10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(8):1156-1167. doi:10.7150/ijbs.5033 This issue Cite
Review
Insights on Foxn1 Biological Significance and Usages of the “Nude” Mouse in Studies of T-Lymphopoiesis
Department of Molecular Biology and Immunology, University of North Texas Health Science Center at Fort Worth, Fort Worth, TX, 76107, USA.
*Current address: Affiliated Tumor Hospital of Guangzhou Medical College, Guangzhou city, Guangdong Province, 510095, China.
Received 2012-8-10; Accepted 2012-9-13; Published 2012-9-24
Abstract
Mutation in the “nude” gene, i.e. the FoxN1 gene, induces a hairless phenotype and a rudimentary thymus gland in mice (nude mouse) and humans (T-cell related primary immunodeficiency). Conventional FoxN1 gene knockout and transgenic mouse models have been generated for studies of FoxN1 gene function related to skin and immune diseases, and for cancer models. It appeared that FoxN1's role was fully understood and the nude mouse model was fully utilized. However, in recent years, with the development of inducible gene knockout/knockin mouse models with the loxP-Cre(ERT) and diphtheria toxin receptor-induced cell abolished systems, it appears that the complete repertoire of FoxN1's roles and deep-going usage of nude mouse model in immune function studies have just begun. Here we summarize the research progress made by several recent works studying the role of FoxN1 in the thymus and utilizing nude and “second (conditional) nude” mouse models for studies of T-cell development and function. We also raise questions and propose further consideration of FoxN1 functions and utilizing this mouse model for immune function studies.
Keywords: FoxN1 gene, T-Lymphopoiesis
Introduction
The nude mutation [1] in the gene (FoxN1, forkhead box N1) [2-4], which encodes a transcriptional factor for the family of forkhead proteins, is responsible for this defect and has been known for a long time. The FoxN1 (former name: Whn or Hfh11) gene, located on chromosome 11 in mice and chromosome 17 in humans [5-9], is mainly expressed in thymic epithelium, distinct keratinocyte populations in the epidermis, and hair follicles. FoxN1 in rodents and FOXN1 in humans are highly conserved in sequence and function [5, 9]. Mutations in FoxN1 cause inborn dysgenesis of the thymus (thymic rudiment and lack of lymphocytes) [10-13] and hairless skin (short and bent hair shafts inside the skin) [9, 13-15], which happen in mice, rats, and humans. The FOXN1 mutation in humans causes human nude (alopecia and nail dystrophy) and results in a primary T-cell deficiency [13, 16-18] related to severe infections, whereas the FoxN1 mutation in mice results in the generation of nude mice, which have been widely used as a model [19, 20] for experimental oncological, immunological, dermatological, and transplantation studies due to their immune deficiency in T-cell development and failure in hair follicle development (nude skin). However, in recent years, comprehensive understanding of the nude gene in the thymus and utility of nude mouse models for immunology and cancer studies are just now emerging. For example, with molecular technology moving forward, such as the development of the loxP-Cre/-CreERT system [21-23], it appears that the precise roles of FoxN1 are just beginning to be unveiled. In this review, we summarize recent findings in ongoing attempts to determine the functions of FoxN1 in the thymus and to utilize nude and “second (conditional) nude” mouse models for studies of T-lymphopoiesis and T-cell function.
1. General roles of FoxN1 in the thymus, skin, and possibly, the neuronal system
Generally, FoxN1, a transcription factor, acts through its target genes in order to regulate the differentiation of epithelial cells. Specifically, FoxN1 regulates keratinocytes to differentiate under proliferating conditions [24, 25]. The typical phenotypes resulting from an inborn null mutation of FoxN1 are developmental failures in the skin and thymic epithelium [15]. Maturation of the thymic epithelial meshwork during thymic organogenesis occurs in two genetic stages [12, 26] - the first stage involves FoxN1-independent induction and outgrowth of the thymic epithelial anlage from the third pharyngeal pouch, controlled by genes such as the Eya1 and Six [27], Hoxa3 [28], and Tbx1 [29, 30]. The second genetic step involves epithelial patterning and differentiation, which is FoxN1-dependent differentiation of the immature epithelial cells into functional cortical TECs (cTECs) and medullary TECs (mTECs). Recent reports emphasize FoxN1 as a powerful regulator that promotes differentiation in both the cTECs and mTECs during thymus organogenesis [31]. FoxN1 expression in the thymus is ambiguously believed to be expressed in all fetal TECs but not in all adult TECs [32, 33], while FoxN1-negative TECs are reported to be derived from FoxN1+ TECs [33]. Therefore, which TEC subsets lose FoxN1 with age, and why these subsets lose FoxN1 with age has yet to be clearly identified. An inborn null mutation in FoxN1 [3] causes a differentiation failure in TECs thereby halting thymic development at a rudimentary stage—the thymic lobe is still present but thymic lymphopoiesis is completely blocked [12, 34]. This causes an alymphoid thymus and severe primary T-cell immunodeficiency in nude mice and humans [8, 35, 36] with congenital alopecia and defective immunity, resulting in death in early childhood from severe infections [37, 38]. Therefore, the FOXN1 mutation is a severe human primary immunodeficiency disease [13, 16-18, 39, 40].
In the skin, FoxN1 is required for normal hair follicle development regulating the initiation of keratinocyte terminal differentiation, which has been well reviewed [9, 15]. FoxN1 expression, mainly in the hair shaft cortex, was reported to peak during anagen—hair growth period, then fall during catagen (destruction) and telogen (rest) [9, 14, 15]. Recently, FoxN1 was also reported to regulate pigmentation in the skin (related to skin darkness), demonstrated by using an engineered—keratin-5-driven FoxN1 (K5-FoxN1) transgenic (Tg) mouse [41]. These authors found that while the FoxN1-null nude mouse completely lacks pigmentation in the hair cortex, K5-FoxN1 Tg confers ectopic acquisition of pigmentation in hair cortical cells. This is said [41] to be due to regulation via the FoxN1-Fgf2 regulatory axis on pigment transfer from melanocytes to keratinocytes.
Although skin and thymus phenotypes resulting from the inborn FoxN1-null mutation are well-known, central nervous system deterioration, such as anencephaly, during the organogenesis resulting from this mutation has only recently been reported [36]. The Amorosi group found that FoxN1 is expressed in the brain choroid plexus of murine embryos by using a FoxN1 heterozygous mouse, in which one copy of FoxN1 bears an inserted β-galactosidase (LacZ) reporter gene [12]. However, this leads us to ask why FoxN1-null gene knockout mice do not show a neural tube defect, and why do not all FoxN1 deficient human fetuses have a neural tube defect [42]? Consequently, whether FoxN1 mutation really causes congenital brain developmental abnormalities remains to be confirmed.
2. Introduction of conditional gene/cell manipulated system into FoxN1 studies
FoxN1 gene-manipulated mouse models, such as loss-of-function [12, 43] and gain-of-function [41, 44] models, have been available in the studies of FoxN1 gene function. However, with molecular and cellular technology moving forward, many new methodologies have been developed. For example, the loxP-Cre/CreERT-mediated conditional FoxN1 gene “loss-resumption or revert” [31, 45] and the conditional FoxN1 gene knockout [46] mouse models have been developed in recent years. This facilitates the determination of the precise roles of FoxN1.
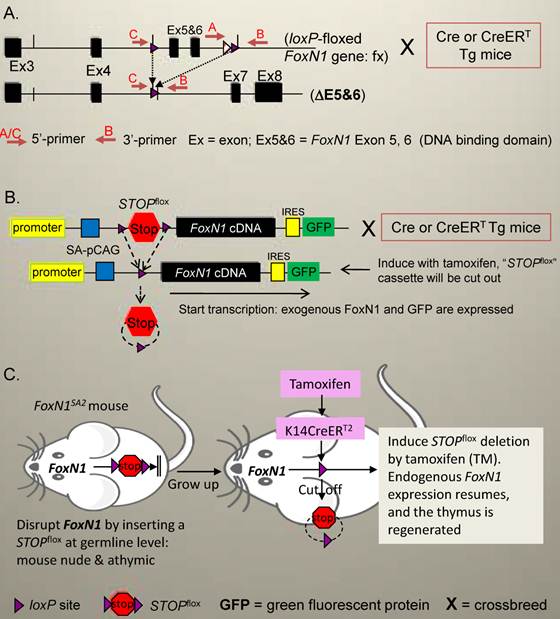
These systems have been used to artificially (conditionally) control gene expression (conditional knockout or over-expression) in the mouse for a couple of decades [21-23]. The loxP-Cre/CreERT system centered on the Cre gene, short for cyclization recombination [47]. The Cre gene encodes a site-specific DNA recombinase, which can recombine DNA at specific sites, which are 34-base pairs long, known as loxP (locus of X-over P1) sequences. These sequences act as magnets for Cre to recombine the DNA fragment in between the two loxP sites, resulting in recombination-excision (deletion) of the loxP-flanked DNA fragment. If this excised DNA fragment is a functional part of a gene, its deletion will cause this gene to become dysfunctional. For example, in the conditional FoxN1 gene knockout mouse (this mouse is now available at the Jackson Laboratory, #012941, http://jaxmice.jax.org/strain/012941.html) [46], the DNA binding domain, i.e. functional domain of transcription factor FoxN1, located on exons 5 and 6 of the FoxN1 locus [48, 49], is flanked by two loxP genes (Fig. 1A). When this domain is deleted (termed ΔE5&6) by Cre or CreERT, the FoxN1 gene loses its function [46]. If this DNA fragment is a loxP-flanked STOP cassette (STOPflox), a roadblock sequence positioned upstream of a functional gene or cDNA, the deletion of this STOPflox will cause the gene to be re-expressed (Fig. 1B) or to resume (Fig. 1C). For example, in our unpublished novel STOPflox -FoxN1 transgenic mouse (Fig. 1B, this mouse is available by request), the flag-FoxN1 cDNA (kindly provided by Dr. Brissette [25]) carried by a composite of CMV-immediate early gene enhancer/chicken β-actin promoter (pCAG) (kindly provided by Dr. McMahon [50, 51]) was inserted into a backbone of the Rosa26 locus. In the front and the end of this fragment a STOPflox cassette and IRES-GFP reporter gene were inserted, respectively. This makes conditional expression of the FoxN1 transgene controlled by Cre/CreERT. Furthermore, in conditional FoxN1 gene “loss-resumption” or reversible mouse models [31, 45], a STOPflox cassette (including two splice acceptors and a hygromycin or neomycine cassette) flanked by loxP sites is inserted into a normal FoxN1 gene, which destroys and silences normal FoxN1 transcription, resulting in an inborn mutant phenotype during organogenesis. After this STOPflox cassette is depleted by introduction of Cre/CreERT, the endogenous FoxN1 expression resumes and the phenotype is reversed (Fig. 1C is an example).
Since Cre can be driven by different tissue-specific promoters, it can be uniquely expressed in certain tissues but not in others. Therefore tissue-specific promoter-driven Cre can achieve tissue-specific loxP-flanked DNA fragment deletion. This is one mechanism of conditional gene expression. The other mechanism is temporally-controlled gene expression in somatic cells rather than in germline cells. This can be achieved by Cre-ERT gene [52, 53], which is the Cre-recombinase fused to a mutated ligand binding-domain of the human estrogen receptor (ER). The estrogen receptor binding-domain represses Cre in an inactive state until de-repressed by Tamoxifen (TM), because the ER binds TM but not estrogens. Therefore, deletion of the loxP-floxed DNA fragment is induced by administration of TM but not mouse or human estrogens [53, 54]. By combining a tissue-specific promoter with CreERT, the loxP-flanked DNA fragment deletion can be controlled in a spatio-temporal fashion, thereby facilitating the introduction of a somatic mutation in a given gene, at a chosen time, in a selected cell type [21-23]. Particularly, this system benefits the study of the later roles of genes whose mutations cause early embryonic lethal phenotypes. Although a mutation of FoxN1 will not cause lethality in embryos, its roles in the developed postnatal thymus and in different keratin-type epithelial cells would have largely remained unknown without the FoxN1flox mouse model.
A new system for cell lineage ablation, based on transgenic expression of a diphtheria toxin receptor (DTR) carried by cell lineage specific gene and induced cyto-ablation via injection of diphtheria toxin (DT) has been developed in recent years [55-58]. Recently, this approach was used in the study of a FoxN1-positive thymic epithelial cell lineage [33]. Dr. Boehm's group clearly showed that after specific FoxN1+ TEC lineage was killed (cytoablation) by induction with DT in early embryogenesis, the orthotopic thymus becomes aplastic, and these TECs cannot fully regenerate.
3. Identical or distinct roles of FoxN1 in the skin and thymus
Although the general role of FoxN1 is to regulate the differentiation of epithelial cells in the thymus and skin, it was largely unknown whether the roles of FoxN1 in the thymus and skin are identical. If not, then how might they differ? The overt differences in FoxN1's roles in the thymus and skin were revealed in a recently published paper [59]. One important difference is that FoxN1 is involved in morphogenesis and maintenance of the three-dimensional (3D) thymic micro-structure, which is important for a functional thymus. As we know, two-dimensional (2D)-monolayer (non-Notch ligand transformed [60]) stromal cells cannot support T-cell development in culture. However re-aggregated stromal cell-constituted 3D pseudo-thymic lobes can fully support T-cell development in a fetal thymic organ culture (FTOC) setting. This is, at least in part, due to the alteration of certain key molecules. For example, dissociated thymic stromal cells lost the Notch ligand Delta-like expression, while re-aggregated thymic stromal cells (3D) regained its expression [61]. However, the normal micro-structure in the skin is two-dimensional or polarized, i.e. the epithelial layer (basal layer) on one side expresses keratin (K)5 and K14, and the epithelial layer (apical layer) on the other side expresses K8 and K18. The other important difference is that FoxN1 determines the pigmentation pattern in the skin [41], but this is inapplicable in the thymus.
Schematic diagram of Cre/CreERT-mediated loxP-deletion system in the FoxN1 gene in mice. (A) FoxN1 conditional gene knockout system [46]: FoxN1 functional domain (exons 5 and 6, a DNA binding domain) is flanked by two loxP sites (termed “fx”). After introduction of Cre or CreERT transgene (Tg) into these mice by crossbreeding (termed as “X”), and induction with tamoxifen (TM, only for CreERT Tg), the loxP-flanked exons 5 and 6 are cut out (termed “ΔE5&6”), and the FoxN1 gene loses its function (knockout). (B) FoxN1 conditional transgenic system (under development): FoxN1 cDNA (exogenous FoxN1) driven by an enhanced promoter and followed by a GFP reporter gene will be targeted into a housekeeping gene, such as Rosa26. Meanwhile, a loxP flanked “STOP” cassette (STOPflox), a roadblock sequence, is placed upstream of FoxN1 cDNA to block FoxN1 expression initially. As soon as tamoxifen is administrated (for CreERT Tg), the roadblock STOPflox is deleted and transcription of exogenous FoxN1 cDNA is turned on, and accompanied by GFP expression. (C) FoxN1 resumption (loss- resumption) system [45]: A STOPflox cassette (including two splice acceptors and a hygromycin cassette flanked by loxP sites) is inserted into the middle of the normal FoxN1 gene, for example, just after exon 6, which destroys normal FoxN1 splicing and silences FoxN1 transcription. After the introduction of CreERT Tg, such as K14-CreERT, into these mice, and induced activation of K14-CreERT with tamoxifen, the STOPflox is cut out, and endogenous FoxN1 expression resumes.
Additionally, using K14Cre transgenic mice [62] to delete FoxN1flox in K14 promoter-driven epithelial cells seems to have a larger impact on the skin than on the thymus [59]. Deletion of FoxN1flox in K14 epithelial cells is sufficient to cause a hair follicle defect resulting in a nude phenotype, similar to that of the natural FoxN1-null mutant mice, but does not induce an alymphoid thymic rudiment, thus differing from the thymic phenotype of the natural FoxN1-null mutant mice. It is unclear whether this phenotype is a result of low versus high expression of K14Cre in the thymus versus the skin. By using a K14Cre-mediated LacZ expression mouse model, which was generated by crossing K14Cre mice with R26-STOPflox-LacZ reporter mice, in which the STOPflox cassette is deleted upon K14Cre expression, thereby subjecting LacZ expression to be controlled by K14 promoter. Jackson Laboratory confirmed that the K14Cre-mediated LacZ is strongly expressed in the postnatal thymus, particularly in the thymic medulla (several images are posted in Jackson Laboratory web site: http://cre.jax.org/Krt14/Krt14-creNano.html). Therefore, the difference observed by Guo et al. should be due to the different impacts of FoxN1 on K14 epithelial cells in the skin and thymus, rather than a result of lower expression of K14Cre in the thymus.
4. Roles of FoxN1 in the prenatal only or both prenatal and postnatal thymus during thymic epithelial cell development and homeostasis
As mentioned previously, there are FoxN1-independet and -dependent genetic stages, during thymic organogenesis and TEC differentiation [12, 26]. Owing to the lack of suitable genetic tools to address it, there was a long-running argument centered on whether FoxN1 continues to maintain a functional thymus following the second genetic stage of thymic organogenesis, especially in the adult thymus. Gordon et al. generated a FoxN1-LacZ mouse model [63], in which a LacZ cDNA cassette was inserted into the 3'UTR of the FoxN1 locus. Chen et al. observed that LacZ has an adverse effect on FoxN1 expression with age, via a supposed methylation mechanism, to induce thymic postnatal involution [64]. Therefore, FoxN1 was experimentally demonstrated to be required in the postnatal thymus. Because this mouse model cannot be spatio-temporally controlled, precise information of defects in timing and TEC subsets is not available, whereas, the inducible FoxN1flox gene knockout mouse model can be used for addressing these question [46].
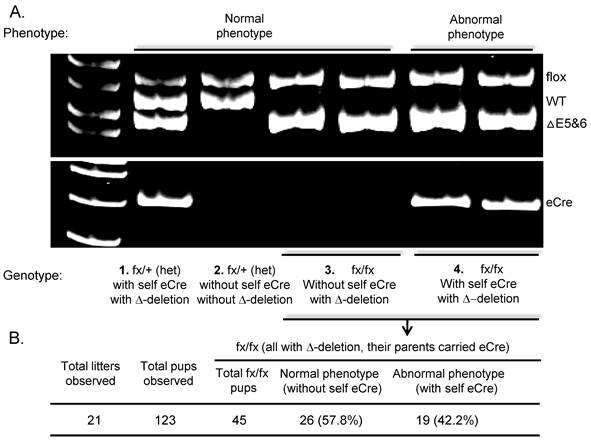
Recently, using FoxN1flox-K14Cre mice, the Guo et al. [59] demonstrated that under certain circumstances the postnatal role of FoxN1 may be even more important than its prenatal role. They found that homozygous FoxN1flox/flox mice without the Cre gene have FoxN1flox deletion (Fig. 2A, genotype case #3) when their mother has the Cre gene. These mice have completely normal phenotypes in the thymus and skin. This deletion record comes from a historic Cre-mediated FoxN1flox deletion and should happen in their prenatal life inside the mother's uterus. However, homozygous FoxN1flox/flox mice carrying their own Cre gene have a FoxN1flox deletion (Fig. 2A, genotype case #4) and display mutant phenotypes in the thymus and skin. This FoxN1flox deletion should happen in both prenatal and postnatal life. This finding demonstrated that FoxN1 deletion happened inside the mother's uterus, driven solely by the parent's K14Cre (no Cre gene in offspring), which does not induce mutant phenotypes in the thymus and skin of the offspring. Instead, only when FoxN1flox is deleted in both prenatal (mediated by mother's Cre) and postnatal (mediated by self Cre) are the mutant phenotypes in the thymus and skin induced. This confirmed the importance of postnatal FoxN1. However, this phenotype could not have been revealed without the loxP-Cre system because in the naturally occurring FoxN1-null mutation the FoxN1 gene cannot be deleted separately in prenatal and postnatal life. Furthermore, this phenotype is not only found in FoxN1flox-K14Cre mice, but also in other FoxN1flox-Cre (resulting in germline deletion) mice, such as in ubiquitous FoxN1flox-Cre mice (EIIa-Cre, Jackson Lab #003724) (Fig. 2). Therefore, FoxN1 is required not only for prenatal epithelial patterning (previously known) but also crucial for postnatal epithelial homeostasis. Prenatal deletion (mediated by Cre inside the mother's uterus) of FoxN1 alone cannot induce mutant phenotypes, but both prenatal and postnatal deletion (via offspring's own Cre) are able to induce thymic and skin mutant phenotypes.
The importance of FoxN1's role in the postnatal thymus, beyond the second genetic stage [12] of thymic organogenesis in the fetal thymus is indisputable. Furthermore, this raises two intriguing issues: 1) FoxN1's probable role in the postnatal thymus is to regulate epithelial cell homeostasis. 2) Since postnatal TECs continue to undergo homeostasis, this process should be supported by tissue-specific stem/progenitor cells in situ. Therefore, adult thymic epithelial stem/progenitor cells probably exist in the postnatal thymus. This is a glaring issue and is discussed in the following section.
Observe mutant phenotypes in mice with prenatal (mediated by parents' Cre, without self Cre) deletion of FoxN1, or with both prenatal and postnatal (mediated by parents' and self Cre) deletion of FoxN1. (A) A representative result of genomic DNA PCR using primers shown in Fig. 1A. Linking this result with the phenotype, we found that homozygous fx/fx (FoxN1flox/flox) mice possessed ΔE5&6 recombination band but did not carry self eCre (EIIaCre - transgenic mouse from Jackson Lab #003724) - genotype #3 in the figure, did not show abnormal phenotypes in the skin and thymus. Therefore, the genotype #3 animals' ΔE5&6 recombination band came from their parents' eCre (their mother carried an eCre transgene). However, homozygous fx/fx mice carrying self eCre with a ΔE5&6 recombination band - genotype #4 in the figure, possessed abnormal phenotypes. (B) Table showing observed total fx/fx mice with/without self eCre Tg, linked to abnormal/normal phenotypes. Although they all have a ΔE5&6 recombination band, the abnormal phenotypes can only be seen in the mice with self eCre Tg.
5. Are all TEC subsets equally FoxN1-dependent in the postnatal thymus?
Based on anatomic, keratin type, and functional criteria, thymic epithelial regions can be divided into the cortex and medulla. The TECs in the cortical region are called cTECs, while those in the medullary region are referred to as mTECs. Both epithelia provide different microenvironments that are responsible for distinct stages in thymocyte development [26, 65]. As we know, FoxN1 is required for the development of both cTECs and mTECs in the fetal stage [12]. However, since FoxN1 is expressed in almost all TECs of the embryonic thymus, but not in all TECs of the adult thymus [32, 33], FoxN1's role in different TEC subsets in the prenatal and postnatal thymus does not seem to be identical. Therefore the question arises, is FoxN1 equally required for homeostasis of both cTECs and mTECs in the postnatal thymus?
In the fetal stage, FoxN1 mainly regulates TEC patterning. Since both cTECs and mTECs arise from the same bi-potential TEC progenitors [45, 66], and FoxN1 regulates the process of differentiation, it is straightforward to conclude that both mTECs and cTECs are equally FoxN1-dependent during fetal thymic organogenesis [67]. However, in the postnatal thymus, mTECs with the keratin type K5+ and K14+, which are similar to epithelial stem cell markers and exhibit progenitor activity in the skin and mammary gland [68-70], were more sensitive to the loss of FoxN1. cTECs possess keratin type K8+ and K18+, with the same marker as mature epithelial cells and terminally differentiated epithelial cells in the apical layer of stratified squamous epithelium of the skin. Additionally, cTECs are not as sensitive as mTECs to the loss of FoxN1 using K5- and K18-CreERT-mediated FoxN1flox-deletion mouse models [46]. This finding in the postnatal thymus may account for the fact that K8+/K18+ cTECs and K5+/K14+ mTECs are not equally FoxN1-dependent in the postnatal thymus. Although it may be due to a long half-life in cTECs compared to that in mTECs, FoxN1 may not be required for mature/differentiated epithelia, which have K8+/18+ markers in the thymic cortical region and skin epithelial apical layer. However, FoxN1 should regulate the immature/undifferentiated TECs (epithelial progenitor cells), which may be a small subpopulation present in the K5+/K14+ TEC populations, located in the medullary region and/or the corticomedullary junction (CMJ) in the postnatal thymus. Specifically, FoxN1 in the adult thymus is required for adult TEC progenitors [33], which express K5+/K14+ markers, and these progenitors support TEC homeostasis in the adult thymus. A recent report further confirms this, showing that thymopoiesis depends on a FoxN1-positive TEC lineage, while FoxN1-negative TECs are descendants of FoxN1-positive TECs. FoxN1-negative TECs do not contribute to thymopoietic function in the adult thymus [33]. Further support of the existence of postnatal TEC progenitors, which are dependent on FoxN1, was made in Osada et al 2010 [71], where premature thymic involution was observed after inhibition of Wnt signaling through conditional expression of Dkk1 resulting in a decline in FoxN1 expression and loss of TEC progenitors.
6. Thymus development is sensitive to FoxN1 dosage: it can neither be insufficient nor excessive
Further progress in the recognition of FoxN1 function in recent years was made by two reports that determined whether thymus development is sensitive to the genetic dosage of FoxN1 and the association with age-related thymic involution. They also determined if heterozygous FoxN1 (a half genetic dose of FoxN1, i.e. FoxN1nu/+), which is known to be sufficient to induce TEC patterning in the thymic organogenesis, is also sufficient to maintain homeostasis for a steady-state normal thymus in the postnatal life. One report [64] showed that the mutant phenotype is dependent on FoxN1 genetic dosage. The thymus in wild type (WT) mice is completely normal, and it is completely abnormal in FoxN1-null mice (natural FoxN1-null nude mouse). The abnormality lies in between these two extremes for FoxN1-null heterozygote (nu/+) and LacZ/nu chimera mice. The degree of severity is: WT < nu/+ < LacZ/nu < null in a genetic dose-dependent manner. Another study [72] using a FoxN1flox mouse carrying a ubiquitous CreERT transgene (uCreERT), that took advantage of a low-dose spontaneous Cre leakage due to incomplete ER blockage in vivo [73], found that spontaneous leakage of uCreERT caused FoxN1flox deletion accompanying a progressive loss of FoxN1+ TECs with accelerated age-related thymic involution. This also occurred in heterozygous FoxN1flox/+ mice (deletion of floxed-FoxN1 in one copy of FoxN1 gene), representing a haplo-insufficient phenotype but related to age, i.e. age-related haplo-insufficiency. This finding extends previous observations in adult natural FoxN1-nu/+ heterozygous mice [74, 75].
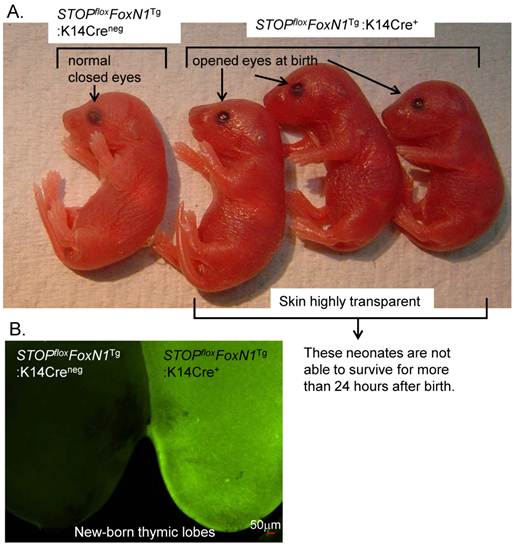
Expression of FoxN1 in the thymi of naturally middle-aged and aged WT mice is significantly reduced [76]. By increasing the dosage of FoxN1 in these thymi via intrathymic administration of exogenous FoxN1-cDNA, a rapid gain-of-function approach, thymic involution and declining thymic function can be partially rescued [72]. Dr. Le's group confirmed this hypothesis at the genetic level with their FoxN1 transgenic mouse model, which showed that an up-regulation of FoxN1 expression in the aged thymus can rejuvenate function of the atrophied thymus [44]. In their experiments, they ingeniously selected the FoxN1 transgenic mice with low copy numbers for their observations. We found that highly over-expressed FoxN1 induces adverse effects on thymus development (data unpublished), and even causes a lethal new-born phenotype (Fig. 3). In our newly generated STOPflox-FoxN1 transgenic mice (Fig. 1B), the FoxN1 cDNA (kindly provided by Dr. Brissette [25]) is driven by the Rosa26 promoter and enhanced expression by a composite of CMV-immediate early gene enhancer/chicken β-actin promoter (pCAG) (kindly provided by Dr. McMahon [50, 51]). This results in high over-expression of FoxN1. In Figure 3, we show that K14Cre-mediated STOPflox-FoxN1 transgenic new-born mice died within 24 hours of birth. These neonatal mice share similar phenotypes with the involucrin promoter-driven FoxN1 transgenic neonatal mice, which have ectopic and enhanced expression of FoxN1 [25], displaying abnormal skin and possessing open eyes at birth.
7. Utilizations of the nude or second nude mouse models in studying a T-cell developmental microenvironment and autoimmunity
The nude mice or second nude (inducible FoxN1 gene knockout) mice provide animal models to facilitate studies of T-cell development and postnatal T-cell function in immunity and autoimmunity related to human disease. Recently, Dr. Boehm's group designed elegant experiments by using a FoxN1-null (nude mouse) model to reveal that thymic epithelia possess synergistic, context-dependent, and hierarchical functions in lymphopoiesis [77]. TECs of FoxN1-null mice were transformed by cDNAs of the chemokines Ccl25 and Cxcl12, the cytokine Scf, and the Notch ligand DLL4 carried by the FoxN1-promoter to generate FoxN1-Ccl25, -Ccl12, -Scf, and -DLL4 transgenic mice under the FoxN1-null background. In these transgenic embryonic thymi, they found precise environmental components that can support mast cells, B progenitor cells, and T progenitor cells, respectively [77]. Another recent work using a tissue-specific FoxN1flox gene knockout mouse model studied influenza infection in aging [78]. They found that K14Cre-mediated FoxN1flox deletion-induced defects in the thymic medulla reduced antigen-specific CD8+ T-cell and IgG responses to influenza virus, combined with increased lung injury, weight loss and mortality. These findings provided the first evidence that defects in the medulla directly causes changes in T-cell function that mimics aging defects during an immune response to an infectious agent [78]. A third recent work using the second nude (conditional FoxN1flox gene knockout) mouse model addressed possible mechanisms of increased autoimmune susceptibility in the elderly [79]. Age-related disruption of steady-state thymic medulla caused by two-dimensional thymic epithelial cysts, primarily generated in the medulla, was found to perturb thymocyte negative selection. Negative selection is the main mechanism for the generation of central immune tolerance [80] necessary to prevent autoimmune diseases. This disruption was confirmed to provoke autoimmune phenotypes, such as inflammatory cell infiltration in multiple organs and the generation of anti-nuclear antibodies [79].
Over-expression of FoxN1 in STOPflox-FoxN1 transgenic mice mediated by K14Cre results in neonatal lethality. (A) Image of eye opening can be seen in STOPfloxFoxN1Tg-K14Cre+ neonates (right three neonates), but not in control STOPfloxFoxN1Tg-K14Cre-neg neonate (left). (B) Green fluorescence is shown in STOPfloxFoxN1Tg-K14Cre+ neonatal thymus (right), but not in control STOPfloxFoxN1Tg-K14Cre-neg neonatal thymus (left).
8. Outstanding questions and future directions
Although much progress has been made in recent years in unveiling the roles of FoxN1 using advanced technology, quite a few questions in this field still exist. For example, if FoxN1 is involved in the regulation of adult thymic epithelial (TE) stem/progenitor cells, more experiments are required to understand the localization and characteristics of individual TE stem/progenitor cells from the postnatal thymus. There is an ongoing debate [81] about whether the adult thymic epithelial stem cells even exist and where they may be located, even though epithelial stem/progenitor cells have been implicated in the FoxN1 “loss-resumption or revertible” (FoxN1SA2) adult thymus [45]. In the FoxN1SA2 mice, a loxP-flanked hygromycin cassette was inserted into FoxN1 introns 6-7 (Fig. 1C). This insertion destroys normal FoxN1 splicing and silences the gene. The mice have a nude phenotype and a defective thymus. Upon hK14-CreERT transgene activation in the adult thymus the insertion is deleted, and FoxN1 is re-expressed in putative adult TE stem/progenitor cells, which can differentiate into normal cortical and medullary TECs and support normal thymic regeneration and function [45]. However, it can be argued that the TEC progenitors in the FoxN1SA2 defective thymus are dormant cells persisting from the fetal stage due to the FoxN1 mutation, which may not represent TE stem/progenitors in the normal adult thymus. Further work is required to resolve the debate.
Another clue for TE stem/progenitors presenting in the natural adult thymus is provided by the FoxN1flox-K5CreERT mouse models, in which the FoxN1 gene was conditionally knocked out in K5+ [46] epithelial cells after TECs fully developed in the adult thymus, resulting in acute thymic atrophy. This is probably due to the disruption of adult TEC homeostasis supported by TE stem/progenitor cells in the adult thymus. Since the K5 and/or K14+ promoters are active in epithelial stem/progenitor cells of the skin and mammary gland, the TE progenitors in the adult thymus may also be present within K5+ and/or K14+ TECs to support TEC homeostasis. However, the proportion and functional characteristics of adult thymic epithelial stem/progenitor cells that are K5+ and/or K14+ is largely unknown. Obtaining direct evidence by lineage-tracking changes in these adult thymic epithelial progenitors is a critical need.
Regulation of TEC homeostasis is possibly co-regulated by FoxN1 and other stem cell-related genes, such as p63. The transcription factor p63, which encodes for multiple isoforms (containing an N-terminal transactivation domain, termed as TAp63, and lacking this domain, termed as ΔNp63) [82], is pivotal for the development of stratified epithelial tissues, including the epidermis, breast, prostate, and thymus [83]. The role of p63 in thymic development may be considered to be essential for the proliferation potential of thymic epithelial stem/progenitor cells [84, 85]. Specifically, thymic development is considered to be regulated by the ΔNp63 isoform through the maintenance of epithelial progenitor “stemness”. By introducing ΔNp63 and TAp63 transgenes into a p63 gene knockout background, ΔNp63, but not TAp63, is able to rescue defective thymus development [84]. Recently, we obtained a clue that the role of TAp63 in the thymus is probably associated with TEC senescence, since it was increased with thymic aging and associated with an age-related increase of senescent cell clusters (Manuscript under preparation). This phenotype may be accelerated by a blockade of TEC differentiation via conditionally knocking out the FoxN1 gene. We suspect that p63 and FoxN1 may form a p63-FoxN1 regulatory axis in TEC homeostasis during aging. However, the mechanism controlling how the proliferation regulator p63 and differentiation regulator FoxN1 work collaboratively in the regulatory axis is still mysterious, and more studies are appreciated.
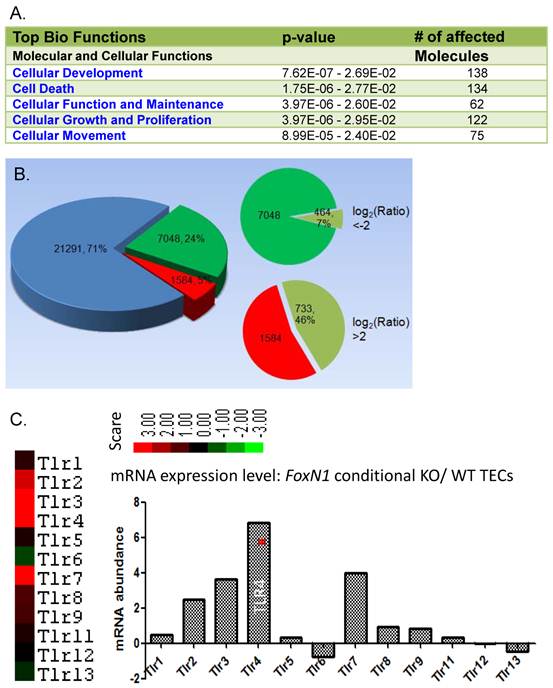
FoxN1 is a transcription factor whose functions are executed by targeting other genes through its DNA binding domain. Therefore, to understand its functional mechanisms in determining its target genes is important. However, the precise target genes that are regulated by FoxN1 remain ill defined, mostly due to technical difficulties in precisely isolating enough physiologically intact TECs at certain developmental stages. One group performed laser-capture micro-dissection to capture TECs from the E12.5 FoxN1-null nude mouse thymi, and found five FoxN1 target TEC genes in their microarray analysis, of which programmed cell death-1 ligand is the only gene of known function [86]. However, emerging studies via immuno-histological methods suggest that changes in these five genes in the FoxN1-null mutant thymus have been undervalued. For example, FoxN1 target Fgf2 has been identified in the skin [41]. FoxN1 may target the Notch ligands, DLL-1 and DLL-4 [87], and the chemokines, CCL25 and CXCL12 [88], in nude thymic anlages. It may also target Notch-1 receptor in the skin as demonstrated in a transgenic mouse model [89]. We also preliminarily analyzed FoxN1 targeting genes by using a microarray assay from FACS sorted CD45- EpCAM+ TECs. One TEC group was derived from ubiquitous-CreERT-mediated FoxN1flox knockout induced by TM in the postnatal thymus [46], the other group was WT mouse TECs. We found that at least 5 groups (Fig. 4A) in over 300,000 genes screened underwent significant changes, either increasing or decreasing. Changes of log2 ratio > 2 or < -2 were observed in 1197 genes (Fig. 4B). The most interesting gene family is toll-like receptor (TLR), in which TLR4 shows a significant increase (Fig. 4C). We are conducting further work on this gene family to determine its physiological significance. Chromatin immunoprecipitation (ChIP) on Chip and ChIP on Sequence approaches may be one of the best ways to determine FoxN1 target genes.
Microarray results of FoxN1 target genes. Total RNAs were isolated from flow cytometry sorted TECs (CD45- MHC-II+) of young mice from ubiquitous-CreERT-mediated FoxN1-deleted (tamoxifen x5 to conditionally delete FoxN1 postnatally) thymic pool and WT thymic pool. (A) Number of affected molecules in top five bio-function groups; (B) Number and % of genes with upward (red) /downward (green) changes in total arrayed genes. Number and % of genes down to log2 ratio < -2 and up to log2 ratio > 2 are given; (C) One of the most involved genetic networks was the Toll-like receptor (TLR) pathway. As shown TLR4 was significantly increased after the FoxN1 was conditionally knocked out.
Concluding remarks
Recent progress using advanced technology to study FoxN1's roles in the thymus shows that FoxN1 regulates not only TEC patterning in the fetal stage but also TEC homeostasis in the postnatal thymus. Comparing the thymus with the skin, FoxN1 has its own distinct roles and impacts on organs in the generation and maintenance of three-dimensional microstructure and pigmentation, respectively. FoxN1's role in the neuron has been brought up, but is still obscure. There is still plenty of room to apply nude and secondary nude (conditional FoxN1 gene knockout) mouse models in studies of immunology, hematology, and tumorgenesis. The functional mechanisms of FoxN1's collaborative roles with other genes during thymic development and aging remain to be further determined.
Acknowledgements
We would like to thank Dr. Yong Zhao (Institute of Zoology, Chinese Academy of Science) for his insightful discussions. We apologize to those colleagues whose work could not be referenced directly due to space constrains.
Funding support: This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health, USA (Grant numbers: R01AI081995) to D-M.S.
Conflict of interest
We do not have conflict of competing financial interest.
References
1. Flanagan SP. 'Nude', a new hairless gene with pleiotropic effects in the mouse. Genet Res. 1966;8:295-309
2. Takahashi Y, Shimizu A, Sakai T, Endo Y, Osawa N, Shisa H. et al. Mapping of the nu gene using congenic nude strains and in situ hybridization. J Exp Med. 1992;175:873-6
3. Nehls M, Pfeifer D, Schorpp M, Hedrich H, Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature. 1994;372:103-7
4. Byrd LG. Regional localization of the nu mutation on mouse chromosome 11. Immunogenetics. 1993;37:157-9
5. Schorpp M, Hofmann M, Dear TN, Boehm T. Characterization of mouse and human nude genes. Immunogenetics. 1997;46:509-15
6. Lisitsyn NA, Segre JA, Kusumi K, Lisitsyn NM, Nadeau JH, Frankel WN. et al. Direct isolation of polymorphic markers linked to a trait by genetically directed representational difference analysis. Nat Genet. 1994;6:57-63 doi:10.1038/ng0194-57
7. Nehls M, Luno K, Schorpp M, Krause S, Matysiak-Scholze U, Prokop CM. et al. A yeast artificial chromosome contig on mouse chromosome 11 encompassing the nu locus. Eur J Immunol. 1994;24:1721-3 doi:10.1002/eji.1830240742
8. Frank J, Pignata C, Panteleyev AA, Prowse DM, Baden H, Weiner L. et al. Exposing the human nude phenotype. Nature. 1999;398:473-4
9. Schlake T. The nude gene and the skin. Exp Dermatol. 2001;10:293-304 doi:100501 [pii]
10. Pantelouris EM. Absence of thymus in a mouse mutant. Nature. 1968;217:370-1
11. Pantelouris EM, Hair J. Thymus dysgenesis in nude (nu nu) mice. J Embryol Exp Morphol. 1970;24:615-23
12. Nehls M, Kyewski B, Messerle M, Waldschutz R, Schuddekopf K, Smith AJ. et al. Two genetically separable steps in the differentiation of thymic epithelium. Science. 1996;272:886-9
13. Pignata C, Fiore M, Guzzetta V, Castaldo A, Sebastio G, Porta F. et al. Congenital Alopecia and nail dystrophy associated with severe functional T-cell immunodeficiency in two sibs. Am J Med Genet. 1996;65:167-70
14. Lee D, Prowse DM, Brissette JL. Association between mouse nude gene expression and the initiation of epithelial terminal differentiation. Dev Biol. 1999;208:362-74
15. Mecklenburg L, Tychsen B, Paus R. Learning from nudity: lessons from the nude phenotype. Exp Dermatol. 2005;14:797-810
16. Albuquerque AS, Marques JG, Silva SL, Ligeiro D, Devlin BH, Dutrieux J. et al. Human FOXN1-deficiency is associated with alphabeta double-negative and FoxP3+ T-cell expansions that are distinctly modulated upon thymic transplantation. PLoS One. 2012;7:e37042
17. Markert ML, Marques JG, Neven B, Devlin BH, McCarthy EA, Chinn IK. et al. First use of thymus transplantation therapy for FOXN1 deficiency (nude/SCID): a report of 2 cases. Blood. 2011;117:688-96
18. Vigliano I, Gorrese M, Fusco A, Vitiello L, Amorosi S, Panico L. et al. FOXN1 mutation abrogates prenatal T-cell development in humans. J Med Genet. 2011;48:413-6
19. Shultz LD, Sidman CL. Genetically determined murine models of immunodeficiency. Annu Rev Immunol. 1987;5:367-403 doi:10.1146/annurev.iy.05.040187.002055
20. Shultz LD. Immunological mutants of the mouse. Am J Anat. 1991;191:303-11 doi:10.1002/aja.1001910310
21. Rajewsky K, Gu H, Kuhn R, Betz UA, Muller W, Roes J. et al. Conditional gene targeting. J Clin Invest. 1996;98:600-3 doi:10.1172/JCI118828
22. Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743-55
23. Kwan KM. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis. 2002;32:49-62
24. Brissette JL, Li J, Kamimura J, Lee D, Dotto GP. The product of the mouse nude locus, Whn, regulates the balance between epithelial cell growth and differentiation. Genes Dev. 1996;10:2212-21
25. Prowse DM, Lee D, Weiner L, Jiang N, Magro CM, Baden HP. et al. Ectopic expression of the nude gene induces hyperproliferation and defects in differentiation: implications for the self-renewal of cutaneous epithelia. Dev Biol. 1999;212:54-67
26. Rodewald HR. Thymus organogenesis. Annu Rev Immunol. 2008;26:355-88
27. Zou D, Silvius D, Davenport J, Grifone R, Maire P, Xu PX. Patterning of the third pharyngeal pouch into thymus/parathyroid by Six and Eya1. Dev Biol. 2006;293:499-512
28. Manley NR, Capecchi MR. The role of Hoxa-3 in mouse thymus and thyroid development. Development. 1995;121:1989-2003
29. Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286-91
30. Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T. et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97-101
31. Nowell CS, Bredenkamp N, Tetelin S, Jin X, Tischner C, Vaidya H. et al. Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet. 2011;7:e1002348
32. Itoi M, Tsukamoto N, Amagai T. Expression of Dll4 and CCL25 in Foxn1-negative epithelial cells in the post-natal thymus. Int Immunol. 2007;19:127-32
33. Corbeaux T, Hess I, Swann JB, Kanzler B, Haas-Assenbaum A, Boehm T. Thymopoiesis in mice depends on a Foxn1-positive thymic epithelial cell lineage. Proc Natl Acad Sci U S A. 2010;107(38):16613-8
34. Blackburn CC, Augustine CL, Li R, Harvey RP, Malin MA, Boyd RL. et al. The nu gene acts cell-autonomously and is required for differentiation of thymic epithelial progenitors. Proc Natl Acad Sci U S A. 1996;93:5742-6
35. Cunningham-Rundles C, Ponda PP. Molecular defects in T- and B-cell primary immunodeficiency diseases. Nat Rev Immunol. 2005;5:880-92
36. Amorosi S, D'Armiento M, Calcagno G, Russo I, Adriani M, Christiano AM. et al. FOXN1 homozygous mutation associated with anencephaly and severe neural tube defect in human athymic Nude/SCID fetus. Clin Genet. 2008;73:380-4
37. Adriani M, Martinez-Mir A, Fusco F, Busiello R, Frank J, Telese S. et al. Ancestral founder mutation of the nude (FOXN1) gene in congenital severe combined immunodeficiency associated with alopecia in southern Italy population. Ann Hum Genet. 2004;68:265-8
38. Pignata C, Fusco A, Amorosi S. Human clinical phenotype associated with FOXN1 mutations. Adv Exp Med Biol. 2009;665:195-206
39. Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4:889-99
40. Jonsson H, Peng SL. Forkhead transcription factors in immunology. Cell Mol Life Sci. 2005;62:397-409 doi:10.1007/s00018-004-4365-8
41. Weiner L, Han R, Scicchitano BM, Li J, Hasegawa K, Grossi M. et al. Dedicated epithelial recipient cells determine pigmentation patterns. Cell. 2007;130:932-42
42. Amorosi S, Vigliano I, Del Giudice E, Panico L, Maruotti GM, Fusco A. et al. Brain alteration in a Nude/SCID fetus carrying FOXN1 homozygous mutation. J Neurol Sci. 2010;298:121-3
43. Su DM, Navarre S, Oh WJ, Condie BG, Manley NR. A domain of Foxn1 required for crosstalk-dependent thymic epithelial cell differentiation. Nat Immunol. 2003;4:1128-35
44. Zook EC, Krishack PA, Zhang S, Zeleznik-Le NJ, Firulli AB, Witte PL. et al. Overexpression of Foxn1 attenuates age-associated thymic involution and prevents the expansion of peripheral CD4 memory T cells. Blood. 2011;118:5723-31
45. Bleul CC, Corbeaux T, Reuter A, Fisch P, Monting JS, Boehm T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature. 2006;441:992-6
46. Cheng L, Guo J, Sun L, Fu J, Barnes PF, Metzger D. et al. Postnatal tissue-specific disruption of transcription factor FoxN1 triggers acute thymic atrophy. J Biol Chem. 2010;285:5836-47
47. Abremski K, Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem. 1984;259:1509-14
48. Schlake T, Schorpp M, Nehls M, Boehm T. The nude gene encodes a sequence-specific DNA binding protein with homologs in organisms that lack an anticipatory immune system. Proc Natl Acad Sci U S A. 1997;94:3842-7
49. Schuddekopf K, Schorpp M, Boehm T. The whn transcription factor encoded by the nude locus contains an evolutionarily conserved and functionally indispensable activation domain. Proc Natl Acad Sci U S A. 1996;93:9661-4
50. Mao J, Barrow J, McMahon J, Vaughan J, McMahon AP. An ES cell system for rapid, spatial and temporal analysis of gene function in vitro and in vivo. Nucleic Acids Res. 2005;33:e155
51. Yu J, McMahon AP. Reproducible and inducible knockdown of gene expression in mice. Genesis. 2006;44:252-61 doi:10.1002/dvg.20213
52. Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93:10887-90
53. Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71-80
54. Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752-7
55. Saito M, Iwawaki T, Taya C, Yonekawa H, Noda M, Inui Y. et al. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol. 2001;19:746-50
56. Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T. et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 2002;17:211-20
57. Cha JH, Chang MY, Richardson JA, Eidels L. Transgenic mice expressing the diphtheria toxin receptor are sensitive to the toxin. Mol Microbiol. 2003;49:235-40
58. Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT. et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419-26
59. Guo J, Rahman M, Cheng L, Zhang S, Tvinnereim A, Su DM. Morphogenesis and maintenance of the 3D thymic medulla and prevention of nude skin phenotype require FoxN1 in pre- and post-natal K14 epithelium. J Mol Med. 2011;89:263-77
60. Zuniga-Pflucker JC. T-cell development made simple. Nat Rev Immunol. 2004;4:67-72
61. Mohtashami M, Zuniga-Pflucker JC. Three-dimensional architecture of the thymus is required to maintain delta-like expression necessary for inducing T cell development. J Immunol. 2006;176:730-4
62. Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775-85
63. Gordon J, Xiao S, Hughes B 3rd, Su DM, Navarre SP, Condie BG. et al. Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev Biol. 2007;7:69
64. Chen L, Xiao S, Manley NR. Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner. Blood. 2009;113(3):567-74
65. Anderson G, Lane PJ, Jenkinson EJ. Generating intrathymic microenvironments to establish T-cell tolerance. Nat Rev Immunol. 2007;7:954-63
66. Rossi SW, Jenkinson WE, Anderson G, Jenkinson EJ. Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature. 2006;441:988-91
67. Shakib S, Desanti GE, Jenkinson WE, Parnell SM, Jenkinson EJ, Anderson G. Checkpoints in the development of thymic cortical epithelial cells. J Immunol. 2009;182:130-7
68. Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc Natl Acad Sci U S A. 1989;86:1563-7
69. Kuraguchi M, Ohene-Baah NY, Sonkin D, Bronson RT, Kucherlapati R. Genetic mechanisms in Apc-mediated mammary tumorigenesis. PLoS Genet. 2009;5:e1000367
70. Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635-48
71. Osada M, Jardine L, Misir R, Andl T, Millar SE, Pezzano M. DKK1 mediated inhibition of Wnt signaling in postnatal mice leads to loss of TEC progenitors and thymic degeneration. PLoS One. 2010;5:e9062
72. Sun L, Guo J, Brown R, Amagai T, Zhao Y, Su DM. Declining expression of a single epithelial cell-autonomous gene accelerates age-related thymic involution. Aging Cell. 2010;9:347-57
73. Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027-32
74. Scheiff JM, Cordier AC, Haumont S. The thymus of Nu/+ mice. Anat Embryol (Berl). 1978;153:115-22
75. Kojima A, Saito M, Hioki K, Shimanura K, Habu S. NFS/N-nu/ + mice can macroscopically be distinguished from NFS/N- +/+ littermates by their thymic size and shape. Exp Cell Biol. 1984;52:107-10
76. Ortman CL, Dittmar KA, Witte PL, Le PT. Molecular characterization of the mouse involuted thymus: aberrations in expression of transcription regulators in thymocyte and epithelial compartments. Int Immunol. 2002;14:813-22
77. Calderon L, Boehm T. Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell. 2012;149:159-72
78. Guo J, Feng Y, Barnes P, Huang FF, Idell S, Su DM. et al. Deletion of FoxN1 in the thymic medullary epithelium reduces peripheral T cell responses to infection and mimics changes of aging. PLoS One. 2012;7:e34681
79. Xia J, Wang H, Guo J, Zhang Z, Coder B, Su DM. Age-Related Disruption of Steady-State Thymic Medulla Provokes Autoimmune Phenotype via Perturbing Negative Selection. Aging Dis. 2012;3:248-59
80. Palmer E. Negative selection--clearing out the bad apples from the T-cell repertoire. Nat Rev Immunol. 2003;3:383-91
81. Rossi SW, Chidgey AP, Parnell SM, Jenkinson WE, Scott HS, Boyd RL. et al. Redefining epithelial progenitor potential in the developing thymus. Eur J Immunol. 2007;37:2411-8
82. Barbieri CE, Pietenpol JA. p63 and epithelial biology. Exp Cell Res. 2006;312:695-706
83. Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol. 2010;5:349-71 doi:10.1146/annurev-pathol-121808-102117
84. Candi E, Rufini A, Terrinoni A, Giamboi-Miraglia A, Lena AM, Mantovani R. et al. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci U S A. 2007;104:11999-2004
85. Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523-36
86. Bleul CC, Boehm T. Laser capture microdissection-based expression profiling identifies PD1-ligand as a target of the nude locus gene product. Eur J Immunol. 2001;31:2497-503
87. Tsukamoto N, Itoi M, Nishikawa M, Amagai T. Lack of Delta like 1 and 4 expressions in nude thymus anlages. Cell Immunol. 2005;234:77-80
88. Bleul CC, Boehm T. Chemokines define distinct microenvironments in the developing thymus. Eur J Immunol. 2000;30:3371-9
89. Cai J, Lee J, Kopan R, Ma L. Genetic interplays between Msx2 and Foxn1 are required for Notch1 expression and hair shaft differentiation. Dev Biol. 2009;326:420-30
Author contact
![]() Corresponding author: Dong-Ming Su, Department of Molecular Biology and Immunology, University of North Texas Health Center at Fort Worth, 3500 Camp Bowie Blvd. Fort Worth, TX, 76107, USA. Tel. 1-817-735-5186, Fax: 1-817-735-2118, E-mail address: dong-ming.suedu
Corresponding author: Dong-Ming Su, Department of Molecular Biology and Immunology, University of North Texas Health Center at Fort Worth, 3500 Camp Bowie Blvd. Fort Worth, TX, 76107, USA. Tel. 1-817-735-5186, Fax: 1-817-735-2118, E-mail address: dong-ming.suedu

 Global reach, higher impact
Global reach, higher impact