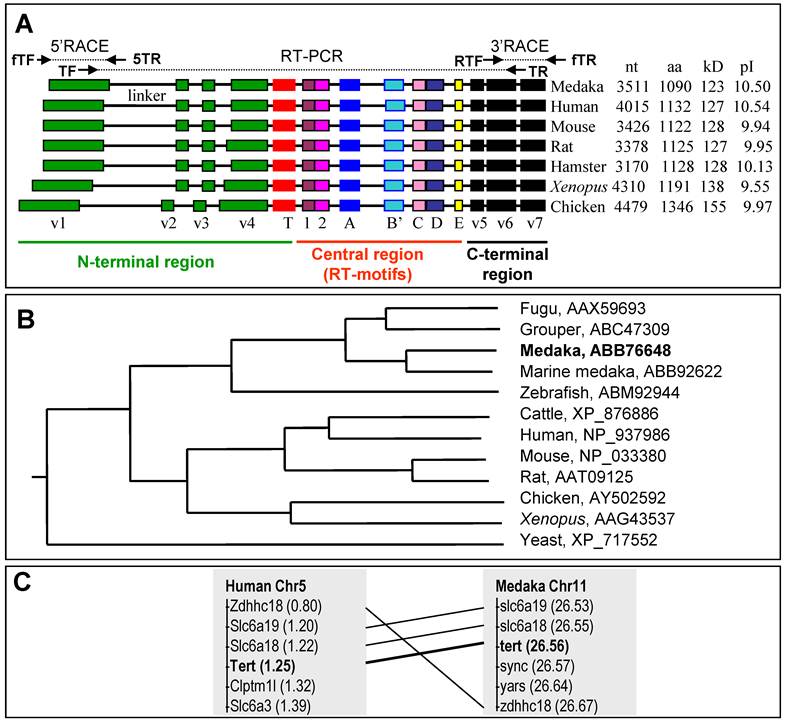
We obtained a total five RNA variants designated terta to terte. The terta is 3511 nt and was obtained from a blastula cDNA library. It encodes the longest isoform TertA with 1,090 amino acid residues (aa), a putative polyadenylation signal upstream of a poly A tail of 27 nt, a 5' untranslated region (UTR) of 101 nt and a 3' UTR of 137 nt (Fig. 1). On a sequence alignment, the medaka TertA shows all structural motifs/domains previously identified in several vertebrate Tert proteins. These are the N-terminal motifs v1-v4 and T, the central RT-motifs 1, 2, A-E, and C-terminal essential motifs v5-v7, which are present in the corresponding regions (Fig. 2).
The predicted medaka TertA shows a 34% maximal identity to the human TERT protein (Fig. 3A). On a phylogenetic tree (Fig. 3B), the medaka TertA together with Tert proteins from four other fish species forms a cluster, while Tert proteins from tetrapod vertebrates forms another cluster, conforming to their organism relationships. Besides a common domain structure, the medaka TertA shares similarities with known vertebrate Tert sequences in predicted molecular mass (123 kDa) and isoelectric point (pI = 10.5) (Fig. 3A).
This cDNA was obtained as illustrated in Fig. 3A. A search on a medaka genome draft sequence database identified scaffold 3719 encoding aa sequences with significant homology to the known vertebrate Tert proteins. Three steps of RT-PCR in combination of 5'- and 3'-RACEs from a blastula cDNA library resulted in the composition of the full length cDNA. The continuity of the assembled cDNA was confirmed by sequencing the cloned RT-PCR products obtained by using a pair of terminal primers.
Medaka tert gene and chromosomal location
A comparison between the medaka tert cDNA and genomic sequence on scaffold 3719 led to the elucidation of its genomic organization. The medaka tert locus consists of 16 exons (Fig. 4A), as in the mouse (ENSMUSG00000021611) and human tert gene (ENSG00000164362). A blast search by using the medaka cDNA as query against the medaka genome sequence led to the location the medaka tert gene as the locus OlPS67_ORYLA on chromosome 11 (http://www.ensembl.org/index.html). Our medaka tert cDNA clone is 99% and 98% identical to the chromosomal locus at nt and aa levels, respectively. Convincingly, the medaka tert gene shows a chromosomal synteny to the human gene (Fig. 3C). Taken together, the medaka tert encodes an ortholog of the human TERT by sequence, gene structure and chromosome synteny.
Medaka tert sequence. Top, Nucleotide sequence of medaka terta. Bottom, Deduced amino acid sequence of medaka TertA. Sequences of different exons are shown alternatively in black and blue color. ATG, TGA and a putative polyadenylation signal are shown in bold. Primer sequences used for RT-PCR analyses are underlined. Sequences absent in tertb and TertB are highlighted in grey.

Tert sequence alignment. Conserved motifs are boxed. Ol, Oryzias latipes (medaka); Fr, Takifugu rubripes (pufferfish); Xl, Xenopus laevis (clawed frog); Mm, Mus musculus (mouse); Hs, Homo sapiens (human). For accession numbers see Figure 3.

Tert protein, gene structure and chromosome context. (A) Phylogenetic tree. The bootstrap value is 100 at all branches. Sequence accession numbers follow organisms. (B) Domain structure of Tert proteins and cloning steps. The N-terminal, central and C-terminal regions are defined. Four conserved vertebrate motifs (v1-v4) and the telomerase-specific motif (T) in the N-terminal region, seven reverse transcriptase motifs (1, 2, A, B, C, D and E) in the central region, and three conserved motifs (v5-v7) in the C-terminal region are boxed. Sizes of cDNAs and proteins, predicted molecular weights and isoelectric point (pI) are shown in boxes. The difference in protein size is due mainly to the variable N-terminal region, in particular the linker between motifs v1 and v2. Note the medaka protein is the smallest among vertebrate proteins so far reported. Cloning steps are illustrated above the medaka Tert protein. Positions of primers used for cloning are indicated, their sequences are listed in Table 1. (C) Comparison of tert gene structure between medaka and human. The primary tert transcript is 4 times larger in human than in medaka. The human introns are not proportional. (D) Chromosome synteny between medaka and human. Chromosomal locations are shown in parentheses.
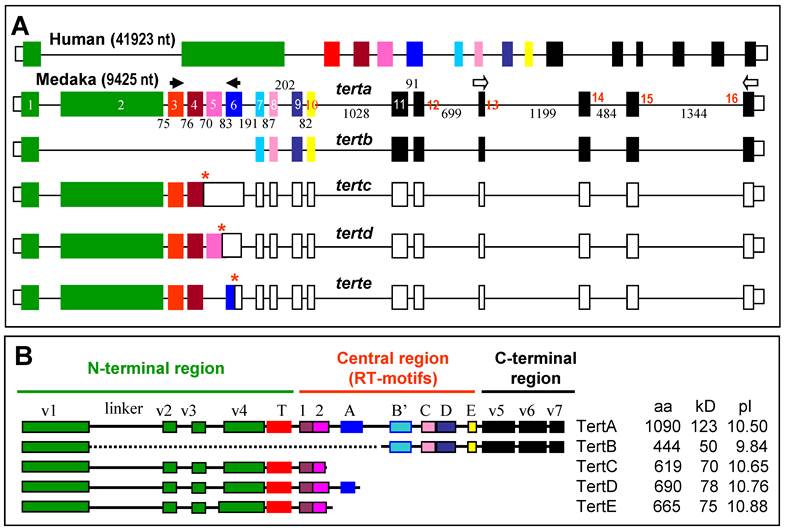
Multiple Tert isoforms are the products of alternative splicing
RT-PCR analysis by using the terminal primer pair led to the identification of 5 variants from the tert locus. Variant tertb was derived from the adult testis, whereas variants tertc to terte were from the embryo. Comparisons between cDNAs and the genomic sequence established that these variants were the products of alternative splicing (Figure 4A). Specifically, tertb results from the exclusion of exons 2~6, tertc and tertd arise from the inclusion of both introns 4~5 and only intron 5, respectively, whereas terte is due to the exclusion of exon 5, respectively (Figure 4A). In tertb, the open reading frame is maintained, leading to isoform TertB of 444 aa, which lacks the region spanning the linker to the central RT motif A (Figure 4B); the variants tertc to terte acquire premature stop codons by frame shifting, predicting isoforms TertC, D and E of 619 aa, 690 aa and 665 aa in length, respectively, which all have an incomplete RT and lack the C-terminal region (Figure 4B). Therefore, all the isoforms TertB~E would act as mutant forms of TertA or functions independently of telomerase activity.
Medaka tert RNA Expression
It has been well documented that telomerase activity is mainly regulated at the transcriptional level, and that tert expression is largely associated with telomerase activity. A salient difference has been reported for tert gene expression: differential expression in human and ubiquitous expression in Xenopus. It has been reported that hTert mRNA is differentially expressed in cancers, stem cells and other telomerase positive cells [26]. The medaka tert transcript was found to be present in all adult somatic tissues examined (Figure 5A), including the brain and heart that are thought to consist largely of postmitotic cells. During embryogenesis, the tert is abundant from the 2-cell stage up to the blastula stage, gradually reduced to a low level at hatch (Figure 5B). Since zygotic transcription takes place after the midblastula transition at the blastula stage, the tert transcript may be inherited at a high level as maternal message, in accordance with its abundance in the adult ovary (Figure 5A).
RNA variants and protein isoforms. (A) Genomic organization. Filled box, translated exon; open box, untranslated exon; thin line between boxes, intron. Exons are numbered within boxes, and sizes of introns are given between boxes. Asterisks denote stop codons. Arrows depict PCR primers. (B) Medaka isoforms TertA to TertE. Different domains are highlighted in different colors as for their encoding exons shown in (A). Sequences have been deposited in the Genbank under accession numbers DQ248968 (TertA) and JF326825-JF326828 (TertB-E).
RNA expression and telomerase activity. (A-C) tert RNA expression in adult tissues (A), developing embryos (B) and cell cultures (C). Numbers of PCR cycles are indicated. MES1, medaka ES cells; SG3, adult medaka spermatogonial stem cells; Or1 and Sok, uncharacterized cell cultures from the adult medaka testis; neg, negative control by using total RNA instead of cDNA as a template. An equal amount of cDNA reaction was used for each sample. β-actin was used as a loading control. (D-F) Telomerase activity in adult tissues (D), developing embryos (E) and cell cultures (F). Positive (pos; +) and negative controls (neg; -) were TRAP reactions by using the commercially supplied extract without and with heat inactivation. S125 and i1, uncharacterized cell cultures from medaka embryos of strain SOK and adult medaka testis of strain i1. (G-J) Phenotypes of representative cell lines. Scale bars, 50 μm.
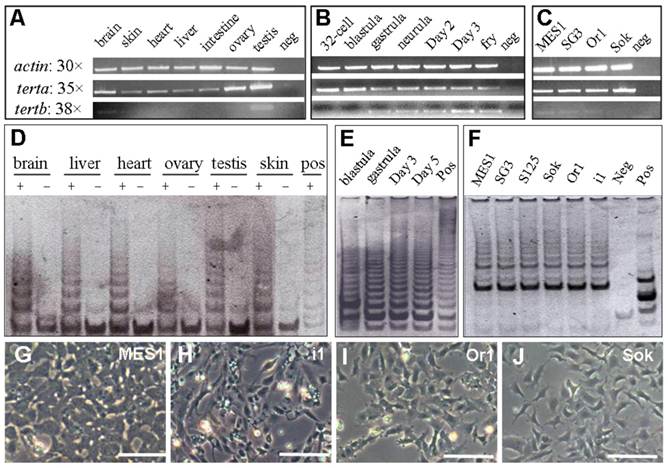
Previously, we have derived three medaka ES cell lines [32]. MES1, one of the three lines, has been shown to be pluripotent, because it is competent for chimera formation [33], induced and directed differentiation [32,38]. We have also established SG3, a normal adult spermatogonial stem cell line capable of sperm production in vitro [34]. As expected, both stem cell lines highly express the tert transcript (Figure 5C). Interestingly, a similarly high level of the tert was also detected in an adult testis-derived cell culture (Sok; Figure 5C).
In contrast, tertb exhibited tissue- and stage-specific expression. In adult tissues, tertb was restricted to the adult testis; in developing embryos, tertb was absent before gastrulation; in cell lines, tertb was barely detectable (Figure 5A-C). Therefore, terta, but not tertb, is ubiquitously expressed in vivo and in vitro.
Expression of telomerase activity
If the terta transcription is the major regulation of telomerase, the ubiquitous terta expression RNA would indicate a universal expression of telomerase activity in the medaka. To address this issue, we performed TRAP assays, a standard for measuring cellular telomerase activity. Telomerase activity is present in adult tissues (Fig. 5D), developing embryos (Figure 5E) and cell cultures (Figure 5F). Heat-inactivation of the tissue or cell extracts abolished the formation of TRAP products. These results demonstrate that telomerase activity, like the ubiquitous terta transcript, is also ubiquitous in vivo and in vitro.
Notably, seven medaka cell lines were used for examining tert expression and/or telomerase activity (Figure 5F). These cell lines represent five different medaka strains and populations (Table 2) and have different phenotypes (Figure 5G-J). Taken together, there is a positive correlation between telomerase activity and terta expression, and ubiquitous terta expression and/or telomerase activity in vitro appears to be independent of strains and cellular phenotypes of stable cell cultures.
Cell lines and their origin
| Cell line | Origin | Strain | Phenotype | Reference |
|---|---|---|---|---|
| MES1 | Fertilization blastulae | HB32C | ES cell | [32] |
| SG3 | Adult testis | i3 | Germ stem cell | [34] |
| HX1 | Gynogenetic blastulae | i1 | ES cell | [35] |
| S125 | Fertilization blastulae | SOK | Unknown | This study |
| Sok | Adult testis | SOK | Unknown | This study |
| Or1 | Adult testis | Orange-red | Unknown | This study |
| i1 | Adult testis | i1 | Unknown | This study |
Up-regulation of tertc expression upon differentiation in vitro and in vivo
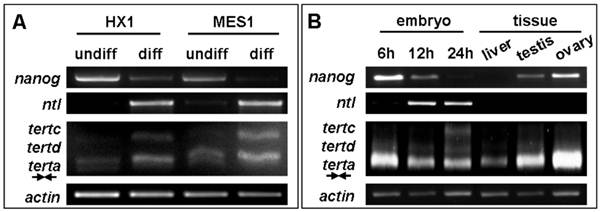
In human, telomerase expression delineates with cellular immortality in vivo and in vitro, and is widely used for monitoring undifferentiated stem cells in culture. We wanted to determine whether tert expression would be also indicative of an undifferentiated state of medaka stem cells in culture. To this end, we made use of diploid ES cell line MES1 and haploid ES line HX1, which have been show to expression a set of seven pluripotency genes including nanog and oct4 under undifferentiated culture condition but dramatically reduce or lost their expression upon induced differentiation[35,39]. We observed that terta expression remained easily detectable in both ES cell lines before and after differentiation. However, tertc expression was absent in undifferentiated ES cells but become evident upon their differentiation (Figure 6A). Successful induction of ES cell differentiation was demonstrated by the down-regulation of nanog expression and the up-regulation of no tail expression (Figure 6A). nanog and no tail are the markers for pluripotency and differentiation of medaka ES cells, respectively[39]. Interestingly, expression of tertc and tertd was barely detectable in early developing embryos until 6 h post fertilization (hpf) when cell differentiation has not yet started, but commenced until 12 hpf and increased further until 24 hpf (Figure 6B). Therefore, unlike mammals in which tert expression or telomerase activity is a stem cell marker[40], the lack of expression of truncated RNA variants such as tertc but not terta expression is associated with pluripotency of undifferentiated ES cells and developing embryos in medaka.
Taken together, the medaka tert produces multiple variants by alternative splicing, which exhibit different expression patterns. On the basis of RNA expression pattern and protein structure, the isoform TertA appears to be the functional homolog of the mammalian Tert, whereas all the isoforms TertB~E may be the dominant-negative mutants and/or possess additional functions independent on the RT activity. Consequently, it appears that in medaka, telomerase expression and activity may be controlled at both the post-transcriptional and post-translational levels. Apparently, the expression of transcript variants for truncated Tert isoforms appears to be a marker of ES cell differentiation.
RNA expression of tert variants during differentiation in vitro and in vivo. (A) RNA expression during ES cell differentiation. Notably, expression of variant tertc increases upon ES cell differentiation. (B) RNA expression in representative stages of embryos and adult tissues. Elevated expression of variant tertc and tertd is seen in embryos at 12 h and 24 h post fertilization when overt cell commitment and differentiation take place. nanog and ntl (no tail) were used as markers of pluripoteny and differentiation, respectively. β-actin was used as a loading control. Arrows define PCR primers as illustrated in Figure 4A.
DISCUSSION
The present study reports the isolation and expression of the medaka tert gene. Four lines of evidence demonstrate that medaka tert encodes an ortholog of the known Tert proteins. First, the medaka Tert predicted from variant terta is similar to its mammalian homologs in the number of aa, molecular weight and isoelectric point. Second, the medaka Tert shows a maximal sequence identity to previously identified Tert proteins, and displays all conserved motifs characteristic of tetrapod Tert proteins. Third, the medaka tert share a common genomic organization with mammalian genes. Finally, the medaka tert shows a chromosomal syntenic relationship with the human gene.
The ability to produce multiple different protein products via differential promoter usage and/or alternative splicing is a hallmark of many eukaryotic genes. In the case of tert gene, seven, three and two isoforms are available in annotated human, chimpanzee and chicken genomes (http://www.ensembl.org/index.html), respectively, which are products of RNA variants that share the first exon. In this study, PCR cloning by using primers flanking the full length open reading frame of the terta leads to a total of five RNA variants, four of which are alternatively splicing products by inclusion of intron 4 and/or 5 and exclusion of exon 5 and are thus encode novel isoforms, as similar isoforms have not yet been reported in other organisms. Because of large deletion (TertB) or significant truncation (TertC~E), the four medaka isoforms TertB to TertE should lack the Tert activity and perhaps function independently of telomerase activity. In mammals, Tert has recently demonstrated to function through a telomere independent mechanism to control stem cell pluripotency and survival [18,19].
In this study, we have made several important observations on medaka telomerase expression. Notably, tert expression and telomerase activity are both ubiquitous in vivo during embryonic and adult life, which conforms to ubiquitous tert expression and/or telomerase activity previously reported in invertebrates [41] and lower vertebrates including fish species such as rainbow trout (Oncorhynchus mykiss)[11], marine medaka (Oryzias melastigma) [42] and more importantly, the medaka [43]. These animals grow continuously throughout life, possess a high capability for regeneration and have little senescence even beyond adulthood. Consequently, telomerase expression occurs in many cells, irrespective of individual age and differentiation status, to support a high proliferation capacity for growth and regeneration. The medaka situation is in sharp contrast to the mammalian situation where telomerase expression is closely associated with immortal cells such as stem cells and cancer cells [7]. Humans and other mammals show active growth in embryonic and juvenile phases and no growth in adult and senescent phases. During their long adult life period, size and weight are kept essentially constant. Therefore, the differential telomerase expression between mammals and non-mammalian animals accompanies differences in cell proliferation and organismal growth.
We have utilized a total of seven cell lines to telomerase expression in vitro. Interestingly, three stem cell lines and four non-stem cell lines show a comparably high level of terta expression and/or telomerase activity. Therefore, terta expression in stable cell lines is also ubiquitous, irrespective of cell origin and phenotype. This observation has an important implication. All the seven medaka cell lines used in this study had undergone ≥50 passages at a 1:3 and thus ≥100 doublings, which indicates the immortality or continuous proliferation capacity. In accordance with the known primary function of telomerase activity for cellular immortality, it is not surprising that non-stem cell cultures also exhibit a high level of tert expression to support active division in vitro, similar to the situation of stem cell cultures. Hence, ubiquitous terta expression in vitro in stable cell lines underscores the role of telomerase in cell growth.
It deserves to note that the medaka tert variants exhibit differential expression. Interestingly, variant terta maintains ubiquitous expression in vivo and in vitro, other variants display tissue- and stage-specific expression (tertb) or differentiation-specific expression (tertc). Therefore, terta expression and the lack of expression of truncated tert variants are indicative of an undifferentiated state of stem cells in culture. This is contrast to the mammalian situation, where a high level of tert expression is a stem cell marker [40]. In this regard, the medaka tert is unique, because it is not its differential transcription but its differential alternative splicing at the post-transcription level that reflects differentiation of ES cells. It has been well-documented that the RNA expression of many pluripotency genes is high in undifferentiated ES cells but dramatically reduced upon ES cell differentiation [35,39].
Stem cell lines have a prominent difference from non-stem cell lines in that stem cells possess pluripotency, namely the potential for differentiation. Our work does not distinguish whether expression of tert variants for truncated isoforms is associated with differentiation per se and/or comprised proliferation, because ES cells upon differentiation exhibit reduced proliferation. Telomerase functions primarily in regulating telomeric length to count the number of cell divisions, and overt replicative senescence due to the absence of telomerase activity appears gradually after ~50 cell cycles in human fibroblast culture [1]. Induced ES differentiation occurs rapidly over a short period of 5~10 days, during which cells could undergo 3~6 cycles of division [32,35]. It is likely that expression of tert variants for truncated isoforms is associated with ES cell differentiation in culture. Future work will determine whether the expression of truncated tert variants is the cause or consequence of stem cell differentiation.
Our findings that the tert can produces multiple isoforms with differential expression in differentiation make medaka a model organism to study the Tert functions in vitro and in vivo.
Acknowledgements
We thank J. Deng for breeding fish, Choy Mei Foong for laboratory management. This work was supported by the Biomedical Research Council of Singapore (R-08-1-21-19-585 & SBIC-SSCC C-002-2007).
Abbreviations
aa: amino acid(s); cDNA: complementary DNA; kDa: kilodalton(s); nt: nucleotide(s); RACE: Rapid Amplification of cDNA Ends; RT: reverse transcriptase; PCR: polymerase chain reaction; Tert: telomerase reverse transcriptase; tert: gene encoding Tert; TRAP: telomeric repeat amplification protocol; UTR: untranslated region.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585-621
2. Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513-522
3. Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458-460
4. Zakian VA. Telomeres: beginning to understand the end. Science. 1995;270:1601-1607
5. Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A. 1989;86:7049-7053
6. Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569-573
7. Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405-413
8. Greider CW. Telomerase activity, cell proliferation, and cancer. Proc Natl Acad Sci U S A. 1998;95:90-92
9. Feng LX, Chen Y, Dettin L, Pera RA, Herr JC. et al. Generation and in vitro differentiation of a spermatogonial cell line. Science. 2002;297:392-395
10. Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y. et al. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet. 1999;21:115-118
11. Klapper W, Heidorn K, Kuhne K, Parwaresch R, Krupp G. Telomerase activity in 'immortal' fish. FEBS Lett. 1998;434:409-412
12. Klapper W, Singh KK, Heidorn K, Parwaresch R, Krupp G. Regulation of telomerase activity in quiescent immortalized human cells. Biochim Biophys Acta. 1998;1442:120-126
13. Kishi S, Uchiyama J, Baughman AM, Goto T, Lin MC. et al. The zebrafish as a vertebrate model of functional aging and very gradual senescence. Exp Gerontol. 2003;38:777-786
14. Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546-552
15. Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA. et al. Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 2001;61:7594-7602
16. Avilion AA, Piatyszek MA, Gupta J, Shay JW, Bacchetti S. et al. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56:645-650
17. Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP. et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349-352
18. Beck S, Jin X, Sohn YW, Kim JK, Kim SH. et al. Telomerase activity-independent function of TERT allows glioma cells to attain cancer stem cell characteristics by inducing EGFR expression. Mol Cells. 2011 to appear
19. Nitta E, Yamashita M, Hosokawa K, Xian M, Takubo K. et al. Telomerase reverse transcriptase protects ATM-deficient hematopoietic stem cells from ROS-induced apoptosis through a telomere independent mechanism. Blood. 2011 to appear
20. Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci U S A. 1997;94:9202-9207
21. Kuramoto M, Ohsumi K, Kishimoto T, Ishikawa F. Identification and analyses of the Xenopus TERT gene that encodes the catalytic subunit of telomerase. Gene. 2001;277:101-110
22. Delany ME, Daniels LM. The chicken telomerase reverse transcriptase (chTERT): molecular and cytogenetic characterization with a comparative analysis. Gene. 2004;339:61-69
23. Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723-1730
24. Guo W, Okamoto M, Park NH, Lee YM. Cloning and expression of hamster telomerase catalytic subunit cDNA. Int J Mol Med. 2001;8:73-78
25. Nasir L, Gault E, Campbell S, Veeramalai M, Gilbert D. et al. Isolation and expression of the reverse transcriptase component of the Canis familiaris telomerase ribonucleoprotein (dogTERT). Gene. 2004;336:105-113
26. Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P. et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785-795
27. Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH. et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955-959
28. Banik SS, Guo C, Smith AC, Margolis SS, Richardson DA. et al. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol Cell Biol. 2002;22:6234-6246
29. Wittbrodt J, Shima A, Schartl M. Medaka--a model organism from the far East. Nat Rev Genet. 2002;3:53-64
30. Li M, Hong N, Xu H, Yi M, Li C. et al. Medaka vasa is required for migration but not survival of primordial germ cells. Mech Dev. 2009;126:366-381
31. Hong N, Li M, Zeng Z, Yi M, Deng J. et al. Accessibility of host cell lineages to medaka stem cells depends on genetic background and irradiation of recipient embryos. Cell Mol Life Sci. 2010Apr;67(7):1189-1202
32. Hong Y, Winkler C, Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes). Mech Dev. 1996;60:33-44
33. Hong Y, Winkler C, Schartl M. Production of medakafish chimeras from a stable embryonic stem cell line. Proc Natl Acad Sci U S A. 1998;95:3679-3684
34. Hong Y, Liu T, Zhao H, Xu H, Wang W. et al. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc Natl Acad Sci U S A. 2004;101:8011-8016
35. Yi M, Hong N, Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430-433
36. Hong Y, Winkler C, Liu T, Chai G, Schartl M. Activation of the mouse Oct4 promoter in medaka embryonic stem cells and its use for ablation of spontaneous differentiation. Mech Dev. 2004;121:933-943
37. Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121:605-618
38. Bejar J, Hong Y, Schartl M. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development. 2003;130:6545-6553
39. Wang D, Manali D, Wang T, Bhat AN, Hong N, Li Z, Wang L, Yan Y, Liu R, Hong Y. Identification of Pluripotency Genes in the Fish Medaka. Int J Biol Sci. 2011;7:440-451
40. Marion RM, Blasco MA. Telomeres and telomerase in adult stem cells and pluripotent embryonic stem cells. Adv Exp Med Biol. 2011;695:118-131
41. Klapper W, Kuhne K, Singh KK, Heidorn K, Parwaresch R. et al. Longevity of lobsters is linked to ubiquitous telomerase expression. FEBS Lett. 1998;439:143-146
42. Yu Chen EX RM, Kong RY, Ng PK, Mok HO. et al. Hypoxia induces telomerase reverse transcriptase (TERT) gene expression in non-tumor fish tissues in vivo: the marine medaka (Oryzias melastigma) model. BMC Mol Biol. 2006;7:27
43. Pfennig F, Kind B, Zieschang F, Busch M, Gutzeit HO. Tert expression and telomerase activity in gonads and somatic cells of the Japanese medaka (Oryzias latipes). Dev Growth Differ. 2008;50:131-141
Author contact
![]() Corresponding author: Prof. Yunhan Hong, Department of Biological Sciences, National University of Singapore, 10 Kent Ridge Crescent, Singapore 119260. Fax: +65 6779 2486; Tel: +65 6516 2915; Email: dbshyhedu.sg
Corresponding author: Prof. Yunhan Hong, Department of Biological Sciences, National University of Singapore, 10 Kent Ridge Crescent, Singapore 119260. Fax: +65 6779 2486; Tel: +65 6516 2915; Email: dbshyhedu.sg

 Global reach, higher impact
Global reach, higher impact