10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(2):219-227. doi:10.7150/ijbs.5380 This issue Cite
Research Paper
Analysis of Core Region from Egg White Lysozyme Forming Amyloid Fibrils
1. Laboratory of Biochemistry and Bioscience, The United Graduate School of Agricultural Sciences, Kagoshima University, Kagoshima 890-0065 Japan;
2. Department of Applied Biochemistry and Food Science, Saga University, Saga 840-8502 Japan.
Received 2012-10-13; Accepted 2012-12-21; Published 2013-2-13
Abstract
Some of the lysozyme mutants in humans cause systemic amyloidosis. Hen egg white lysozyme (HEWL) has been well studied as a model protein of amyloid fibrils formation. We previously identified an amyloid core region consisting of nine amino acids (designated as the K peptide), which is present at 54-62 in HEWL. The K peptide, with tryptophan at its C- terminus, has the ability of self-aggregation. In the present work we focused on its structural properties in relation to the formation of fibrils. The K peptide alone formed definite fibrils having β-sheet structures by incubation of 7 days under acidic conditions at 37°C. A substantial number of fibrils were generated under this pH condition and incubation period. Deletion and substitution of tryptophan in the K peptide resulted in no formation of fibrils. Tryptophan 62 in lysozyme was suggested to be especially crucial to forming amyloid fibrils. We also show that amyloid fibrils formation of the K peptide requires not only tryptophan 62 but also a certain length containing hydrophobic amino acids. A core region is involved in the significant formation of amyloid fibrils of lysozyme.
Keywords: lysozyme, amyloid fibril formation, core region, tryptophan, egg white.
Introduction
Abnormal unfolding and accumulation of proteins are thought to provoke various amyloidosis such as Alzheimer's disease, Perkins disease and Creutzfeldt-Jakob disease [1-3]. The proteins related to these diseases do not have a common amino acid sequence or tertiary structure, but possess similar properties for forming amyloid fibrils consisting of a cross β-sheet structure, which accumulate in various tissues and induce abnormality [4-6]. However, the occurrence of non-toxic amyloid fibrils from some proteins have been observed, and this complicates understanding of the relationship between amyloid fibrils and disease [7,8].
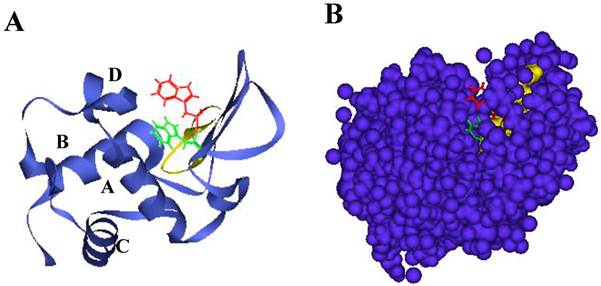
Lysozyme is a ubiquitous enzyme present in many organisms that attacks the peptidoglycan cell wall of certain microorganisms, selectively hydrolyzing the β-1,4 glycosidic linkages between the N-acetylmuramic acid and N-acetylglucosamine [9]. Hen egg white lysozyme (HEWL), which belongs to the c-type class of lysozymes [10], has two domains called α and β; the former has four α-helices (A to D) and the latter consists mainly of an antiparallel β-sheet. A long loop and enzyme active site are located in a cleft that is formed between these two domains (Figs. 1A and B) [11]. HEWL is known to form amyloid fibrils under denaturing conditions in organic solvent, detergent, chemical denaturant or acid pH with high temperature [12-14]. The properties of such fibrils and the process of fibrillation have been studied in considerable detail [15,16]. It is generally accepted that the formation of amyloid fibrils is implicated in particular regions of protein as well as the entire protein [17]. Verma et al. verified that most point mutations have a significant effect on human lysozyme dynamics, many of which lead to local unfolding and trend to form amyloid fibrils [18]. We recently found during analysis of the interaction between ovalbumin and HEWL that a peptide from HEWL has the ability of self-fibrillation, and designated it as the K peptide; we think that the region might work as a core for the amyloidogenesis of HEWL [19]. This peptide, corresponding to the residues 54-62 (GILQINSRW) of HEWL and being located in the hydrophobic cluster 3 [20-22], contains tryptophan 62, which is thought to be crucial to forming fibrils [21,23,24]. It is shorter than the previously reported fibril-forming peptide of 16 amino acids from the residues 49-64 (GSTDYGILQINSRWWC) of HEWL [25]. The K peptide region is found as a pocket in the junction connecting the α- and β-domains, and is probably buried partially within the lysozyme molecule (Figs. 1A and B). Here, we reconfirmed the self-fibrillation ability of the K peptide and compared it to those of some K peptide mutants. In each of these mutants, a certain residue was replaced by another amino acid. For the aggregation and fibrillation of HEWL, not only the length of the K peptide region but also the presence of aromatic amino acids, especially at the 62nd position, is assumed to be critical.
Materials and Methods
Materials
HEWL was purchased from Seikagaku Kogyo (Japan). The K peptide, the residues 54-62 (GILQINSRW) of HEWL, and its derivatives (GILQINSRG etc., see Table 1), as well as the peptide STDYGILQINSRW, i.e., the residues 50-62 of HEWL, all with more than 90% purity, were synthesized by Greiner Bio-one GmbH (Germany). The latter peptide, named the STDY-K peptide, was applied as a positive control of fibril formation. The reagents used were of a grade for biochemical use.
Summary of competent peptides on fibrils formation.
| Peptide | (pI) | pH2 | pH4 | pH7 | pH9 |
|---|---|---|---|---|---|
| STDYGILQINSRW | (5.55 ) | +++ | ++ | - | - |
| GILQINSRW | (9.75) | + | ++ | + | - |
| GILQINSRG | (9.75) | - | - | - | - |
| GILQINSGW | (5.52) | ++ | + | - | - |
| GILQINSR | (9.75) | - | ± | - | - |
| GILQINS | (5.52) | - | - | - | - |
| ILQINSRW | (9.75) | ± | + | - | - |
| LQINSRW | (9.75) | - | - | - | - |
| ILQINSR | (9.75) | - | - | - | - |
+++, much; ++, surely; +, somewhat; ±, very few; -, non.
Structure of HEWL and location of the K peptide region. A. Illustration by ribbon representation. Red color represents the K peptide region with the side chain of tryptophan 62. B. Illustration by space-filling model. Red and yellow show tryptophan 62 and other amino acids of the K peptide, respectively. Green and dark violet illustrate tryptophan 63 and other amino acids except the K peptide region. Amino acids in the K peptide are largely buried in the molecule.
Preparation of aggregates and fibrils
The solutions containing each of the K peptide and other peptides prepared by dissolving with 100% dimetylsulfoxide (DMSO) were diluted to the peptide concentration of 2 mg/ml in different pH buffers, and incubated at 37°C for the indicated time periods. The buffers used were 20 mM glycine-HCl for pH 2, 20 mM sodium acetate for pH 4, 20 mM potassium phosphate for pH 7 and 20 mM Tris-HCl for pH 9. The occurrence and features of the fibrils were monitored using atomic force microscopy (AFM) and a transmission microscope (TEM) as detailed below, using the incubated peptide preparations. These were further diluted to 80 μg/ml by each pH buffer used and subjected to biophysical analyses. Care was taken to lower the final concentration of DMSO in the final preparations to about 0.08% in order to avoid the adverse influence of DMSO on the biophysical data. HEWL was dissolved in 50 mM glycine-HCL buffer, pH 2, to the concentration of 2 mg/ml and treated under the conditions as reported previously (19).
Observation of morphology by TEM and AFM
For TEM analysis, the samples incubated at 37°C for 2 weeks and those for HEWL incubated at 55°C for 2weeks were prepared by coating a copper grid (200 mesh covered with Formvar/carbon) with a thin layer of diluted particle suspension. After negative staining with 2% (w/v) phosphotungstic acid for 2 min, the copper grid was then dried at room temperature. The grids were then examined using a 670 electron microscope (Hitachi, Japan) at 80 kV. For AFM analysis, the samples were diluted with dH20 after formation of fibrils. They were prepared by placing 5 μl of the pre-incubated solution on freshly cleaved mica (Ted Pella Inc., Redding, CA) at room temperature for 60s, washed twice with 50 μl of deionized water, and blow-dried with air. AFM images were acquired in different regions of each sample in tapping mode using a JEOL NanoWizard, JSPM-5410 (Japan).
Procedures for biophysical analyses
ThT fluorescence was assayed in 25 μM ThT for emission at 482 nm at an excitation of 440 nm [26] using a Corona Microplate reader fluorescence spectrophotometer (Hitachi, Japan). Congo red binding assay was made according to the previously reported procedures [26], wherein the absorbance spectra were scanned from 400 to 700 nm at 25°C on a spectrophotometer. The far UV CD spectra were recorded using a Jasco J-820 spectrophotometer in the 200-250 nm range. For the three analyses described above, the peptide preparations were diluted by using 20 mM of each buffer.
The emission spectra of ANS were measured between 400 and 600 nm at an excitation of 380 nm. For this, peptide solutions diluted by 20 mM glycine-HCl buffer, pH 2.2, was used after mixing with 100 μM ANS. Excitation and emission slit widths were both set at 5 nm. The integrated fluorescence is the average area of three independent emission spectra after subtracting the blank emission. The values reported are averaged over multiple sets of experiments. Changes in tryptophan fluorescence intensity were determined by measuring the fluorescence emission at 320-440 nm at an excitation wavelength of 295 nm using peptide solutions diluted in 20 mM of each buffer.
Acrylamide quenching of tryptophan fluorescence was carried out by measurement of fluorescence intensity after serial addition of small aliquots of freshly prepared acrylamide solution (0-0.5 M) in 10 mM 2-(N)-cyclohexylamino)-etanesulfonic acid (CHES)-NaOH, pH 9.0, to a sample (peptide solution diluted by 20 mM potassium phosphate buffer, pH 7.0) taken in a cuvette, followed by mixing and incubation for 5 min in the sample compartment in the dark. Excitation wave-length was set to 295 nm for each sample and emission intensity was scanned from 300 to 500 nm. Quenching data were analyzed by fitting to the Stern-Volmer equation [27].
F0/F=1+Ksv[Q]
Where F0 and F are the fluorescence intensities in the absence and presence of the quencher, respectively. [Q] is the molar quencher concentration and Ksv is the Stern-Volmer quenching constant.
These experiments described above were repeated three times.
Results
Time dependency of fibrils formation and their morphology
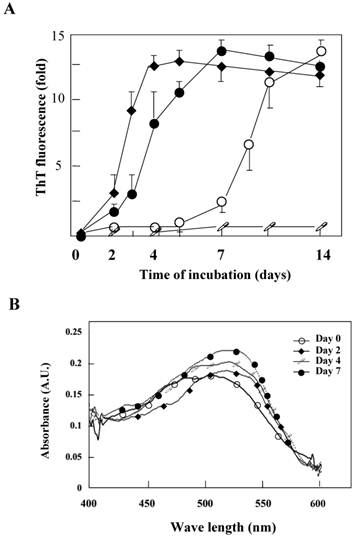
The K peptide was dissolved with a small amount of DMSO, and the resulting solution was diluted with each buffer at pH 2.0, 4.0, 7.0 or 9.0 as detailed in Materials and Methods. The prepared preparations were then incubated at 37°C for 1 to 14 days. The K peptide tends to aggregate itself and was dissolved first with DMSO. When DMSO-solubilized K peptide preparations were diluted with high concentration buffer, these produced substantial amounts of amorphous aggregates and obscured the analyses. Accordingly, pH buffers of a comparatively lower concentration (20 mM) were employed. The concentrations of the peptide and DMSO during incubation in the pH buffers were 2 mg/ml and 2%, respectively. The fibrils from K peptide at pH 4 were formed faster, and more were formed, than those at pH 2. The aggregation kinetics was monitored by the increase in the ThT fluorescence emission (Fig. 2A) as a fibrillation index. The results for the K peptide incubated at pH 4 fit a sigmoid curve, indicating the presence of an initial lag phase of about 24 h, followed by a rapid growth phase that reached a plateau on day 7 (solid circles). On the other hand, the K-peptide preparation incubated at pH 2 exhibited a retarded increase of fluorescence intensity reaching a plateau on day 10, while those incubated at pHs 6 and 9 gave hardly any changes (patterns not shown). We inferred that the most effective way to generate ThT-positive products from the K peptide is the incubation at pH 4 for 7 days at 37°C. The rate of increase in fluorescent intensity by the K peptide was higher than that by HEWL itself incubated at pH 2 (open circles). The STDY-K peptide (STDYGILQINSRW, corresponding to the residues 50-62 of HEWL) was confirmed to aggregate at pH 2 more rapidly than the K peptide (diamond patterns), whereas the K-peptide derivative GILQINSRG (the mutant with glycine substituted for tryptophan) gave no signal of aggregation (open squares). To further verify the occurrence of fibrils, Congo red binding to the incubating K peptide samples was performed. As shown in Fig 2B, a time-dependent increase in absorption signal, accompanied by a red shift of the spectral maximum, was observed.
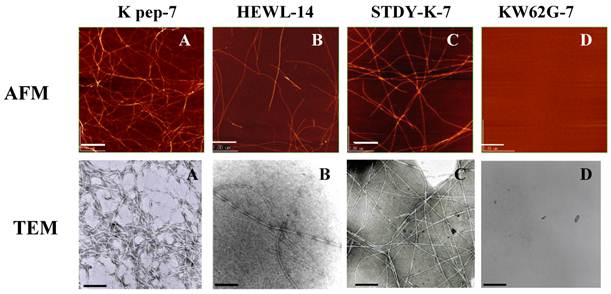
Inspection by AFM and TEM (Fig. 3) indicated that the fibrils found in the K peptide preparation on day 7 of incubation at pH 4 had a typical amyloid morphology (panels A). The fibrils produced from the K peptide at pH 2 showed similar patterns (not shown). These contained short and twisting fibrils with a diameter of ca. 5-10 nm, resembling the amyloid fibrils of HEWL observed after incubation for 14 days at pH 2 (panels B). The STDY-K peptide (see above) after incubation in acidic milieu also produced amyloidal patterns, while the K peptide mutant GILQINSRG gave no aggregation after incubation at all pHs tested (panels C and D, respectively; both show pH 2 preparations after day 7 of incubation).
CD spectrum of fibrils originated from the K peptide etc
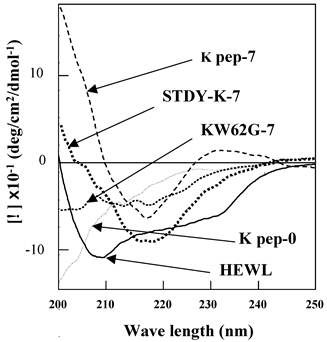
Far UV-CD spectroscopy was conducted to explore the secondary structural features, in particular those of the β-sheet, of the present preparations (Fig. 4). The K peptide incubated at 37°C for 7 days at pH 4 showed a spectrum bearing a minimum at 218 nm (dashed line), which is typical of a β-sheet. The calculation by Yang's method [28] from the spectrum apparently indicated that the content of the β-structure (sum of β-sheet and β-turn) was 87% with a scant random coil, but without an α-helix. As expected, the STDY-K peptide exhibited the signal of the β-sheet (bold dotted line), whose content was calculated to be approximately 70%. Both peptides showed a similar pattern. In contrast, the K peptide before incubation presented an inconstant spectrum having no minimum in the vicinity of 218 nm (long dashed dotted line), indicating that it scarcely had β-conformation. No typical spectrum of CD for the β-structure was given by the peptide GILQINSRG (dotted line), although it was incubated as described above. The solid line in Fig. 4 shows a CD spectrum of HEWL.
Aggregation kinetics monitored by ThT or Congo red binding. A. The aggregation kinetics of the K peptide (˜) monitored by the increase in the ThT fluorescence emission in comparison with those for HEWL (™), the STDY-K peptide (¿) and the W-substituted K peptide GILQINSRG (□). Each peptide sample was incubated at 37°C for up to 14 days in 20 mM glycine-HCl buffer at pH 4, 2, 2 and 2, respectively, prior to mixing with the ThT probe. B. Absorption spectra of Congo red with the K peptides. The peptides were incubated at 37°C in 20 mM glycine-HCl buffer at pH 4 on days 0 (™), 2 (¿), 4 (□) and 7 (˜). Preparations were subjected to these analyses at the concentration of 80 μg/ml as described in Materials and Methods.
AFM and TEM images of the K peptide in comparison with other substances. A (K pep-7), the K peptide incubated for 7 days at pH 4.0; B (HEWL-14), HEWL incubated for 14 days at pH 2.0; C (STDY-K-7), the STDY-K peptide incubated for 7 days at pH 2.0; D (KW62G-7), GILQINSRG (the W-substituted K peptide) incubated for 7 days at pH 2.0. The concentration of each peptide and HEWL during analysis was 2 mg/ml (cf. Materials and Methods).
Far-UV CD spectra of the K peptide and other substances. Samples indicated in the legend to Fig. 3 were used. As a control, non-incubated K peptide preparation was also analyzed (K pep-0). Before being subjected to the analysis, each peptide and HEWL was diluted to 80 μg/ml as described in Materials and Methods.
Features of ANS binding and tryptophan fluorescence
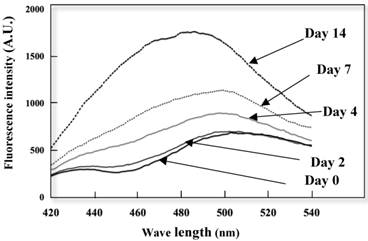
Conformation alterations of the K peptide fibrils in terms of the exposed hydrophobic surface were investigated by the ANS binding assay. As illustrated in Fig. 5, extensive augmentation (approx 50%) of ANS fluorescence occurred as the incubation time of the K peptide proceeded. Moreover, there was a blue shift in the λ max from 515 nm (before incubation) to 490 nm (through incubation). This suggests that the formation of an exposed hydrophobic surface on the K peptide fibrils proceeded during incubation at pH 4.
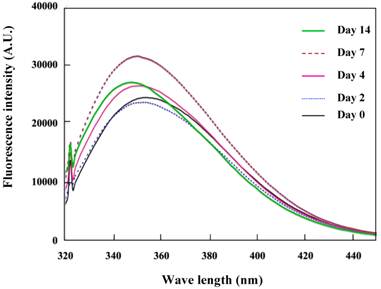
The K peptide consists of hydrophobic amino acids including tryptophan that is sited at the C-terminal. Therefore, the fluorescence intensity of tryptophan is also likely to mirror the configuration features of this peptide. The K peptide during incubation at pH 4 was analyzed for its intrinsic tryptophan fluorescence spectrum, which increased gradually and attained the maximum on day 7 (Fig. 6). It was around this time when ThT fluorescent reached a plateau and fibrils were clearly observed in the K peptide preparation (see Figs. 2A and 3). Comparable data were obtained in the K-peptide incubated at pHs 2, 7 and 9, although the fluorescence intensity was low in these cases (data not shown).
Fluorescence quenching measurements
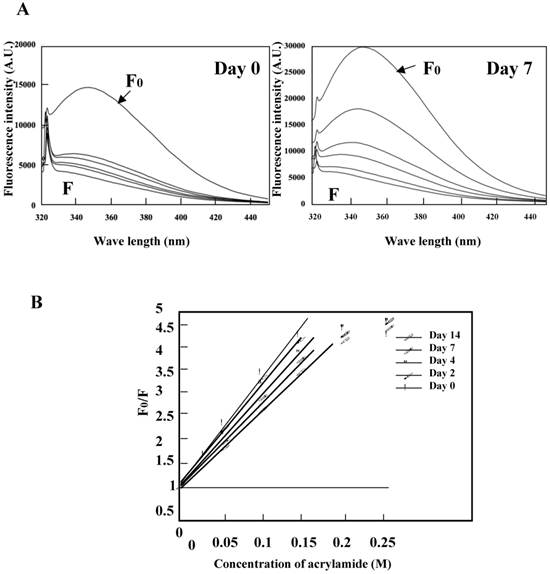
Acrylamide quenching of tryptophan fluorescence was conducted for the K peptide preparations before and after incubation at pH 4 (Fig. 7A, left and right panels, respectively). Tryptophan fluorescence in the day-0 K peptide was quenched by 62% with 0.05 M of acrylamide, whereas that in the day-7 K peptide was quenched only by 41% under the same conditions as above. These facts seemed to imply that the fibrils-forming K peptide requires more acrylamide for quenching tryptophan fluorescence than that in the intact state. Stern-Volmer plots were drawn for the K peptide preparations (Fig. 7B), and from the data the Ksv (Stern-Volmer constant) values were reduced from 19.3, 18.5 18.1 17.1 and 15.3 for the preparations on days 0, 2, 4, 7 and 14, respectively.
Changes in ANS fluorescence spectra of the K peptide during incubation. The K peptide was incubated for 0 to 14 days at pH 4.0 at the concentration of 2 mg/ml as detailed in Materials and Methods and after dilution to 80 μg/ml, subjected to the analysis of ANS fluorescence emission. Excitation wavelength was set to 380 nm and emission wavelength to 420-540 nm.
Intrinsic tryptophan fluorescence due to the K peptide. Samples were prepared as described in the legend to Fig. 5 and analyzed for the tryptophan fluorescence emission. Excitation wavelength was set to 295 nm and emission wavelength to 320-450 nm.
Quenching of tryptophan fluorescence by acrylamide. The K peptide preparations were incubated as described in the legend to Fig. 5 and carried to the quenching test. A. Fluorescence spectra on day 0 (left) and day 7 (right) of incubation. Spectra stand for 0 (F0), 0.05, 0.1, 0.15, 0.02 and 0.25 M of acrylamide from top to bottom. B, Stern-Volmer plots against the quencher concentration. Ksv constant was calculated from the equation of F0/F=1+Ksv[Q] and the results are discussed in the text.
Significance of amino acid residue in the K peptide
The K peptide, consisting of nine amino acids, is abundant in the hydrophobic side chains. In particular, tryptophan located in the C-terminus, which is the 62th amino acid of lysozyme protein, seems to be crucial to forming fibrils. Therefore, all the K-peptide derivatives prepared in this study were observed to see whether or not, or to what extent, they had the ability to contribute to fibrillation. The STDY-K peptide was run in parallel as a positive control. All the peptides were dissolved with a small amount of DMSO, then diluted with various pH buffers as described above, incubated for up to 14 days at 37°C and observed by TEM. Degrees of fibril formation were roughly scored and the results obtained are summarized in Table 1. The K peptide formed fibrils, but GILQINSRG, the C-terminal substitution mutant of the K peptide, had no ability to form fibrils. Distinct fibrils bearing a minimum at 218 nm of the CD spectra were observed by GILQINSGW (data not shown). Fibrils were formed only slightly by the peptide GILQINSR in which the C-terminal tryptophan was simply depleted from the K peptide. The peptide ILQINSRW, the K peptide ablated with the N-terminal glycine, formed fibrils, albeit at lower levels. The K peptide was completely deprived of its fibrillation ability when deleted with any two residues as far as tested. We conclude that a certain length of peptide and the presence of hydrophobic amino acids including the C-terminal tryptophan are important for forming fibrils.
Discussion
The characterization of the process of protein misfolding that occurs as a preface to the formation of fibrils has provided insight into fibril deposits that are associated with neurodegenerative amyloid diseases [29,30]. Amyloid fibrils are ordered aggregates of a normally soluble peptide or protein. Furthermore, knowledge of the conversion of protein from normal to abnormal will help conquer profound protein degenerative diseases. For this purpose, HEWL is a good model protein as it has been well-characterized for amyloid fibril generation in vitro [31]. A specific region in the lysozyme molecule is thought to be involved in the formation of amyloid fibrils. It has been reported that HEWL has six hydrophobic clusters, whose cluster 3 is involved in long-range interaction of disulfide-reduced lysozyme in the formation of amyloid fibrils (20-23). The K peptide including tryptophan 62, which is a crucial amino acid on fibrils formation (21,23), corresponds to the cluster 3. It was previously reported that the fragment 49-101 (or 50-87/88-101) of HEWL has a high tendency to aggregate [11,32]. Krebs et al. reported that the fragment 49-64 in HEWL readily forms amyloid fibrils in acid solution [33]. We have shown that the nonapeptide GILQINSRW named the K peptide, corresponding to 54G-62W of HEWL, forms aggregates and amyloid-like fibrils by itself (see above), and also by interacting with ovalbumin [19]. Although the K peptide is much shorter than the fragment 49-64 and may have lost the well-structured conformation, it forms fibrils, suggesting that it is a core part triggering the amyloidogenesis. The STDY-K peptide, which is more soluble in water than the K peptide, showed faster elevation of ThT fluorescence than the K peptide. A longer peptide-bearing core region may be more liable to form fibrils than the K peptide. This region is in agreement with the portion called the core of amyloid formation.
In spite of its highly hydrophobic nature with a pI of 9.75, the K peptide could be dissolved in 20 mM buffers of pH 2 to 4. After a certain period of incubation in such an aqueous milieu, it formed morphologically definite fibrils observable under AMF and TEM, producing a fluorescence-enhancement of ThT and a red shift in the absorption spectra of Congo red, both of which are known to interact with amyloid fibrils having a cross-β-sheet structure [34,35]. We suppose that the fibrils generated from the K peptide were of the bona fide amyloid type and the inference was supported by the CD data which implied the presence of a second structure with a β-sheet. The fibrils are likely to be comparable in structure to those produced by HEWL and the STDY-K peptide. ANS is known to preferentially bind to hydrophobic clusters giving rise to an enhancement in fluorescence emission accompanying a blue-shift of the spectral maximum [36,37]. In our case, the rise of ANS fluorescence emission with K peptide was clear until the incubation of 14 days, and the blue-shift was also observed in the emission spectra with the K peptides incubated for longer times (see Fig. 5). At any rate, we speculate that the hydrophobicity of the K peptide aggregate or fibrils was increased in time by the exposure of the hydrophobic amino acids (other than tryptophan) to the surface while fibril clusters are formed. The results of acrylamide quenching of tryptophan fluorescence, which serves as an effective tool for monitoring the tryptophan environment in proteins or peptides [38,39], indicate that fibrils produced from the K peptide configure to a regular structure by transition to orchestrate a fairly hydrophobic surface, making array-like amino acid stacking. We infer that the decrease in the Stern-Volmer constant (Ksv) during incubation of the K peptide is interpreted by assuming that the tryptophan residue is in more internal and has a propensity to be non-polarized. The possible involvement of hydrophobic amino acids to the process of fibril formation was additionally suggested by the fact that fibrillation of the K peptide was inhibited by a comparatively high concentration of SDS >5 mM (unpublished data). In native HEWL, the region containing the K peptide, which is partially embedded inside the lysozyme molecule and located in the active site cleft (see Figs. 1A and B), might possibly keep the conformational stability of protein and thus repress aggregation. It is inferred that exposure of the K peptide region to the surface causes it to act as a core for aggregation/fibrillation and in turn amyloidosis.
The significance of the K peptide in amyloid formation is verified by previous observations that single-point mutants I56T, F57I, W64R and D67H, as well as double mutants F57/T70 and W112R/T70N, of human lysozyme bring about non-neuropathic hereditary amyloidosis (the single-point mutants were less stable than the wild-type protein) [15, 40-45]. These critical amino acid residues are mostly located in the region 55-63 (GIFQINSRY) [44], which is the counterpart of the K peptide of HEWL and was shown to also lie at the interface between the α- and β-domains [44]. In addition, the 64 residue, although located in the long loop of the β-domain, is present in the circumjacent region of the human K peptide. The human counterpart of the HEWL-K peptide, while differing from the latter in the third and the last residues (phenylalanine and tyrosine instead of leucine and tryptophan, respectively), forms strong aggregation and fibrils in a similar manner to that for the present K peptide under the same conditions as those applied for the latter (unpublished data), and this may also be a core of amyloidosis. Although additional studies are needed to confirm the relationships between the peptide structure and fibril formation, we believe that the findings obtained in this research will contribute to a better understanding of the mechanisms underlying the formation of self-assembly, amorphous aggregates and arrayed fibrils in proteins and peptides implicated in amyloid diseases.
Acknowledgements
We are grateful to Dr. H. Yagi of Osaka University and Mr. Sogawa of Kagoshima University for their technical support. This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Abbreviations
HEWL, hen egg white lysozyme; ThT, thioflavin T; ANS, 8-anilino-1-naphthalenesulfonic acid; CD, circular dichroism; AFM, atomic force microscopy; TEM, transmission electron microscope
Competing Interests
The authors have declared that no competing interest exists.
References
1. Veerhuis R, Boshuizen RS, Familian A. Amyloid associated proteins in Alzheimer's and prion disease. Curr. Drug Targets CNS Neurol. Disord. 2005;4:235-248
2. Aarsland D, Perry R, Brown A, Larsen JP, Ballard C. Neuropathology of dementia in Parkinson's disease: a prospective, community-based study. Ann. Neurol. 2005;58:773-776
3. Chakraborty C, Nandi S, Jana S. Prion disease: a deadly disease for protein misfolding. Curr. Pharm. Biotechnol. 2005;6:167-177
4. Kelly JW. Alternative conformations of amyloidogenic proteins govern their behavior. Curr. Opin. Struct. Biol. 1996;6:11-17
5. Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 1998;8:101-106
6. Dobson CM. Protein folding and misfolding. Nature. 2003;426:884-890
7. Frandrich M, Fletcher MA, Dobson CM. Amyloid fibrils from muscle myoglobin. Nature. 2001;410:165-166
8. Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009;5:15-22
9. Callewaert L, Michiels CW. Lysozymes in the animal kingdom. J. Biosci. 2010;35:127-160
10. Lesnierowsli G, Kjowski J. Lysozyme. In: (ed.) Huopalahti R, Lopez-Fandino R, Anton M, Schade R. Bioactive Egg Compounds. Berlin Heiderberg: Springer-Verlag. 2007:34-42
11. Mishra R, Sorgjerd K, Nystrom S, Nordigarden A, Yu YC, Hammarstrom P. Lysozyme amyloidogenesis is accelerated by specific nicking and fragmentation but decelerated by intact protein binding and conversion. J. Mol. Biol. 2007;399:1029-1044
12. Morozova-Roche LA, Zurdo J, Spencer A, Noppe W, Receveur V, Archer DB, Joniau M, Dobson CM. Amyloid fibril formation and seeding by wild-type human lysozyme and its disease-related mutational variants. J Struct Biol. 2000;130:339-3351
13. Goda S, Takano K, Yamagata Y, Nagata R, Akutsu H, Maki S, Namba K, Yutani K. Amyloid protofilament formation of hen egg lysozyme in highly concentrated ethanol solution. Protein Sci. 2000;9:369-375
14. Cao A, Hu D, Lai L. Formation of amyloid fibrils from fully reduced hen egg white lysozyme. Protein Sci. 2004;13:319-324
15. Swaminathan R, Ravi VK, Kumar S, Kumar MV, Chandra N. Lysozyme: a model protein for amyloid research. Adv. Protein Chem. Struct. Biol. 2011;84:63-111
16. Xu M, Shashilov VA, Ermolenkov VV, Fredriksen L, Zagorevski D, Lednev IK. The first step of hen egg white lysozyme fibrillation, irreversible partial unfolding, is a two-state transition. Protein Sci. 2007;16:815-832
17. Frare E, Polverino De Laureto P, Zurdo J, Dobson CM, Fontana A. A highly amyloidogenic region of hen lysozyme. J. Mol. Biol. 2004;340:1153-1165
18. Verma D, Jacobs DJ, Livesay DR. Changes in Lysozyme Flexibility upon Mutation Are Frequent, Large and Long-Ranged. PLoS Comput Biol. 2012;8:1-18
19. Sugimoto Y, Kamada Y, Tokunaga Y, Shinohara H, Matsumoto M, Kusakabe T, Ohkuri T, Ueda T. Aggregates with lysozyme and ovalbumin show features of amyloid-like fibrils. Biochem Cell Biol. 2011;89:533-544
20. Wirmer J, Schlorb C, Klein-Seetharaman J, Hirano R, Ueda T, Imoto T, Schwalbe H. Modulation of compactness and long-range interactions of unfolded lysozyme by single point mutations. Angew. Chem. Int. Ed. Engl. 2004;43:5780-5785
21. Ohkuri T, Shioi S, Imoto T, Ueda T. Effect of the structure of the denatured state of lysozyme on the aggregation reaction at the early stages of folding from the reduced form. J. Mol. Biol. 2009;347:159-168
22. Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Long-range interactions within a nonnative protein. Science. 2002;295:1719-1722
23. Zhou R, Eleftheriou M, Royyuru AK, Berne BJ. Destruction of long-range interactions by a single mutation in lysozyme. Proc Natl Acad Sci U S A. 2007;104:5824-5829
24. Mishima T, Ohkuri T, Monji A, Imoto T, Ueda T. A particular hydrophobic cluster in the residual structure of reduced lysozyme drastically affects the amyloid fibrils formation. Biochem. Biophys. Res. Commun. 2007;356:769-772
25. Pertinhez TA, Bouchard M, Smith RA, Dobson CM, Smith LJ. Stimulation and inhibition of fibril formation by a peptide in the presence of different concentrations of SDS. FEBS Lett. 2002;529:193-197
26. Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151-160
27. Molla AR, Maity SS, Ghosh S, Mandal DK. Organization and dynamics of tryptophan residues in tetrameric and monomeric soybean agglutinin: studies by steady-state and time-resolved fluorescence, phosphorescence and chemical modification. Biochimie. 2009;91:857-867
28. Yang JT, Wu CSC, Martinez HM. Calculation of protein conformation from circular dichroism. Methods in Enzymol. 1986;130:208-269
29. Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333-366
30. Moore RA, Taubner LM, Priola SA. Prion protein misfolding and disease. Curr. Opin. Struct. Biol. 2009;19:14-22
31. Trexler AJ, Nilsson MR. The formation of amyloid fibrils from proteins in the lysozyme family. Curr. Protein Pept. Sci. 2007;8:537-557
32. Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim. Pol. 2004;51:299-321
33. Krebs MR, Wilkins DK, Chung EW, Pitkeathly MC, Chamberlain AK, Zurdo J, Robinson CV, Dobson CM. Formation and seeding of amyloid fibrils from wild-type hen lysozyme and a peptide fragment from the beta-domain. J. Mol. Biol. 2000;300:541-549
34. Blake C, Serpell L. Synchrotron X-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous beta-sheet helix. Structure. 1996;4:989-998
35. Xu S. Cross-beta-sheet structure in amyloid fiber formation. J. Phys. Chem. B. 2009;113:12447-1255
36. Demarest SJ, Fairman R, Raleigh DP. Peptide models of local and long-range interactions in the molten globule state of human alpha-lactalbumin. J. Mol. Biol. 1998;283:279-291
37. Demarest SJ, Horng JC, Raleigh DP. A protein dissection study demonstrates that two specific hydrophobic clusters play a key role in stabilizing the core structure of the molten globule state of human alpha-lactalbumin. Proteins. 2001;42:237-242
38. Eftink MR, Ghiron CA. Fluorescence quenching studies with proteins. Anal. Biochem. 1981;114:199-227
39. Vanhove M, Lejeune A, Pain RH. Beta-lactamases as models for protein-folding studies. Cell Mol. Life Sci. 1998;54:372-377
40. Pepys MB, Hawkins PN, Booth DR, Vigushin DM, Tennent GA, Soutar AK, Totty N, Nguyen O, Blake CC, Terry CJ. et al. Human lysozyme gene mutations cause hereditary systemic amyloidosis. Nature. 1993;362:553-557
41. Valleix S, Drunat S, Philit JB, Adoue D, Piette JC, Droz D, MacGregor B, Canet D, Delpech M, Grateau G. Hereditary renal amyloidosis caused by a new variant lysozyme W64R in a French family. Kidney Int. 2002;61:907-912
42. Yazaki M, Farrell SA, Benson MD. A novel lysozyme mutation Phe57Ile associated with hereditary renal amyloidosis. Kidney Int. 2003;63:1652-1657
43. Rocken C, Becker K, Fandrich M, Schroeckh V, Stix B, Rath T, Kahne T, Dierkes J, Roessner A, Albert FW. ALys amyloidosis caused by compound heterozygosity in exon 2 (Thr70 Asn) and exon 4 (Trp112Arg) of the lysozyme gene. Hum. Mutat. 2006;27:119-120
44. Kumita JR, Johnson RJ, Alcocer MJ, Dumoulin M, Holmqvist F, McCammon MG, Robinson CV, Archer DB, Dobson CM. Impact of the native-state stability of human lysozyme variants on protein secretion by Pichia pastoris. FEBS J. 2006;273:711-720
45. Granel B, Valleix S, Serratrice J, Cherin P, Texeira A, Disdier P, Weiller PJ, Grateau G. Lysozyme amyloidosis: report of 4 cases and a review of the literature. Medicine (Baltimore). 2006;85:66-73
Author contact
![]() Corresponding author: Yasushi Sugimoto, Ph.D., Laboratory of Biochemistry and Bioscience, The United Graduate School of Agricultural Sciences, Kagoshima University, Kagoshima 890-0065 Japan. Tel.: and Fax: 81+ 99 285 8781; E-mail address: yasushiagri.kagoshima-u.ac.jp.
Corresponding author: Yasushi Sugimoto, Ph.D., Laboratory of Biochemistry and Bioscience, The United Graduate School of Agricultural Sciences, Kagoshima University, Kagoshima 890-0065 Japan. Tel.: and Fax: 81+ 99 285 8781; E-mail address: yasushiagri.kagoshima-u.ac.jp.

 Global reach, higher impact
Global reach, higher impact