10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(5):496-508. doi:10.7150/ijbs.4668 This issue Cite
Research Paper
Differential Evolution of Duplicated Medakafish mitf Genes
Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Received 2012-5-29; Accepted 2013-5-8; Published 2013-5-24
Abstract
Gene duplication is a major force of evolution. One whole genome duplication (WGD) event in the fish ancestor generated genome-wide duplicates in all modern species. Coloration and patterning on the animal body surface exhibit enormous diversity, representing a mysterious and ideal system for understanding gene evolution. Surface colors and patterns are determined primarily by pigment cells in the skin and eye. Thus, microphthalmia-associated transcription factor (Mitf) as a master regulator of melanocyte development is excellent for studying the evolution of WGD-derived gene duplicates. Here we report the evolution of mitf duplicate, mitf1 and mitf2, in the fish medaka (Oryzias latipes), which encode medaka co-homologs Mitf1 and Mitf2 of the mouse Mitf. Compared to mitf1, mitf2 exhibits an accelerated sequence divergence and loses melanocytic expression in embryos at critical developmental stages. Compared to a Xiphophorus counterpart, the medaka Mitf2 displayed a reduced activity in activating melanogenic gene expression by reporter assays and RT-PCR analyses. We show that the medaka Mitf2 has the ability to induce melanocyte differentiation in medaka embryonic stem cells but at a remarkably reduced efficiency compared to the Xiphophorus counterpart. Our data suggest differential evolution of the medaka mitf duplicate, with mitf1 adopting conservation and mitf2 employing degeneration, which is different from the duplication-degeneration-complementation proposed as the mechanism to preserve many gene duplicates in zebrafish. Our finding reveals species-specific variations for mitf duplicate evolution, in agreement with enormous diversity of body coloration and patterning.
Keywords: mitf, gene duplicate, melanocyte, neural crest, pigmentation.
Introduction
Gene duplication is a major force of evolution and plays a critical role in increasing complexity, and two rounds of whole genome duplication (WGD) have been proposed to occur during vertebrate evolution [1]. Duplicated genes may adopt three major modes of evolutionary divergence: loss by deleterious mutations (nonfunctionalization), acquisition of a new adaptive function (neofunctionalization) or division of an ancestral function (subfunctionalization). The role of gene duplication in evolution is perhaps best illustrated by the growth hormone (GH)-prolactin (PRL) family [2]. This family consists of GH, PRL, somatolactin (SL) and placental lactogen (PL), which has arisen from a single ancestor by gene duplication followed by extensive functional divergence. Although GH retains the original role in growth control in all vertebrate species, the other members have distinctive functions between fish and mammals: PRL is important for the mammary gland in mammals but for osmoregulation in fish [3], SL is fish-specific and selectively regulates proliferation and morphogenesis of pigment cells [4], and PL is specific to mammals and essential for placental development. They also exhibit differences in expression pattern: GH, PRL and SL retain the pituitary expression, whereas PL expression occurs in the placenta.
Fish models provide an ideal system to study the modes and mechanisms of duplicated genes' evolution in vertebrates. After split from the tetrapod lineage, fish has undergone a third WGD event in the common ancestor of all modern-day teleosts [5]. Interestingly, the resultant genome-wide gene duplicates usually coexist, leading to more genes in fish [6]. The duplication-degeneration-complementation (DDC) mode was originally proposed in zebrafish to explain the preservation of duplicated genes: Duplicated genes undergo partial and complementary retention and degeneration to perform complementary functions that jointly match the function of a single ancestral gene [7]. This DDC mode emphasizes that complementary mutations in the regulatory sequences of gene duplicates lead to the retention rather than the loss of gene duplicates following WGD [7].
Fishes are the most diversified group of vertebrates and comprise 32,400 known species (as of Feb 6th, 2013 at http://fishbase.mnhn.fr/search.php). The enormous species diversity and presence of numerous gene duplicates make fishes an excellent system for studying gene evolution. It will be no surprising that diverse species may use other modes besides subfunctionalization. Neofunctionalization has recently been reported for the evolution of an antifreeze protein under escape from adaptive conflict in an antarctic zoarcid fish [8]. In the case of neofunctionalization, one of the duplicated genes must retain the original expression and function, and the other undergoes degeneration in expression and/or sequence to acquire an emerging new function. Therefore, the duplication-conservation-degeneration (DCD) may represent a transitory status from gene redundancy to neofunctionalization. The DCD mode is elusive to study, because it is often difficult to determine whether a gene under degeneration is indeed a degenerating gene: its remnant expression and/or activity could obscure observation and experimentation. Consequently, compelling evidence for the DCD mode has been absent, and the mechanism underlying this mode has remained poorly understood.
External coloration in animals including fish is a distinctive character for ornamental industry and evolutionary biology [9]. The body coloration is controlled by the development and patterning of pigment cells. Genes involved in fish pigmentation is of special interest to study the mechanisms underlying the evolution of duplicated genes in various species[10]. One such gene is mitf encoding microphthalmia-associated transcription factor (Mitf), a member of the basic helix-loop-helix leucine zipper (bHLH-Zip) protein family [11,12]. Mitf plays a critical role in the development of melanocytes and retinal pigment epithelium (RPE) [13-16] and in human pigment diseases [17,18].
In mammals, a single mitf gene produces multiple isoforms by differential usage of alternative promoters and exons [15,19-21]. In fish, a genomic approach has been evolved to produce two Mitf proteins corresponding to the mouse MitfA and MitfM [22,23]. Through the ancient WGD in the fish lineage 450 million years ago [24], many fish species have two distinct genes that are homologous/paralogous to single copy genes in tetrapod vertebrates including mammals and birds. In fish, mitf has been identified in ~40 species, and the presence of two mitf genes has been documented in several model fish species including fugu, pufferfish, Xiphophorus[22] and zebrafish [25,26]. However, there is the lack of a nomenclature system for fish mitf genes. As a result, fish mitf genes have been named as mitfa, mitfb, mitfm, mitf1 and mitf2 on the basis of sequence homology and/or N-terminus. For example, the two zebrafish mitf genes are called mitfa and mitfb, with their protein products corresponding to the mouse MitfM and MitfA isoforms [25,26]. A mitfa mutation called nacre affects the development of pigment cells on the skin but not in the eye, leading to cutaneous albinism [26], whereas mitfb, which is mainly expressed in the eye, provides necessary function for RPE development in the absence of mitfa activity [25]. Interestingly, ectopic expression of mitfb can rescue the pigment phenotype of mitfa mutant [25,26]. Therefore, in zebrafish, the two mitf genes have undergone subfunctionalization to provide complementary expression patterns and functions in pigmentation [22,25].
Medaka (Oryzias latipes) is a fish model to study pigment cell biology and the evolution of duplicated mitf genes in fish. This small laboratory fish is an excellent lower vertebrate model for analyzing development [27] and pigmentation biology. In this organism, there are more than 40 inbred lines and mutant strains showing diverse pigmentation patterns and abnormalities [28]. In fact, medaka has been the first teleost from which the tyrosinase gene was identified as the first fish gene involved in pigmentation [29]. In addition, medaka is a unique lower vertebrate that has male germ stem cells [30], diploid [31,32] and even haploid embryonic stem (ES) cell lines [33] for studying pigment cell development in vitro. For example, we and others have previously demonstrated that forced expression of a heterologous mitf gene from Xiphophorus is sufficient for directing differentiation into melanocytes from medaka ES cell [34] and germ stem cells [35]. The medaka genome has been fully sequenced [36] and increasingly annotated (http://www.ensembl.org/index.html).
Here we show that medaka has two mitf genes, mitf1 and mitf2, which encode Mitf1 and Mitf2 that correspond to the mouse MitfA and MitfM, respectively, on the basis of gene structure, chromosome synteny and phylogenetic sequence comparisons. We analyzed the embryonic and adult expression patterns of the mitf duplicate by RT-PCR and in situ hybridization, and determined the pigmentary activity of Mitf2 by reporter assay, real-time PCR analysis and melanocyte induction. We propose that medaka uses the DCD mode for evolution of its duplicated mitf genes through differential retention or loss of melanocytic expression in developing embryos.
Results
Medaka Mitf1 and Mitf2 correspond to mammalian MitfA and MitfM
The sequenced medaka genome has two annotated mitf genes called mitf1 and mitf2 (http://www.ensembl.org). We followed this annotated gene nomenclature. The sequence appears to be partial for mitf1 and complete for mitf2. A sequence comparison led to a putative full length of open reading frame (ORF) for mitf1. Cloning and sequencing led to the full length cDNA sequences for mitf1 and mitf2, which predict proteins Mitf1 and Mitf2 of 512 and 406 amino acid residues, respectively (Supplementary Material: Figure S1). On a sequence alignment, both medaka Mitf proteins are highly similar to the known Mitf homologues from other fish species and human (Supplementary Material: Figure S2).
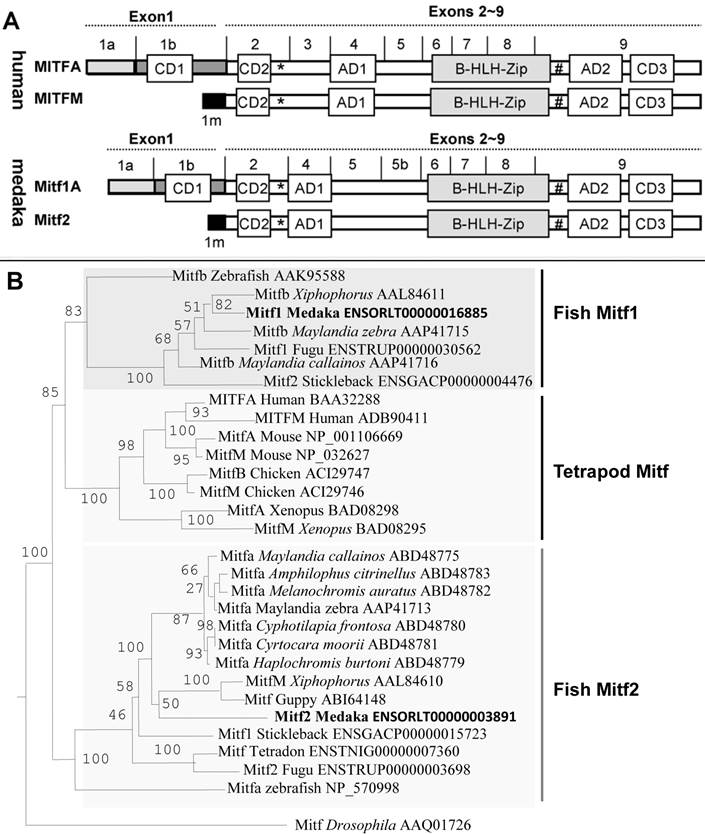
A single gene in birds and mammals produces multiple Mitf isoforms with distinct N-terminal sequences. Two major isoforms are MitfA and MitfM, which in human have 118 and 11 amino acids in the N-terminus, respectively (see below). Together with a comparison in protein domain structure, the medaka Mitf1 corresponds to the human isoform MitfA, and the Mitf2 is equivalent to the human MitfM (Figure 1A). Therefore, the two major fish Mitf proteins encoded by two distinct genes resemble the two major Mitf isoforms encoded by a single gene in higher vertebrates.
So far the fish mitf genes and their protein products have controversially been named in the literature. For example, fish genes whose protein products are similar to the mammalian MitfM isoform have been called mitfa in zebrafish, fugu and tetraodon or mitfm in Xiphophorus, whereas those whose protein products are similar to the mammalian MitfA isoform have been referred to as mitf1 or mitfb [22,25,37]. All the five sequenced genomes of fish species, namely zebrafish, medaka, fugu, tetradon and stickleback, contain two separate mitf genes. The gene pairs are annotated as mitf1 and mitf2 in both medaka and stickleback (http://www.ensembl.org/), which were followed in this study. On the basis of protein domain structure, fish Mitf1 and Mitf2 proteins are more similar to the mammalian MitfA and MitfM, respectively. Hence, mitfa or mitfm genes previously described in other fish species are equivalents of the medaka mitf2 gene, and previous mitfb genes correspond to the medaka mitf1. In this regard, the annotated stickleback Mitf1 and Mitf2 appears to be equivalents of medaka Mitf2 and Mitf1, respectively (Figure 1B). Designation of fish mitf genes as mitf1 and mitf2 will allows to similarly nomenclature fish Mitf isoforms, a salient feature of the mitf gene as has been demonstrated in mammals [38,39].
By using an invertebrate Mitf protein from Drosophila [40] as the out-group, phylogenetic trees were constructed on the basis of amino acid sequences. In the phylogenetic tree constructed by using the neighbor-joining algorithm on the DNAMAN package, all vertebrate Mitf proteins fall into two major clades. One clade consists of fish Mitf2 proteins only, and the other comprises fish Mitf1 proteins and all mammalian Mitf proteins, as represented by the human MitfA and MitfM isoforms (Figure 1B). However, fish Mitf2 but not Mitf1 proteins may clustered with tetrapod counterparts on phylogenetic trees constructed by the maximum likelihood (Supplementary Material: Figure S3) and minimum evolution methods (Supplementary Material: Figure S4). In either case, the cross-species Mitf1 sequence similarity is generally higher than within-species comparisons between Mitf protein pairs in the fish species, suggesting that fish mitf duplicates may have shared an ancient WGD event in the common ancestor.
Medaka Mitf1 and Mitf2 differ in sequence divergence rate
A total of ~40 fish Mitf sequences are available in public databases (http://www.ncbi.nlm.nih.gov; http://www.ensembl.org). This allowed us to compare the sequence divergence of Mitf duplicates in various fish species by using tetrapod Mitf proteins as a reference (Table 1). Seven Mitf pairs of representative species fall into two categories. The first include Xiphophorus, fugu, two cichlid species and zebrafish and exhibits an apparent difference of 6% in sequence divergence between Mitf1 and Mitf2: On average, the 5 Mitf1 proteins are 73.5% identical to the tetrapod Mitf proteins, whereas this value decreases to 67.6% for the 5 Mitf2 proteins (Table 1). Medaka also belongs to this category but represents an extreme. Notably, the medaka Mitf1 is 72.2% similar to the tetrapod reference. However, this value for the medaka Mitf2 sharply declines to 60.2%, leading to a difference of up to 12% between the medaka Mitf1 and Mitf2 (Table 1). The second category is represented by stickleback, in which Mitf1 and Mitf2 display 67.1~69.7% and 67.6~72% identity to the tetrapod reference, respectively, and thus a similar rate of sequence divergence. Taken together, medaka Mitf1 exhibits a slower, and perhaps basal, evolutionary rate of change, whereas Mitf2 shows a significant acceleration in divergence rate.
Phylogenetic comparisons. (A) Schematic protein domain structure. Alternative exon 1 (1a, 1b and 1m) and common exons 2-9 are shown above the protein. Also shown are protein domains: AD, activation domain; CD, conserved domain; bHLH-Zip, basic helix-loop-helix leucine zipper domain; asterisk, phosphorylation site by MAPK; hash, phosphorylation site by GSK3β. (B) Phylogenetic tree by the NJ algorithm. The Drosophila Mitf serves as the out-group. Bootstrap values are given. Accession numbers follow organisms and original names of Mitf proteins.
Percentage Mitf sequence identities between fishes and tetrapods1)
| Fish Mitf (aa) | Tetrapod Mitf (aa) | |||||||
|---|---|---|---|---|---|---|---|---|
| New name | Species | Current name | human | mouse | chicken | |||
| MitfA (520) | MitfM (419) | MitfA (526) | MitfM (419) | MitfA (468) | MitfM (413) | |||
| Mitf1 | Xiphophorus | Mitfb (462) | 72.6 | 73.5 | 70.9 | 71.2 | 73.1 | 72.6 |
| Fugu | Mitf1(478) | 73.7 | 72.8 | 71.6 | 69.6 | 71.9 | 70.1 | |
| Maylandia callainos | Mitfb (402) | 77.4 | 77.1 | 76.1 | 74.7 | 77.8 | 76.0 | |
| Maylandia zebra | Mitfb (512) | 75.1 | 73.9 | 72.7 | 70.7 | 74.5 | 72.7 | |
| Zebrafish | Mitfb (427) | 74.6 | 75.8 | 73.0 | 72.3 | 73.6 | 72.4 | |
| Average | 74.7 | 74.6 | 72.9 | 71.7 | 74.2 | 72.8 | ||
| 73.5 | ||||||||
| Medaka | Mitf1 (523) | 73.3 | 73.8 | 71.2 | 70.6 | 72.6 | 71.8 | |
| Average | 72.2 | |||||||
| stickleback | Mitf2 (493) | 69.7 | 69.0 | 68.1 | 67.1 | 68.9 | 68.9 | |
| Mitf2 | Xiphophorus | MitfM (402) | 65.3 | 68.7 | 63.9 | 66.1 | 65.3 | 64.9 |
| Fugu | Mitf2 (394) | 66.9 | 70.6 | 65.7 | 67.9 | 66.4 | 66.0 | |
| Maylandia callainos | Mitfa (401) | 68.5 | 71.4 | 66.8 | 69.0 | 68.2 | 67.9 | |
| Maylandia zebra | Mitfa (401) | 68.2 | 71.1 | 66.5 | 68.7 | 67.9 | 67.6 | |
| Zebrafish | Mitfa (412) | 67.8 | 71.5 | 65.6 | 67.8 | 68.0 | 67.7 | |
| Average | 67.3 | 70.7 | 65.7 | 67.9 | 67.2 | 66.8 | ||
| 67.6 | ||||||||
| Medaka | Mitf2 (406) | 59.5 | 63.0 | 58.9 | 60.9 | 59.5 | 59.1 | |
| Average | 60.2 | |||||||
| stickleback | Mitf1 (404) | 68.8 | 72.0 | 66.8 | 69.0 | 67.9 | 67.6 | |
1) For gene accession numbers see Figure 1B.
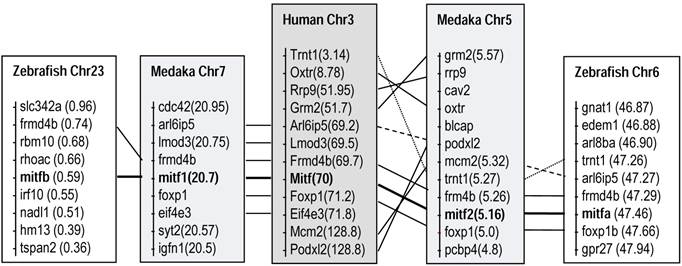
Fish mitf1 and mitf2 are co-homologs of the human Mitf by chromosome synteny. Medaka and zebrafish are compared to human. Data are compiled from http://www.ensembl.org/index.html. Chr, chromosome; numerals in parentheses, chromosomal positions.
Medaka mitf1 and mitf2 genes are the products of WGD
Comparisons of chromosome locations revealed clear syntenic relationships between the mitf-bearing regions of human chromosome 3 and those of fish chromosome pairs, chromosomes 5 and 7 in medaka, and chromosomes 6 and 23 in zebrafish, respectively (Figure 2). These data strongly suggest that the presence of mitf gene pairs in diverse fish species is the consequence of WGD, which has been dated ~450 million years ago [5,41,42]. The chromosome syntenic relationship also demonstrates that the medaka mitf1 and mitf2 are co-homolog of the human Mitf gene.
Medaka mitf1 and mitf2 differ in the level of expression
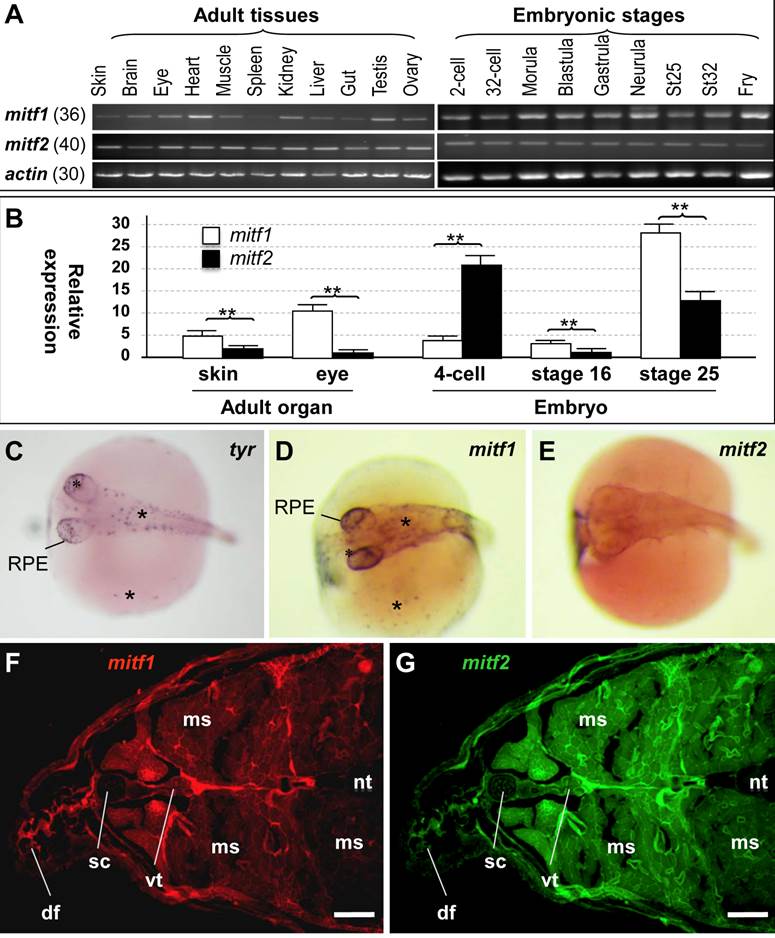
In mammals, a single mitf gene shows differential expression from alternative promoters and exons, and thereby producing multiple isoforms [15,19-21]. We performed RT-PCR analysis to examine mitf expression (Figure 3A). In adult tissues, the transcripts of both genes were detected in all samples examined, including the ectodermal skin, brain and eye, mesodermal muscle, heart, kidney and spleen, endodermal liver and gut, and male and female gonads (testis and ovary). There was no apparent difference between mitf1 and mitf2 in a ubiquitous adult expression pattern. However, the level of mitf2 expression appeared to be substantially lower than mitf1. After 36 cycles of PCR, mitf1 was easily detectable (Figure 3A), whereas mitf2 was barely detectable (data not shown). mitf2 became detectable only after 40 cycles (Figure 3A). In developing embryos, the transcripts of mitf1 and mitf2 were detected as early as 2-cell stage (Figure 3A), suggesting their maternal inheritance. The expression of mitf1 and mitf2 occurs throughout embryogenesis. Notably, the RNA is clearly more abundant for mitf1 than for mitf2 also in developing embryos. Real-time RT-PCR analyses (Figure 3B) revealed that the mitf1 RNA was more abundant than mitf2 by 3 and 8 fold in the adult skin and eye, and 3 and 2 fold at embryonic stages 16 and 25, respectively. However, at the 4-cell stage, the mitf2 RNA was more abundant than mitf1 by 5 fold. Taken together, mitf1 and mitf2 show different levels of RNA expression in embryogenesis and adult tissues.
Medaka mitf1 retains and mitf2 loses melanocytic expression in embryos
To study the expression patterns of the medaka mitf1 and mitf2 in more detail, we performed in situ hybridization (ISH) in developing embryos at stage 29 (3 days post fertilization; dpf) [43], when pigment cells from the neural crest are clearly visible on the body and yolk sac, and pigmented RPE is evident in the eye [44]. To prevent black pigmentation in melanocytes, we treated embryos with phenylthiourea and used them for ISH. We made use of the tyr transcript as a melanocyte marker, which encodes tyrosinase, the key enzyme for melanin synthesis in maturing/mature melanocytes [29], In stage-29 embryos, tyr-positive cells were found as individual cells on the embryonic body, yolk sac and in RPE (Figure 3C). We found that mitf1 exhibited a similar expression pattern (Figure 3D). However, mitf2 expression was barely detectable at this stage (Figure 3E). Taken together, the medaka mitf1 resembles the mouse mitfm in predominant expression in the pigment lineage, whereas the medaka mitf2 did degenerate its expression in embryonic melanocytes. This suggests that medaka mitf1 but not mitf2 might be a major player for embryonic melanocytes at this critical stage.
We then performed fluorescence ISH to examine the adult expression on cross-sections of the posterior trunk. Both genes exhibited a ubiquitous expression in a wide variety of tissues, including the skin, muscle, neural tube and vertebra (Figure 3F and G). We noticed that the mitf2 signal was relatively stronger than mitf1 in the spinal cord and muscle. Hence, mitf1 and mitf2 show a generally overlapping expression pattern in adult tissues.
Medaka Mitf2 possesses a reduced melanogenic activity
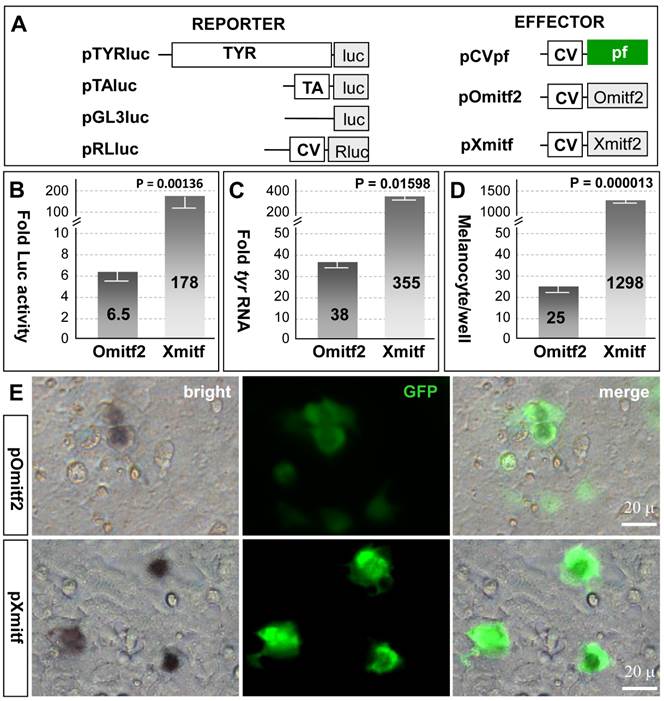
Mitf controls pigmentation through two distinct aspects: One is to activate the expression of enzymes (e.g. tyrosinase) involved in melanin synthesis (melanogenesis) and in maturing/mature melanocytes and the other is to regulate melanocyte formation. The observation that the medaka mitf2 exhibits an accelerated sequence divergence rate and a lower level of expression provokes us to examine whether this gene has maintained activity. To perform transfection experiments in medaka ES cell line MES1, which is capable of directed melanocyte differentiation following transfection by pXmitf that expresses the Xiphophorus mitfm [34], we constructed several expression vectors (Figure 4A). pTYRluc contains the 3.3-kb medaka tyrosinase gene promoter (TYR) ahead of the luciferase [45], and pOmitf2 constitutively expresses the medaka mitf2 cDNA. In undifferentiated MES1 cells, mitf expression is not detectable by RT-PCR analysis [33], but can be activated by forced XmitfM expression [34]. Transfection with pTYRluc alone or together with pCVpf as a control led to a low reporter activity, which was 6 times that of transfection with pTAluc that contains a TATA-box minimal promoter. When pOmitf2 was used for cotransfection with pTYRluc, a 6.5-fold increase in luciferase activity was observed (Figure 4B), demonstrating that the medaka Mitf2 maintains melanogenic activity. However, when pXmitf was used for cotransfection, an increase by up to 178 fold was obtained (Figure 4B). Therefore, the melanogenic activity of the medaka Mitf2 is reduced by 27 fold compared to the Xiphophorus Mitf2. We then examined the induction of endogenous tyr expression by ectopic mitf expression in MES1 cells. Upon transfection with pOmitf2 and pXmitf, the tyr RNA expression was increased by 38 and 355 fold, respectively (Figure 4C). Compared to the Xiphophorus Mitf2, the medaka Mitf2 displays a 9.3-fold reduction in its activity to induce the endogenous tyr expression. Taken together, the medaka mitf2 retains melanogenic activity that has considerably been degenerated compared to the Xmitf.
RNA expression. (A) RT-PCR analysis in adult tissues and embryos. The number of PCR cycles is given in parenthesis for each gene. (B) Real-time PCR analysis. β-actin was used as the control for data calibration. Data are means ±s.d (error bars) of five determinations. **, p ≤ 0.01. (C-E) Chromogenic ISH with antisense riboprobes on whole mount embryos at stage 29. Melanocytes (asterisks) in the body, yolk sac (sc) and RPE are positive for tyr and mitf1 but not mitf2. (F and G) Cross-section of the adult posterior trunk after two-color fluorescence ISH with mitf1 (F) and mitf2 (G) antisense riboprobes. df, dorsal fin; ms, muscle; nt, neural tube; sc, spinal cord; sk, skin; vt, vertebra. Scale bars, 100 µm.
Medaka Mitf2 possesses a reduced melanocyte-inducing activity
We then addressed whether the medaka Mitf2 possessed melanocyte-inducing activity, the original function of the prototype mouse MitfM. To this end, we took the advantage of an in vitro system for melanocyte induction again by using the medaka ES cell line MES1, in which Xmitf expression is sufficient to direct melanocyte differentiation [34]. When pOmitf2 was used for transfection, 25 melanocytes were formed per well of a 12-well plate. This number increased to 1298 for pXmitf transfection (Figure 4D). This difference by 52 fold is not ascribed to variations in transfection efficiency, because many GFP-positive transgenic cells following pOmitf2 transfection did not become melanocytes, whereas the majority of GFP-positive cells following pXmitf transfection did (Figure 4E). Convincingly, the medaka Mitf2 possesses a reduced activity for melanocyte induction in vitro compared to the Xiphophorus counterpart.
In summary, the medaka mitf duplicates adopt two distinct directions of evolution: While mitf1 displays conservation, mitf2 shows significant degeneration in protein sequence, expression and activity.
Medaka Mitf2 has a reduced activity. MES1 cells were used for cotransfection and analyses. (A) Vectors. (B) Luciferase reporter assay. Each of reporter plasmids pGL3luc, pTAluc and pTYRluc in combination with pRLluc was cotransfected with effector plasmids pOmitf2 or pXmitf into MES1 cells, and Luc activity was determined at day 3 dpt. pRLluc was used as the internal control for calibration of different samples and transfection efficiencies. pGL3luc was used as the negative control, whose activity was considered as background. pTAluc was used as the standard for normalization, whose activity was defined as unit activity. Fold activity was obtained by compared to control cotransfection with pCVpf. Data are means ±s.d of three experiments with triplicates each. (C) Real-time RT-PCR analysis of induced endogenous tyr expression. Data are means ±s.d of five determinations each. (D) Melanocyte formation. Data are means ±s.d of triplicate determinations in 12-well plates. P values are given in (B-D). (E) Micrographs showing the number of melanocytes relative to the number of GFP-positive transgenic cells.
Discussion
We have recently identified Mitf as a transcriptional activator of medaka germ genes in vitro [46], and both mitf1 and mitf2 mRNAs as the component of the Balbiani body [47], a structure resembling germ plasm that exists transient in growing oocytes [48]. In this study, we have identified the medaka mitf1 and mitf2 gene duplicate as the co-homologs of the mouse mitf. Strikingly, the medaka mitf1 retains expression in the embryonic pigmentary lineage including both melanocytes and RPE, in which the medaka mitf2 lacks detectable expression. We propose that medaka makes use of the DCD mode for the evolution of duplicated mitf genes: In the developing embryo, mitf1 has retained the original melanocytic expression and perform the ancestral mitf function, whereas mitf2 has undergone degeneration to such a degree that its melanocytic expression becomes hardly detectable at the critical stages. In contrast, zebrafish utilizes the DDC mode for the evolution of its duplicated mitf genes, where both genes have undergone partial retention and degeneration to jointly display complementary expression and thus execute complementary subfunctions of a single ancestral mitf in both melanocytes and RPE. Whether mitf2 degeneration is towards neofunctionalization or nonfunctionalization remains open.
Concurrent retention and loss of expression in the embryonic pigmentary lineage is a key feature of the DCD mode. Since various cell lineages are formed during embryogenesis, loss of expression at critical stages of embryogenesis may exclude an essential role for the degenerating mitf2 in the pigment lineage establishment. Notably, the mitf2 (also named mitfa) promoter can drive transgene expression in adult pigment cells [49].
Besides the loss of expression in the embryonic pigmentary lineage, the medaka mitf2 also shows an expression level that is much lower than the conserved mitf1 in both embryos and adult tissues. We show that reduced mitf2 expression is not compensated by increased melanocyte-inducing and melanogenic activity, but instead, accompanied by a remarkably degenerated melanogenic activity. These facts together with a higher divergence rate of the Mitf2 sequence strongly suggest that mitf2 has already been released from the constraint for pigment development and is in a transitory status towards neofunctionalization or nonfunctionalization. The mouse mitf is also expressed in several non-pigmentary lineages in the form of various variants. A substantially lower expression level would imply that mitf2 has degenerated its role to some extent also in non-pigmentary lineages. It deserves to note that in the case of medaka mitf gene pair, the DCD mode operates at multiple aspects, including sequence divergence rate, expression and activity of the protein product. It is unknown whether these aspects act independently or collectively.
Expression of the Xiphophorus mitf gene pair has been examined by RT-PCR analysis in adult tissues: mitfb shows ubiquitous expression, whereas mitfm is expressed only in the eye, skin and melanomas [22]. This suggests that the Xiphophorus mitf duplicates also adopt the DCD mode: mitfm has retained its pigmentary expression, whereas mitfb has acquired non-pigmentary expression in the adulthood. It is unknown whether the mitf genes also use the DCD mode during Xiphophorus embryogenesis.
To compare the relative protein activity, we have chosen the medaka Mitf2 and its Xiphophorus counterpart Xmitf, the latter being the only fish Mitf that has been studied both for expression in vivo [37] and activity in vitro [34]. This comparison has revealed a great reduction in the activity of the medaka Mitf2 protein product, consistent with its accelerated protein sequence divergence indicative of degeneration. An ideal situation is to include the medaka Mitf1 also in the experiment, which is, however, not practical, at the present. In contrast to the mitf2 that produces only one form, the mitf1 is able to produce multiple isoforms, which isoform is functionally equivalent to mitf2 remains to be determined.
Our finding that the wide adult expression of medaka mitf2 accompanies the retention of melanocyte-inducing activity by its protein product in combination with the fact that Mitf is a melanocyte master regulator raises an interesting question as to why many mitf2-expressing cells do not become melanocytes. A low level of expression and a dramatically reduced activity for melanocyte induction may account for an insufficient activity for melanocyte induction in non-pigment cells. This notion is supported by the fact that the expression of highly active Xmitf is limited to the pigment cell-containing tissues in Xiphophorus [22]. It deserves to note that the medaka mitf2 has been compared for melanogenic activity and melanocyte induction with the Xmitf in this study. The medaka mitf1 can produce multiple isoforms (data not shown). Future work will elucidate which isoform corresponds to the Mitf2 protein product to allow for a direct comparison between mitf1 and mitf2.
Our observation that both mitf1 and mitf2 are maternal factors in medaka is intriguing, because in all other vertebrate species examined so far, the mitf gene does not show maternal inheritance and its expression does not commence until late stages of embryogenesis when the pigment cell lineage starts to develop. For example, zygotic mitf expression occurs at the 16-somite stage in zebrafish [25,26], stage 21/22 in Xenopus [50], the 5-day-old embryo in chicken [51] and the 9.5-day-old embryo in mouse [52]. Interestingly, mitf maternal inheritance has been described in diverse invertebrate species such as Drosophila [40] and ascidians [53].
Our data illustrate that the mitf gene duplicates may adopt different ways of evolutionary divergence in different species. This species-specific difference has recently reported also to tyrp1 encoding tyrosinase-related protein 1 (Tyrp1). In medaka embryos, tyrp1a expression occurs in the RPE and melanophores while tyrp1b is expressed only in melanophores. In zebrafish embryos, the duplicated tyrp1 genes exhibit overlapping expression in the RPE and melanophores, and knockdown of both tyrp1 genes leads to pigmentary defects [54]. In this case, medaka adopts the DCD mode, and zebrafish uses the DDC mode as usual. It will be interesting to determine whether non-pigmentary genes also exhibit such a species-specific difference in evolutionary divergence.
Approximately 30% of zebrafish genes are duplicates [5,55], their current co-existence may well be explained by the DDC mode [7]. Evolution of many other gene duplicates must follow a different way, so that one copy can no longer be identified as a member of the gene pair. This copy may have undergone neofunctionalization or nonfunctionalization. During the past process of neofunctionalization or nonfunctionalization, the DCD mode must have operated. Therefore, the DCD mode used by the medaka mitf gene may represent an intermediate step for gene loss or neofunctionalization in the past, and towards gene loss or neofunctionalization in the future. During early stages of evolution, the DCD mode might be insufficiently distinguishable to the DDC mode because of remnant expression and/or function.
Methods
Fish
Work with fish followed the guidelines on the Care and Use of Animals for Scientific Purposes of the National Advisory Committee for Laboratory Animal Research in Singapore (permit number 27/09). Medaka strains HB32C, orange, i1 and af were maintained under an artificial photoperiod of 14-h/10-h light/darkness at 26°C as described [56]. Embryogenesis was staged according to the description [44].
Sequence Analysis
Multiple sequence alignment was conducted by using the Vector NTI suite (version 11, Invitrogen). A phylogenetic tree was constructed by using the DNAMAN software (version 4.15, Lynnon Biosoft) from a matrix of pairwise genetic distances according to the neighbor-joining (NJ), maximum likelihood [57] and minimum evolution (ME) methods, and 1,000 trials of bootstrap analyses were used to provide confidence estimates for tree topologies. Genomic organization and chromosomal locations were investigated by comparing the cDNA and corresponding genomic sequence (http://www.ensembl.org).
Plasmids
The medaka tyrosinase (tyr) cDNA is available (accession number NM_001104802) and annotated under ENSORLG00000004539. Putative medaka mitf1 (ENSORLG00000013461) and mitf2 (ENSORLG00000003123) are available in the genome sequence (http://www.ensembl.org/index.html). Their cDNA sequences were PCR-amplified from a cDNA library of 9-day-old embryos by using primer sets tyr882f (CGCTGACGTAGAGTTCACCAT) plus tyr1621R (TAGCCGTCATTGTGGCCAA) for tyra, GAATTCATGCAGTCCGAATCCGGAATAG plus ATCGATACAGCCCTGCTCGTTTTCATCCAT for mitf1, and ttgAGATCTGAATTCATGTTGGAGATGTTGGAATACAGC plus ttgATCGATACAGCCATTCTCTTTTTCCTCCATGC for mitf2. The PCR products were of 740 bp, 1230 bp and 1182 bp in length and cloned into pGEM-T, producing pGEMtyr, pGEMmi1 and pGEMmi2. Correct cloning was confirmed by sequencing.
Four luciferase [45] reporter plasmids were used in this study. pGL3-basic is promoterless and contains the firefly luc (Promega). pTAluc was derived by inserting the TATA-containing minimal promoter of the herpes simplex virus thymidine kinase upstream of luc in pGL3-basic. To construct pTYRluc, the 3.3-kb promoter of a medaka tyrosinase gene (tyra; accession number AB010101) was obtained by PCR from genomic DNA with primer TTACGCGTAGTGAGTGATGCACCTCTGC and TCTCGAGATTCACCCACATCTGTCC (restriction sites are underlined) and inserted between Mlu I and Xho I ahead of the luc in pGL3-Basic. pRL-CMV expresses the Renilla Luc and used as internal reference to calibrate transfection efficiency (Promega).
Two effector plasmids used were pXmitf (originally called pCMVTKmitf-M) expressing the Xiphophorus mitf-M [34] and pOmitf2 expressing the medaka mitf2 , the latter being constructed by inserting the medaka mitf2 cDNA in frame to the cherry sequence in pCS2+. In addition, pCVpf was constructed by inserting in frame the puromycin acetyltransferase gene to gfp in pEGFP-N1 (Clontech). These three plasmids use the human cytomegalovirus early enhancer/promoter (CV) to drive transgene expression.
RNA isolation and RT-PCR
Total RNA from medaka embryos, adult tissues and cell culture was isolated by using the Trizol reagent (Invitrogen) and used for cDNA synthesis by using oligo(dT)25 and the MMLV reverse transcriptase (Invitrogen) as described [30]. PCR was performed by using primers GTAGATTACATCCGCAAGCTG and ACAGCCCTGCTCGTTTTCATCCAT for mitf1, AGCTCCTGTGCATCTGGCATT and GTTGAACCTTCGCCTTCGTT for mitf2. As a control, β-actin was amplified from the same set of cDNA samples using primers TTCAACAGCCCTGCCATGTAC and CCTCCAATCCAGACAGAGTATT. PCR was run for 30 cycles (β-actin) and 36 or 40 cycles (mitf1 or mitf2) of 10 s at 94°C, 15 s at 58°C and 60 s at 72°C. The PCR products were separated on 1.0% agarose gels and documented with a bioimaging system (Synoptics, Cambridge, UK).
Real-time RT-PCR
Real-time PCR was performed with SYBR® Green qPCR SuperMix (Invitrogen) on the IQ5 real-time PCR detection system (BioRad). The primers for detecting were tyr1514F (TCACCATGCTTTCATTGACAGC) and tyr1621R for the medaka tyra, mitf1-QF (TCTCCTGCCATCATGCACA) and mitf1-QR (AGCGCTCTTACCTCAGCTTCA) for the medaka mitf1, mitf2-QF (CAACTTGATTGAACGAAGGCG) and mitf2-QR (TGTTCCAGCGCATATCTGGAT) for the medaka mitf2, actinqs (AGAAGAGCTATGAGCTGCCTG) and actinqr (AACATCACACTTCATGATGCTG) for the medaka β-actin as a loading control. The relative fold change of tyr quantities was calculated with 2(-delta delta Ct) method [58].
In situ hybridization
Antisense RNA probes were synthesized from linearized pGEMtyr, pGEMmi1 and pGEMmi2 by using the DIG RNA labeling kit [59] as described [60]. Embryos at stage 29 were subjected to in situ hybridization (ISH) with DIG-labeled probe followed by chromogenic staining with NBT/BCIP as described [61]. Sections were subjected to two-color fluorescence in situ hybridization [20] by using the tyramide signal amplification (TSATM) Plus Fluorescence Systems according to the manufacturer's instruction (NEL756, PerkinElmer Life Science) as described [60]. Briefly, after hybridization and blocking as described above, the samples were incubated with the POD-conjugated anti-FITC-antibody (Sigma) at a 1:2000 dilution for 2 hours at room temperature to detect the FITC-labeled probes. Following six PBST washes the samples were incubated for 30 minutes in the TSA-Fluorecein at a 1:100 dilution in the TSA Amplification Buffer (0.004% H2O2 in 0.1 M borate buffer, pH8). The samples were then subjected to detection of the DIG-labeled probe: They were treated for 1 hour in 1% H2O2 and incubated for two hours with the horseradish peroxidase (POD)-conjugated anti-DIG antibody (Sigma) at a 1:2000 dilution, followed by 30-min incubation in TSA-Cy3 for 30 min. The samples were finally stained for nuclei by using DAPI (µg/ml) and embedded in the Gold Antifade reagent (Invitrogen) for microscopy. In this FISH procedure, two differently labeled antisense RNA probes are co-incubated with a sample in the hybridization step and subjected to sequential color development into green and red fluorescence [62].
Cell culture and transfection
Plasmid DNA was prepared by using the Qiagen Midi- and Maxi-preps kit (Qiagen). The medaka ES cell line MES1 was maintained on gelatin-coated tissue culture plastic ware in medium ESM4 containing 15% fetal bovine serum [63] and transfected as described [64]. In this cell line it has previously been shown that pXmitf transfection is sufficient to direct melanocyte differentiation [34]. Cells in 24-, 12- and 6-well plates were transfected with 0.5, 1 and 3 μg of plasmid DNA by using the Gene Juice reagent (Novagen) or the DNAfectinTM 2200 with a ratio of 1 μg DNA to 4 μl of Gene Juice (Novagen) or the DNAfectin reagent (Applied Biological Materials Inc.) according to the manufacturers' instructions. pCVpf was cotransfected with pOmitf2 or pXmitf at a 1:4 ratio. Control transfection received only pCVpf. Appearance of black pigmented melanocytes was monitored from day 3 post transfection (dpt) onwards. In some experiments, cell transfection was followed by drug selection to enrich for transgenic cells. For this, cells following cotransfection with pCVpf were grown in the presence of puromycin (final 1 μg/ml) for 2 days before harvest for RNA isolation and RT-PCR analysis.
Luciferase reporter assay
For reporter assays, cells were transfected in 96-well plates with modifications [46,65,66]. Briefly, ~30,000 cells were seeded in 96-well plates (Biostar) and transfected with 102 ng of plasmid DNA. The following day, cells were at ~70% of confluence and transfected. For cotransfection, plasmid DNA for each well of 96-well plate contains 2 ng of pRL-CMV as the internal reference, 50 ng of a firefly luciferase reporter (pGL3-basic, pTAluc or pTYRluc), 25 ng of pOmitf2 or pXmitf, and 25 ng of pCVpf. For control transfection, each well received 2 ng of pRL-CMV, 50 ng of a firefly luciferase reporter plasmid and 50 ng of pCVpf. Transfection efficiency was monitored by GFP expression from pCVpf in a duplicate 96-well plate to be at ~10% and ~30% for the Gene Juice and DNAfectinTM 2200 reagents, respectively. At day 3 or 4 dpt, cells were measured for luciferase activity by using the Dual-GloTM Luciferase assay kit (Promega) and measured on GlomaxTM 96 Microplate Luminometer (Promega). Briefly, each well was replaced by 20 µl of fresh medium containing only 9% FBS. The plate was equilibrated at room temperature (RT) for 10 min. For measuring firefly luciferase activity, 20 µl of Dual-Glo™ Luciferase reagent was added to each well and mixed. After incubation at RT for 20 min, the firefly luminescence was measured in the GlomaxTM 96 Microplate Luminometer (Promega), Subsequently, 20 µl of Dual-Glo™ Stop & Glo® Reagent (1:100 dilution of substrate to buffer) was added to each well, mixed, incubated at RT for 10 min and measured similarly for the Renilla luciferase activity. The ratio between firefly luciferase activity and Renilla luciferase activity was used for comparisons. The reading from pGL3-basic was considered as background, and that from pTAluc treated as unit activity. Relative luciferase activity from pTYRluc alone or in combination with pOmitf2 or pXmitf was normalized to that of pTAluc.
Microscopy
Microscopy was done as described [43]. Briefly, Cells were observed and photographed on Zeiss Axiovert2 invert microscope and Axiovert200 upright microscope equipped with a Zeiss AxioCam MRc digital camera and AxioVision 4 software, and embryos and fry were visualized using a Leica MZFLIII stereo microscope equipped with a Fluo III UV-light system and photographed by using a Nikon E4500 digital camera (Nikon Corp).
Supplementary Material
Fig.S1 - S4.
Acknowledgements
We thank Jiaorong Deng for breeding fish, and Dr. J. Altschmied (Düsseldorf, Germany) for plasmid pXmitf. We acknowledge Yovita Ida Purwanti and Haobin Zhao for their participation in part of work. This work was supported by the National Research Foundation Singapore (NRF-CRP7-2010-03).
Abbreviations
mitf, microphthalmia-associated transcription factor; RPE, retinal pigment epithelium; WGD, whole genome duplication; dpt, day(s) post transfection
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ohno S, Wolf U, Atkin NB. Evolution from Fish to Mammals by Gene Duplication. Hereditas-Genetiskt Arkiv. 1968;59:169-187
2. Forsyth IA, Wallis M. Growth hormone and prolactin-molecular and functional evolution. Journal of Mammary Gland Biology and Neoplasia. 2002;7:291-312
3. Sakamoto T, McCormick SD. Prolactin and growth hormone in fish osmoregulation. Gen Comp Endocrinol. 2006;147:24-30
4. Fukamachi S, Sugimoto M, Mitani H, Shima A. Somatolactin selectively regulates proliferation and morphogenesis of neural-crest derived pigment cells in medaka. Proc Natl Acad Sci U S A. 2004;101:10661-10666
5. Amores A, Force A, Yan YL. et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711-1714
6. Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Current Opinion in Cell Biology. 1999;11:699-704
7. Force A, Lynch M, Pickett FB. et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531-1545
8. Deng C, Cheng CHC, Ye H, He XM, Chen LB. Evolution of an antifreeze protein by neofunctionalization under escape from adaptive conflict. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21593-21598
9. Leclercq E, Taylor JF, Migaud H. Morphological skin colour changes in teleosts. Fish and Fisheries. 2010;11:159-193
10. Braasch I, Schartl M, Volff JN. Evolution of pigment synthesis pathways by gene and genome duplication in fish. Bmc Evolutionary Biology. 2007;7:74
11. Hodgkinson CA, Moore KJ, Nakayama A. et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395-404
12. Vance KW, Goding CR. The transcription network regulating melanocyte development and melanoma. Pigment Cell Research. 2004;17:318-325
13. Steingrimsson E, Moore KJ, Lamoreux ML. et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256-263
14. Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757-3767
15. Hallsson JH, Favor J, Hodgkinson C. et al. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics. 2000;155:291-300
16. Tsujimura T, Morii E, Nozaki M. et al. Involvement of transcription factor encoded by the mi locus in the expression of c-kit receptor tyrosine kinase in cultured mast cells of mice. Blood. 1996;88:1225-1233
17. Smith SD, Kelley PM, Kenyon JB, Hoover D. Tietz syndrome (hypopigmentation/deafness) caused by mutation of MITF. J Med Genet. 2000;37:446-448
18. Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251-255
19. Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365-411
20. Hershey CL, Fisher DE. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347:73-82
21. Bharti K, Liu W, Csermely T, Bertuzzi S, Arnheiter H. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development. 2008;135:1169-1178
22. Altschmied J, Delfgaauw J, Wilde B. et al. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics. 2002;161:259-267
23. Braasch I, Brunet F, Volff J-N, Schartl M. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol Evol. 2009;1:479-493
24. Yamanoue Y, Miya M, Inoue JG, Matsuura K, Nishida M. The mitochondrial genome of spotted green pufferfish Tetraodon nigroviridis (Teleostei: Tetraodontiformes) and divergence time estimation among model organisms in fishes. Genes Genet Syst. 2006;81:29-39
25. Lister JA, Close J, Raible DW. Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev Biol. 2001;237:333-344
26. Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757-3767
27. Wittbrodt J, Shima A, Schartl M. Medaka--a model organism from the far East. Nat Rev Genet. 2002;3:53-64
28. Kelsh RN, Inoue C, Momoi A. et al. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech Dev. 2004;121:841-859
29. Koga A, Hori H. Albinism due to transposable element insertion in fish. Pigment Cell Res. 1997;10:377-381
30. Hong Y, Liu T, Zhao H. et al. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc Natl Acad Sci USA. 2004;101:8011-8016
31. Hong Y, Winkler C, Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes). Mech Dev. 1996;60:33-44
32. Hong Y, Winkler C, Schartl M. Production of medakafish chimeras from a stable embryonic stem cell line. Proc Natl Acad Sci U S A. 1998;95:3679-3684
33. Yi M, Hong N, Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430-433
34. Bejar J, Hong Y, Schartl M. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development. 2003;130:6545-6553
35. Thoma EC, Wagner TU, Weber IP. et al. Ectopic expression of single transcription factors directs differentiation of a Medaka spermatogonial cell line. Stem Cells Dev. 2010
36. Kasahara M, Naruse K, Sasaki S. et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714-719
37. Delfgaauw J, Duschl J, Wellbrock C. et al. MITF-M plays an essential role in transcriptional activation and signal transduction in Xiphophorus melanoma. Gene. 2003;320:117-126
38. Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163-1176
39. Vance KW, Goding CR. The transcription network regulating melanocyte development and melanoma. Pigment Cell Res. 2004;17:318-325
40. Hallsson JH, Haflidadottir BS, Stivers C. et al. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics. 2004;167:233-241
41. Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699-704
42. Postlethwait JH, Woods IG, Ngo-Hazelett P. et al. Zebrafish comparative genomics and the origins of vertebrate chromosomes 10(12). Genome Res. 2000;10:1890-1902
43. Xu H, Li Z, Li M, Wang L, Hong Y. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka. PLoS One. 2009;4(6):e6097
44. Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121:605-618
45. Falleni A, Lucchesi P, Ghezzani C, McDonald JC, Jones HD. The female gonad in two species of Microplana (Platyhelminthes, Tricladida, Rhynchodemidae): ultrastructural and cytochemical investigations. J Morphol. 2009;270:1042-1054
46. Zhao H, Li M, Purwanti YI. et al. Mitf is a transcriptional activator of medaka germ genes in culture. Biochimie. 2012;94:759-767
47. Li M, Yuan Y, Hong Y. Identification of the RNAs for transcription factor Mitf as a component of the Balbiani body. J Genetics Genomics. 2013;40:75-81
48. Xu H, Li M, Gui J, Hong Y. Fish germ cells. Sci China Life Sci. 2010;53:435-446
49. Schartl M, Wilde B, Laisney JA. et al. A mutated EGFR is sufficient to induce malignant melanoma with genetic background-dependent histopathologies. J Invest Dermatol. 2010;130:249-258
50. Kumasaka M, Sato H, Sato S, Yajima I, Yamamoto H. Isolation and developmental expression of Mitf in Xenopus laevis. Dev Dyn. 2004;230:107-113
51. Mochii M, Mazaki Y, Mizuno N, Hayashi H, Eguchi G. Role of Mitf in differentiation and transdifferentiation of chicken pigmented epithelial cell. Dev Biol. 1998;193:47-62
52. Nakayama A, Nguyen MT, Chen CC. et al. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech Dev. 1998;70:155-166
53. Yajima I, Endo K, Sato S. et al. Cloning and functional analysis of ascidian Mitf in vivo: insights into the origin of vertebrate pigment cells. Mech Dev. 2003;120:1489-1504
54. Braasch I, Liedtke D, Volff JN, Schartl M. Pigmentary function and evolution of tyrp1 gene duplicates in fish. Pigment Cell Melanoma Res. 2009;22:839-850
55. Postlethwait JH, Yan YL, Gates MA. et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345-349
56. Hong N, Li Z, Hong Y. Fish stem cell cultures. Int J Biol Sci. 2011;7:392-402
57. Lee HC, Li L, Gu W. et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803-814
58. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408
59. Padmanabhan PK, Dumas C, Samant M. et al. Novel Features of a PIWI-Like Protein Homolog in the Parasitic Protozoan Leishmania. PLoS One. 2012;7:e52612
60. Li M, Shen Q, Xu H. et al. Differential conservation and divergence of fertility genes boule and dazl in the Rainbow Trout. PLoS One. 2011;6:e15910
61. Liu L, Hong N, Xu H. et al. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells. Gene Expr Patterns. 2009;9:451-458
62. Li M, Shen Q, Wong FM. et al. Germ cell sex prior to meiosis in the rainbow trout. Protein Cell. 2011;2:48-54
63. Hong Y, Winkler C, Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes). Mech Dev. 1996;60:33-44
64. Hong Y, Chen S, Gui J, Schartl M. Retention of the developmental pluripotency in medaka embryonic stem cells after gene transfer and long-term drug selection for gene targeting in fish. Transgenic Res. 2004;13:41-50
65. Li M, Guan G, Hong N, Hong Y. Multiple regulatory regions control the transcription of medaka germ gene vasa. Biochimie. 2013;95:850-7
66. Zhao H, Hong N, Lu W. et al. Fusion gene vectors allowing for simultaneous drug selection, cell labeling, and reporter assay in vitro and in vivo. Anal Chem. 2012;84:987-993
Author contact
![]() Corresponding author: Professor Yunhan Hong, Department of Biological Sciences, National University of Singapore, Science Drive 4, Singapore 117543. E-mail: dbshyhedu.sg. Tel: +65-65162915; Fax: +65-67792486.
Corresponding author: Professor Yunhan Hong, Department of Biological Sciences, National University of Singapore, Science Drive 4, Singapore 117543. E-mail: dbshyhedu.sg. Tel: +65-65162915; Fax: +65-67792486.

 Global reach, higher impact
Global reach, higher impact