10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(9):917-933. doi:10.7150/ijbs.7224 This issue Cite
Review
The Many Faces of C/EBPδ and their Relevance for Inflammation and Cancer
Laboratory of Cell and Developmental Signaling, Center for Cancer Research, National Cancer Institute, Frederick, MD-21702-1201, U.S.A.
Received 2013-7-22; Accepted 2013-8-27; Published 2013-9-20
Abstract
The CCAAT/enhancer binding protein delta (CEBPD, C/EBPδ) is a transcription factor that modulates many biological processes including cell differentiation, motility, growth arrest, proliferation, and cell death. The diversity of C/EBPδ's functions depends in part on the cell type and cellular context and can have opposing outcomes. For example, C/EBPδ promotes inflammatory signaling, but it can also inhibit pro-inflammatory pathways, and in a mouse model of mammary tumorigenesis, C/EBPδ reduces tumor incidence but promotes tumor metastasis. This review highlights the multifaceted nature of C/EBPδ's functions, with an emphasis on pathways that are relevant for cancer and inflammation, and illustrates how C/EBPδ emerged from the shadow of its family members as a fascinating “jack of all trades.” Our current knowledge on C/EBPδ indicates that, rather than being essential for a specific cellular process, C/EBPδ helps to interpret a variety of cues in a cell-type and context-dependent manner, to adjust cellular functions to specific situations. Therefore, insights into the roles and mechanisms of C/EBPδ signaling can lead to a better understanding of how the integration of different signaling pathways dictates normal and pathological cell functions and physiology.
Keywords: C/EBP, transcription factor, cell signaling, inflammation, cancer.
Introduction
The CEBPD gene encoding the C/EBPδ protein, a member of the C/EBP transcription factor family, was cloned by sequence homology more than 20 years ago. Several reviews have been written about the C/EBP family and have emphasized other C/EBP proteins [1-6]. This article will focus specifically on C/EBPδ. Initially, C/EBPδ was mostly used as a marker for adipocyte or myeloid differentiation and of inflammatory activation. Early work on C/EBPδ was conducted primarily in cell lines and attributed a variety of functions to C/EBPδ that were not necessarily fully supported by studies in vivo. For example, C/EBPδ is not essential for adipocyte or myeloid differentiation in mice. Intriguingly, studies of C/EBPδ in different systems have led to it being labeled as a “tumor suppressor” or also as a tumor-promoting “mesenchymal master regulator” [7, 8]. From these seeming contradictions, and from more recent studies using Cebpd knockout mice, it has become apparent that C/EBPδ in fact functions as a modulator of many processes. This review summarizes and attempts to integrate some of our current knowledge about C/EBPδ, with an emphasis on findings that have relevance for cancer biology and inflammatory signaling.
Normal cellular functions occur through the orchestrated temporal and spatial interactions of molecules, whose roles have evolved and been selected for over millions of years. Genetic mutations and epigenetic changes that disturb these interactions may result in dedifferentiation, loss of growth control, or escape from cell death mechanisms, and eventually lead to cancer. Most cancer-related deaths are due to metastatic disease, which is the result of a series of complex steps in which malignant cells disseminate from the primary tumor and begin to establish growth in other organs [9]. It is also now apparent that most cancers can thrive only by corrupting the normal host environment, at both the cellular microenvironment and systemic levels [10].
Proteins whose aberrant activity can drive tumor development are termed oncogenes. Proteins whose normal activity primarily inhibits tumor development have been termed tumor suppressors. However, since the emergence of the “TGF-β paradox” [11, 12], it has become ever more clear that this nomenclature is often inaccurate or, at best, incomplete, and that this limitation in conceptualizing protein functions applies not only to cancer but to most paradigms in cell and organismal biology. Indeed, such labels can hinder our appreciation of the full complexity of molecular functions and of the diversity of molecular interactions that underlie the great variety of cellular and physiological outcomes. Our current knowledge of C/EBPδ suggests that it is one of the molecules that defy labeling, because it takes on multiple, context-dependent functions. The detailed study of such molecules presents an opportunity to gain deeper insights into the diversity and complexities of signaling pathways and their adaptations that govern normal and pathological conditions.
C/EBPδ as a versatile modulator of gene transcription
The C/EBP family of transcription factors is defined by a highly homologous basic region-leucine zipper (bZIP) domain that mediates its dimerization and DNA binding. C/EBPδ is encoded by the single-exon CEBPD gene, and only one full-length protein isoform is known to date. C/EBP factors can bind DNA only as dimers by virtue of the leucine zipper scissors' grip mechanism [1]. C/EBPδ's confirmed heterodimerization partners consist of all the other C/EBP proteins, including C/EBPζ /CHOP as well as ATF4 [13]. However, the specific roles of such heterodimeric interactions in C/EBPδ's physiological functions are completely unknown. Interestingly, both CHOP and the ATFs are stress response factors, with well-characterized roles in hypoxia and the unfolded protein response [14]. These proteins are prime candidates for physiological heterodimer formation with C/EBPδ, especially under hypoxia (see section below). This notion awaits further investigation.
C/EBP proteins exhibit largely identical DNA-binding specificities, at least in vitro [2, 13, 15]. Therefore, it is impossible to predict from DNA sequences which C/EBP member may target a particular gene. For example, the aromatase promoter is activated by C/EBPβ but not by C/EBPδ in human adipose fibroblasts [16]. Furthermore, target gene regulation may also depend on interactions with other regulatory factors. The prolactin gene promoter is repressed by C/EBPδ and activated by C/EBPα, in each case through interaction with Pit-1 [17, 18]. Comprehensive surveys of C/EBPδ's target genes by chromatin immunoprecipitation (ChIP) followed by subsequent validation using cells with C/EBPδ knockdown or knockout have been reported for a mouse mammary epithelial cell line [19], human keratinocytes [20], and mouse macrophages [21]. In addition, C/EBPδ ChIP data from K562 and HepG2 cells are now available from the ENCODE project (encodeproject.org).
C/EBPδ has a bona fide transcription transactivation domain [1], and most of C/EBPδ's target gene promoters are activated by it [19-21]. Figure 1 illustrates C/EBPδ's protein structure, highlights its protein interactions that have been confirmed with endogenous proteins, and shows the amino acids that are required for functional modifications. Target gene transactivation can be achieved by C/EBPδ's interaction with the CBP co-activator: that is, C/EBPδ triggers CBP phosphorylation, which depends on residues Leu81 and Phe82 on C/EBPδ [22]. However, C/EBPδ can also inhibit gene expression, depending in part on its protein modification or selective protein interactions. Sumoylation of C/EBPδ, which requires its K120 residue, abolishes its interaction with the CBP/p300 co-activator, and may be one mechanism by which C/EBPδ becomes a transcription repressor instead of an activator [23]. The K120 residue is also required for the acetylation of C/EBPδ by p300/CBP in vitro and may contribute to target gene transactivation [23, 24].
Schematic of the C/EBPδ protein, and mapped modifications and interactions. Only interactors that have been mapped to a sub-domain of C/EBPδ and whose interactions were confirmed at the level of endogenous proteins have been included in this Figure: SIAH2 [56] , FBXW7 [57], FANCD2 [41], IPO4 [41]. Amino acids (aa) that are important for protein modifications are indicated: #phosphorylation by GSK-3β; *K120 is necessary for acetylation and sumoylation [23] and for ubiquitination by SIAH2 [56] but not by SCFFBXW7 [57]. K120 was not confirmed as the acceptor site for these modifications. Transactivation Domain: aa 41-108 [60, 178] Basic Region/Leucine Zipper: aa 195-222/226-254 [6].
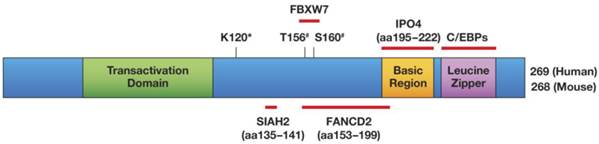
The physical interaction of C/EBPδ with SP1 [25] or ZNF638 [26] leads to synergistic gene activation, whereas C/EBPδ's interactions with Rad [27] or DIPA [28] inhibit the activation of target genes. C/EBPδ represses the expression of IGF-1 in osteoblasts [29], CD-RAP in chondrocytes [30], apoC-III in hepatoma cells [31, 32], manganese superoxide dismutase [33], and FBXW7α [34]. C/EBPδ's interaction with HDAC1 contributes to the repression of the haptoglobin [35] and of the PPARγ (PPARG) promoter [36], whereas its interaction with FOXO3a activates PPARγ expression [37]. C/EBPδ's association with GATA4 [38] or Runx 2 [39] can modulate target gene activation versus repression. Furthermore, its interaction with SMAD3 inhibits the DNA-binding function of C/EBPδ, which may be part of the mechanism by which TGF-β inhibits adipocyte differentiation [40]. Although far from complete, the diverse effects illustrated in the above list suggest that almost anything can result from C/EBPδ binding to a target promoter. Furthermore, a non-transcriptional function for C/EBPδ has also been discovered: it chaperones the FANCD2 DNA repair protein into the nucleus [41]. FANCD2 has also been shown to interact with the transcription factor NFκB and to inhibit as well as activate gene expression [42, 43]. Therefore, it is quite plausible that FANCD2 also modulates target gene regulation by C/EBPδ, in addition to the role of their interaction in nuclear translocation of FANCD2.
Most of the above-mentioned data were generated in cultured cell lines. Insights into physiologically relevant functions have been derived from mice with a null mutation of the Cebpd gene. Analysis of Cebpd knockout (KO) mice revealed that the protein is not essential for normal development or the adult life of mice in a pathogen-free environment [44, 45]. This is consistent with C/EBPδ's expression being low to undetectable in most organs and cell types. However, C/EBPδ expression is rapidly but transiently induced by a variety of stimuli [1, 6], which may be indicative of C/EBPδ's function as a modulator for specific physiological adaptations. Table 1 summarizes the phenotypes that have been characterized in Cebpd null mice to date.
Regulation of C/EBPδ expression and activity
As mentioned above, C/EBPδ expression is typically low to undetectable. However, a number of extracellular signals, including cytokines, bacterial lipopolysaccharide (LPS), and corticosteroid, can induce it, suggesting that C/EBPδ has a role in stress responses [6]. Hypoxia also induces C/EBPδ in cultures of various cell types, and in certain tissues, including brain and tumors [7, 34, 47, 48]. Table 2 summarizes some of the reports on the regulation of C/EBPδ expression, and highlights cases in which inflammatory regulators and hormones induce its expression. In addition, Runx factors have been described as either activating or inhibiting the CEBPD promoter activity [39, 49]. Finally, C/EBPδ can activate its own gene expression [1, 21].
Induced C/EBPδ expression is typically transient. In macrophages, LPS induces CEBPD through NFκB , which is subsequently silenced by ATF3 [21]. This feedback loop does not exist in kidney fibroblasts, where CEBPD is induced through c-Jun and maintained by a mechanism requiring IRAK-1 [50]. Interestingly, the histone deacetylase inhibitor trichostatin A interrupts c-Jun's binding to SP-1 and the Cebpd promoter, and prevents LPS-induced C/EBPδ expression in RAW264.7 macrophages [51]. Thus, CEBPD expression can also be modulated through modification of the c-Jun protein. In keratinocytes, CEBPD gene expression is inhibited by ΔNp63α, while C/EBPδ augments ΔNp63α expression, providing a mechanism for the transient C/EBPδ induction during differentiation [52]. The transient nature of C/EBPδ expression hints that persistently elevated C/EBPδ levels might lead to an adverse outcome. This concept will be discussed in more detail below, in the section on Inflammation.
At the level of mRNA stability, C/EBPδ expression can be increased by the binding of HuR to its 3'UTR. This mechanism can be activated by UV radiation through p38MAPK, and by prostaglandin E2 (PGE2), which trigger the nuclear to cytoplasmic translocation of HuR [53, 54]. Ectopic microRNA let-7c downregulates the CEBPD mRNA level in macrophages by a mechanism that requires the 3'UTR, which suggests that the CEBPD mRNA is directly targeted by let-7c [55].
At the level of protein stability, C/EBPδ is downregulated by the SIAH2 E3 ubiquitin ligase [56] and the SCF-FBXW7α polyubiquitination complex [57]. SIAH2 can act as an oncogene [58], while FBXW7α is a tumor suppressor in epithelial cancers [59]. Hence, two potentially antagonizing pathways both downregulate C/EBPδ expression. In turn, C/EBPδ inhibits the expression of FBXW7α [34], resulting in a negative feedback loop [57]. The degradation of C/EBPδ by FBXW7 depends on C/EBPδ's prior phosphorylation on threonine T156 (and possibly serine S160) by GSK-3β kinase. C/EBPδ is therefore stabilized by signals that inhibit GSK-3β activity, such as TLR4 activation [57].
Summary of C/EBPδ knockout mouse phenotypes. Only reports with in vivo studies (i.e., not those using only cultured cells) are included.
| Primary organ and cell type(s) | Main loss of function phenotype | Affected Gene(s)# | Strain background | References |
|---|---|---|---|---|
| Adipocytes | Exacerbated impairment of adipocyte differentiation due to C/EBPβ null mutation | Ucp1↓ | Mixed C57BL6x 129/Ola | [44] |
| Adipocytes | Reduced body and fat mass index specifically in females* | - | Mixed 129S1xC57BL6 (F2) | [81] |
| CNS, dendritic cells | Less severe clinical score for experimental autoimmune encephalomyelitis | Il10↑ | C57BL/6 | [126] |
| Brain, glia | Reduced LPS response | Tnfa↓ Il6↓ Il1b↓ | C57BL/6 | [127] |
| Brain | Enhanced learning by contextual fear conditioning | - | Mixed 129S1xC57BL6 | [45] |
| Peripheral motor neurons | Impaired regeneration after sciatic nerve injury | Sprr1a↓ | C57BL/6 | [163] |
| Tumor implant, lymphendothelial cells | Impaired lymphangiogenesis in grafted tumors | Vegfc↓ Vegfr3↓ | C57BL/6 | [48] |
| Systemic inflammatory, macrophages | Decreased collagen-induced arthritis | Ccl20↓ Cxcl1↓ Il23a↓ Tnfaip6↓ | C57BL/6 | [164] |
| Systemic, inflammatory | Reduced LPS responses and increased survival | Systemic: IL-6↓** TNFα↓ | C57BL/6 | [122] |
| Systemic, macrophages | Impaired clearance, specifically of high titer E. coli infections | Il6, Saa3, Cxcl2 and others↓ | C57BL/6 | [21] |
| Lung, macrophages | Reduced macrophage infiltration and clearance of Klebsiella infection** | Systemic Il-6↑ TNF-α↑ | C57BL/6 | [75] |
| Lung, epithelial and inflammatory cells | Relative resistance to pneumococcal pneumonia** | Pafr↓ | C57BL/6 | [120] |
| Lung, macrophages | Reduced LPS-induced lung injury | Lung: Il-6, TNF-α, MIP-2 ↓ | C57BL/6 | [82] |
| Kidney | Reduced kidney damage after renal ischemia/reperfusion injury** | Systemic TNFα↓ IL-6↓ | C57BL/6 | [122] |
| Kidney, tubular cells | Reduced renal absorption of lactate and urate | Slc5a8↓ Slc5a12↓ | C57BL/6 | [165] |
| Kidney, mesangial cells | Reduced experimental glomerulonephritis | Acta2↓ | 129 | [166] |
| Mammary gland | Delayed cell death and involution after lactation## | Tp53↓ Bcl2a1↓ Igfbp5↓ Ccnd1↑ Survivin↑ | C57BL/6 | [70] |
| Mammary gland, tumor | Increased MMTV-Neu mammary tumor multiplicity and reduced lung metastases, impaired hypoxia adaptation | Fbxw7a↑ Cxcr4↓ | FVB/N | [34] |
| Mammary gland, tumor; peritoneal macrophages | Reduced LPS response | Fbxw7a↑ Tlr4↓ | FVB/N | [57] |
| Mammary gland | Increased ductal branching in aged mice *** | - | Mixed 129S1xC57BL6 | [90] |
| Negative Results: | ||||
| Ovary, theca/interstitial cells | Not essential despite LH-induced expression | - | 129S1 | [71] |
| Bone | Normal bone density | - | Mixed 129S1xC57BL6 (F2) | [81] |
| Mammary gland | Morphologically normal involution## | Trpm2↓ Igfbp5↓ | Mixed 129S1xC57BL6 | [90] |
| Lung, macrophages | Normal inflammatory response to IgG | - | C57BL/6 | [83] |
#This column highlights some of the genes/proteins whose expression is regulated directly or indirectly by C/EBPδ. See citations for details.
## Note contradictory results of [90] and [70], possibly due to strain background.
*This phenotype was not observed previously in a study using the same strains [45], nor in studies using mice backcrossed to C57BL/6 [70], 129S1 [71], or FVB/N strains [34]. We speculate that the reduced body mass of C/EBPδ null female mice may be due to a maternal/paternal effect, because only this study [81] used compound C/EBPδ and C/EBPβ heterozygous breeders.
** Controls were not from the same cross as mutants.
***This phenotype was not observed in mice backcrossed to the C57BL/6 or FVB/N strain background ([70], and unpublished data) and may therefore be specific to the mixed strain background.
Regulation of C/EBPδ expression: Compilation of some reports describing activators or inhibitors of C/EBPδ expression at the RNA and/or protein level. See text for additional examples and further discussion. Exogenous Agent: Experimental treatments. Mediator(s): Molecules that regulate C/EBPδ expression directly or indirectly by pharmacological inhibition/activation, overexpression, RNA interference, or gene deletion. Parentheses indicate inferred mediators. Cell type(s): Predominant cell type used in the cited studies (h, human; m, mouse; r, rat; MEF, mouse embryo fibroblast; VSMCs, vascular smooth muscle cells). Associated Process: Cellular response to agent/mediator(s) whether functional relevance for C/EBPδ in that process was demonstrated or not. Reference(s): Examples are given with preference for most recent publications, as earlier literature on the subject is typically discussed in these papers.
| Exogenous Agent | Mediator(s) | Cell type(s) | Associated Process | References |
|---|---|---|---|---|
| Induction of C/EBPδ expression | ||||
| EGF | MAPK, CREB | A431 epidermoid carcinoma (h) | Growth Arrest | [167] |
| Hypoxia | HIF-1 | lymphendothelial cells (h) | Lymphangiogenesis | [48] |
| LPS | NFkB (REL) | Bone marrow-derived macrophages (m) | Inflammatory signaling | [21] |
| LPS | IRAK1, AP-1 | Kidney fibroblasts (m) | Chronic inflammation | [50] |
| LPS | IRAK1, IKKε (IKBKE) | Bone marrow-derived macrophages (m), MEF (m) | Inflammatory signaling | [61, 118] |
| Testosterone | CWR22 prostate cancer cells | Proliferation/progression | [94] | |
| Testosterone (DHT) | Androgen receptor | Rv1 and R1 prostate cancer cell lines (h) | ? | [168] |
| Progesterone | Progesterone receptor B | MDA-MB-231 breast tumor cells (h) | Inhibition of proliferation | [169] |
| Dexamethasone | Glucocorticoid receptor | C2C12 myotubes, muscle (m); A549 lung epithelial cells (h) | Muscle atrophy; glucocorticoid response | [132] [131] |
| IL-1 | JNK, CK2, p50/p65 NFκB | Hep3B hepatoma (h) | Inflammatory signaling | [170] |
| IL-1β+IFN-γ | STAT1 | INS-1E insulinoma cells (r) | Inflammation, cell death | [112] |
| IL-6 | STAT3, SP1 | HepG2 hepatoma cell line (h) | (Inflammatory signaling) | [171] |
| IL-6 | STAT3 | LNCaP prostate cancer cells (h) | Growth arrest | [92] |
| Oncostatin M | JAK2/STAT3 | HC11 mammary epithelial cells (m) | Growth arrest | [88] |
| Insulin | PI3K | VSMC | Proinflammatory gene expression | [172] |
| Runx2 | Osteoblasts (r) | Proliferation | [39] | |
| cAMP | Mdm2/CREB/Crtc2 | MEF | Adipogenesis | [173] |
| Vasoactive intestinal peptide, pituitary adenylate cyclase activating protein, noradrenalin, | (cAMP) | Cortical astrocytes (m) | Glycogen synthesis | [174] |
| 1-(2-hydroxy-5-methylphenyl)-3-phenyl-1,3-propanedione (HMDB) | p38, CREB | A431 (epidermoid) and HeLa (cervical) carcinoma cell lines (h) | Cell death | [99] |
| 1,25-Dihydroxyvitamin D3 | (VDR) | LNCaP prostate cancer cells; MCF-7, T47D breast cancer cells (h) | Growth inhibition | [93] |
| Inhibition of C/EBPδ expression | ||||
| M-CSF | Let-7c | Bone marrow-derived macrophages (m) | M2 polarization | [55] |
| Src (family) kinase | SIAH2 | Breast tumor cell lines (h) | C/EBPδ degradation, “transformation” | [56] |
| GSK3β, FBXW7α | Peritoneal macrophages | C/EBPδ degradation, inhibition of LPS- response | [57] | |
| Hsp70 | TLR2, IL-10 | Bone marrow-derived dendritic cells | Anti-inflammatory | [175] |
| LPS | ATF3 | Bone marrow-derived macrophages (m) | Feed-back inhibition of inflammatory signaling | [21] |
| LPS, TNFα | Miz1(pS178) | Lung tissue and MLE-12 lung epithelial cells (m) | Attenuation/inhibition of inflammatory signaling | [135] |
| Myc/Max/Miz1 | HC11 mammary epithelial cells (m) | (proliferation) | [134] | |
| Chrysin (Flavonoid) | ? | Embryonic cortical neurons, microglia cultures (m), BV2 microglial cell line (m) | Inhibition of inflammation | [128] |
| Troglitazone | Inhibition of STAT3 by PPARγ | VSMCs | Inhibition of IL-1β response | [136] |
| TGF-β | ? | VSMC | Inhibition of IL-1β-induced expression | [162] |
| SUZ12/YY1 | A431 (epidermoid) and HeLa (cervical) carcinoma cell lines (h) | (proliferation, tumor progression) | [142] | |
| ΔNp63α | HaCaT cells and primary keratinocytes (h) | Keratinocyte differentiation | [52] | |
| Pref-1 | Sox9 | MEF, 3T3-L1 pre-adipocytes | Inhibition of adipocyte differentiation | [176] |
| Tunicamycin | IREα | Liver, hepatocytes | Unfolded protein response, prevention of steatosis | [177] |
| Testosterone | (indirect) | Prostate tissue (r) | (tissue maintenance) | [94] |
Although no other phosphorylation sites on C/EBPδ have been mapped to date, several additional reports point to a role of phosphorylation in the regulation of C/EBPδ activity. The serine residue S191 in the extended basic region of C/EBPδ is necessary for the PGE2-mediated activation of C/EBPδ in osteoblasts, although the kinase has not been identified, and S191 has not been confirmed as a phosphorylation site [60]. IL-1 treatment of HepG2 cells induces the tyrosine-phosphorylation of C/EBPδ [32], Casein Kinase II phosphorylates C/EBPδ in vitro [15], and IKKi/IKKε (IKBKE) is essential for the LPS-induced phosphorylation of C/EBPδ [61]. In all cases, the phosphorylation events stimulated C/EBPδ's DNA-binding activity [15, 32, 61]. Experiments with phsophatase inhibitors also support that C/EBPδ needs to be phosphorylated to bind DNA [62]. Future investigations will have to unravel how IL-1, LPS, CaKII, IKBKE, and also GSK-3β intersect to regulate the C/EBPδ pathway. In addition to phosphorylation, the C/EBPδ protein can be modified by acetylation and sumoylation. These modifications require an intact K120 lysine residue, and, as mentioned above, serve to switch its transactivator function on or off [23]. Interestingly, the same K120 lysine residue is also required for the ubiquitination and degradation of C/EBPδ by SIAH2 [56].
By virtue of nuclear localization signals in its DNA-binding domain, C/EBPδ is primarily a nuclear protein, which is translocated at least in part by the IPO4 importin [41]. However, C/EBPδ's nuclear localization can be regulated. For example, TNF-α not only induces C/EBPδ expression, but also promotes its nuclear accumulation in the adult rat hepatocyte cell line RALA255-10G [63]. C/EBPδ's nuclear translocation is augmented by the DNA-damaging agent mitomycin C [41]. Billiard et al. demonstrated that a flag-tagged C/EBPδ protein is constitutively nuclear in rat calvarial osteoblasts, but the endogenous protein is cytoplasmic in the absence of PGE2 [64]. This last report highlights the potential for misconstruing physiological roles by relying too heavily on overexpression studies.
C/EBPδ constitutively resides in the nucleus of primary vascular smooth muscle cells (VSMCs) from the hypertensive rat, but not in those from normal rats [65]. In VSMCs, IL-1β triggers the relocation of C/EBPδ from its perinuclear localization to the nucleus [66]. The perinuclear localization of C/EBPδ has also been observed in a mammary epithelial cell line ectopically expressing the SUMO E3 ligase PIASy [67]. In differentiating adipocytes, during the clonal expansion phase, C/EBPδ localizes to the centromere when cells enter the S phase [68]. Thus, C/EBPδ's activity can be regulated at several levels by the modulation of its subcellular localization. However, the mechanistic details of these pathways and/or their physiological relevance remain to be determined.
Nuclear localization is not equivalent with activity. In a cell culture model of adipocyte differentiation, the DNA-binding activity of C/EBPδ is delayed compared to its induced expression [68]. In human primary osteoblasts, cytoplasmic PKA is necessary for the nuclear localization of C/EBPδ, while further activation of C/EBPδ requires nuclear PKA to mediate PGE2-induced IGF-1 promoter transactivation. However, PKA does not phosphorylate C/EBPδ in vitro, suggesting that the mechanism may be indirect [69].
Analyses of C/EBPδ expression in tissues have so far highlighted its primarily nuclear localization, which has been reported in the involuting mouse mammary gland [70], hypoxic/necrotic areas of mouse mammary tumors and human glioblastoma [7, 34], theca/interstitial cells of the mouse ovary [71], human breast epithelial cells [72], meningioma tumor cells [73], and lung bronchial epithelial and alveolar type II cells [74, 75]. However, even when specific, diffuse immunohistochemical staining in the cytoplasm can be difficult to distinguish from background, leaving open the possibility that C/EBPδ is more widely expressed in the cytoplasm than is currently recognized.
To conclude, much remains to be learned about how the various signaling pathways that regulate C/EBPδ expression and activity at different levels and through subcellular localization are integrated to control C/EBPδ's functions and target gene responses.
Integration of C/EBPδ signaling with other C/EBP family members
All five members of the C/EBP family can heterodimerize [13] and therefore could give rise to 15 different dimers with potentially unique or also redundant functions. The existence of several isoforms of C/EBPα, C/EBPβ and C/EBPε [1] may lead to even greater complexity. If and how this diversity is realized in nature is largely unknown. While many studies have described either synergistic or antagonistic regulation of expression of certain C/EBP family members, few have explored the functional consequences of such expression patterns. All full-length C/EBP proteins have been largely associated with growth arrest and differentiation functions [76]. However, these can be cell type-specific and exceptions exist, especially when considering different isoforms of the same protein [3, 4, 76]. The subsequent sections will present examples for C/EBPδ that highlight its multifaceted nature.
As mentioned above, certain gene promoters may be regulated differentially by specific C/EBP factors in cultured cells. However, the in vivo relevance of these observations has not been established. Most compellingly, gene replacement studies in mice have illustrated that C/EBPβ can substitute for C/EBPα in hematopoiesis and adipocyte differentiation [77, 78]. However, the mitochondrial biogenesis in fat cells is significantly altered in such mouse mutants [78]. It is conceivable that in many cases, C/EBP factors have redundant functions and act synergistically as suggested for C/EBPβ and C/EBPδ in inflammatory signaling [79, 80] and adipocyte differentiation [44]. Variations in expression between C/EBP members could be merely due to the divergence in the regulation of expression while preserving similar functions. For example, the inducible C/EBPδ protein may support the role of C/EBPβ that exhibits a more constitutive expression pattern [81]. Furthermore, similar functions may have been channeled into different signaling pathways. For example, C/EBPβ but not C/EBPδ is essential for immune complex-induced lung injury in mice, whereas C/EBPδ - not C/EBPβ - mediates LPS-induced lung injury, although both proteins are induced by the treatments [82, 83]. On the other hand, antagonistic roles of C/EBPα and C/EBPδ have been observed with respect to HIF-1α regulation. C/EBPα inhibits HIF-1 function by competing with HIF-1α for HIF-1β binding [84]. In contrast, C/EBPδ enhances HIF-1 activity by supporting HIF-α expression [34]. If and how these opposing functions are utilized in the same cell to modulate cell biology has not been explored. To fully understand the role of C/EBPδ functions compared to other C/EBP members, more systematic functional comparisons will be necessary including in-cell analysis of heterodimers and gene replacement studies that allow for temporal and spatial control of expression.
C/EBPδ and the regulation of cell proliferation and differentiation
C/EBPδ was first cloned by homology to other C/EBP factors and was subsequently recognized as a gene that is induced during growth arrest and the differentiation of certain cell types. For example, C/EBPδ expression is induced during G0 growth arrest, promotes contact inhibition in mammary epithelial cells [85, 86] and fibroblasts [87], and mediates the growth arrest of mammary epithelial cell lines in response to Oncostatin M [88]. However, the effect of C/EBPδ on proliferation can be cell-type dependent, and may reflect its role in the expression of secreted factors and/or intracellular regulators of the cell cycle.
Ectopic C/EBPδ reduces the growth rate of mammary epithelial cell lines but not of HEK293 cells [89]. These findings are in agreement with the observation that the loss of C/EBPδ leads to hyperproliferation and excess branching of the mammary gland epithelium in aged mice of a mixed 129B6 strain background [90]. In cultured mammary and myeloid tumor cell lines C/EBPδ promotes the downregulation of cyclin D and/or cyclin E and p27CIP2 expression [85, 89, 91], which is consistent with the suppression of cell growth. C/EBPδ also mediates growth inhibition by IL-6 or 1,25-dihydroxyvitamin D3 in the androgen-dependent LNCaP prostate cancer cell line [92, 93]. In contrast, another study described high basal levels of C/EBPδ expression in androgen-dependent human prostate cancer cells (three primary lines and CWR22 cells), which are further enhanced by testosterone [94]. Thus, more studies are needed to clarify if C/EBPδ regulates prostate cancer cell growth or differentiation in vivo, and how its functions may be modulated by hormone-receptor signaling.
In addition to regulating the expression of intrinsic cell-cycle regulators, C/EBPδ also modulates cell proliferation and growth arrest indirectly through the expression of secreted factors. In pituitary prolactinoma cells, C/EBPδ inhibits proliferation by suppressing prolactin expression [18]. On the other hand, C/EBPδ promotes the proliferation of cultured osteoblasts by directly activating the expression of IGF-1 [95]. Similarly, C/EBPδ promotes the proliferation of VSMCs in response to IL-1β by inducing expression of the PDGFA-receptor [66]. These cases in which C/EBPδ stimulates proliferation indirectly through secreted factors demonstrate clearly that the expression of C/EBPδ does not necessarily inhibit cell proliferation and can be compatible with cell proliferation.
In addition to the regulation of proliferation, C/EBPδ has also been implicated in the differentiation of certain cell types. For example, C/EBPδ expression is associated with early stages of pre-adipocyte differentiation [96, 97]. The loss of C/EBPδ alone in vivo impairs the development of brown fat and further exacerbates an adipogenesis defect observed in C/EBPβ KO mice [44]. In addition, studies of adipogenic differentiation in cell culture have provided molecular evidence for a direct role of C/EBPδ in tumor suppressor pathways. That is, C/EBPδ (like C/EBPβ and C/EBPα) specifically associates with the hypophosphorylated form of the retinoblastoma protein, Rb, which predominates in the G0/G1 phase of the cell cycle [98]. For C/EBPβ it was determined that the interaction with Rb potentiates promoter transactivation and promotes adipocyte differentiation; this interaction must be regulated by additional factors or specific modifications, because it is only seen in differentiating cells [98]. On the other hand, Rb interferes with reporter gene transactivation by C/EBPδ in the HeLa and A431 cancer cell lines where C/EBPδ attenuates the activity of the Rb partner E2F-1 and promotes apoptosis [99]. In breast cancer cell lines, C/EBPδ promotes phosphorylation of Rb, which is associated with Rb's tumor suppressor function, through downregulation of the kinase inhibitor cyclin D1 [56]. In a BCR-ABL-positive cell line (KCL22), C/EBPδ also interacts with Rb and E2F-1 and induces growth arrest and myeloid differentiation [91]. In summary, while the reported details vary, C/EBPδ's intersection with the Rb pathway has been correlated with cell death, differentiation, and inhibition of oncogenic transformation.
Consistent with these observations, a number of reports point to a tumor-suppressor function of C/EBPδ in the myeloid lineage. The CEBPD promoter is silenced in 35% of 260 human acute myeloid leukemia samples [8], and its expression is downregulated in the blast crisis phases of chronic myelogenous leukemia (CML) [100]. The overexpression of C/EBPδ in primary mouse hematopoietic progenitor cells or a cell line model of acute myelogenous leukemia (AML) leads to myeloid differentiation accompanied by upregulation of the G-CSF receptor and downregulation of c-myc expression [8, 101], as also seen in CML cell lines [91]. Indeed, C/EBPδ is expressed in differentiated granulocytes/neutrophils [91, 102]. In macrophages, C/EBPδ regulates many genes associated with functions of differentiated cells [21] and is specifically associated with M1 macrophage polarization [55]. While direct experimental evidence from mouse models is still missing, the potential tumor suppressor function of C/EBPδ in AML and possibly CML is most likely due to its function in stimulating differentiation, which may or may not be correlated with its function in cell cycle arrest.
The antithesis of the differentiated cell is the undifferentiated stem cell, with its self-renewal capacity. Very little information exists about C/EBPδ expression or functions in stem cells. One report describes C/EBPδ expression in limbal stem cells of the eye, where it maintains p63 expression, prolongs the cell cycle, and maintains stemness [103]. In addition, C/EBPδ expression was observed in cultured long-term self-renewing neural stem cells, dependent on the stem-cell maintenance factor LIF [104]. A role for C/EBPδ in stem-cell maintenance is consistent with its ability to downregulate drivers of the cell cycle. Interestingly, FBXW7α, which inhibits and is inhibited by C/EBPδ [34, 57], has been implicated in promoting stem-cell differentiation, pluripotency, and quiescence, depending on the cell type [105-107]. Therefore, the potential roles and mechanisms of C/EBPδ signaling in stem/progenitor cells are a promising but currently under-explored area of research.
C/EBPδ and the regulation of cell death and survival
The first evidence for C/EBPδ's ability to promote cell death came from the observation that ectopic C/EBPδ exacerbates the death of growth factor-deprived mammary epithelial cells [85]. C/EBPδ also promotes the cell death required for lumen formation when transformed mammary epithelial cells are cultured as 3D acini on matrigel [56]. Indeed, the loss of C/EBPδ delays cell death at the onset of the mammary gland involution that follows lactation [70]. Furthermore, in the context of this physiological cell death, C/EBPδ is induced by STAT3 and is necessary for the downregulation of cyclin D1, upregulation of p53 and IGFBP5, and proper regulation of several members of the Bcl family [70]. C/EBPδ's ability to promote cell death in mammary epithelial cells suggests that it has potential as a tumor suppressor for breast cancer. In support of this notion, C/EBPδ expression is downregulated with breast cancer progression [108], and the loss of C/EBPδ increases tumor multiplicity in the MMTV-Neu mammary tumor model [34].
Interestingly, the transcription factor STAT3, which promotes cell death during mammary gland involution [109] is, on the other hand, frequently activated in breast cancer [110], and promotes metastasis in the MMTV-Neu mouse model, along with expression of Cebpd and an inflammatory and proangiogenic signature [111]. Indeed, despite the increased number of tumors in Cebpd KO mice, the loss of Cebpd results in a significantly reduced incidence of lung metastasis, which correlates with C/EBPδ's role in supporting HIF-1α expression and the consequent cell survival under hypoxia [34]. In addition, C/EBPδ can promote cell survival in response to DNA-damaging agents (see next section for more detail). Given that growth arrest is necessary for DNA repair and adaptation to hypoxia, it is conceivable that the growth arrest functions of C/EBPδ contribute to its promotion of survival under stress conditions.
In another example of C/EBPδ's role in cell death, the β-diketone anti-cancer drug HMDB induces its expression, and C/EBPδ augments cervical tumor cell death and the activation of PPARG2 and GADD153/CHOP (CHOP) transcription in response to HMDB [99]. On the other hand, C/EBPδ protects pancreatic β cells from cytokine-induced cell death by attenuating the expressions of GADD153/CHOP and STAT1 [112]. These are examples of the cell type- and context-dependent functions of C/EBPδ, as the same target gene(s) may be associated with opposite outcomes in different cell types.
Collectively, these reports indicate that C/EBPδ by itself is not a cell death inducer. Rather, C/EBPδ can promote cell death or survival, perhaps as a cell type- and context-dependent interpreter of extracellular signals.
C/EBPδ and genome integrity
Several reports indicate a role for C/EBPδ in the DNA damage response and genomic integrity. C/EBPδ renders urothelial bladder carcinoma cell lines resistant to cisplatin, due to its role in expression of the Cu/Zn-superoxide dismutase SOD1, which scavenges reactive oxygen species [113]. The loss of C/EBPδ results in centrosome amplification and genomic instability in cultured mouse embryo fibroblasts (129S1 strain) [87]. This may be due in part to C/EBPδ's role in promoting the degradation of cyclin D1 [89] and in the Fanconi Anemia (FA) replication-associated DNA repair pathway. C/EBPδ promotes the nuclear import of FANCD2, a critical protein for the FA pathway, by physically interacting with FANCD2 and the importin IPO4. This so-far unique non-transcriptional function of C/EBPδ may underlie its role in promoting cell survival in response to the DNA-damaging agent mitomycin C [41]. However, the loss of C/EBPδ is not sufficient to cause tumors in mice. Hence, the role of C/EBPδ in genomic stability may be compensated for in vivo and/or is only critical when cells are challenged by stress conditions such as in vitro or microenvironmental stressors, drugs, or mutations. On the other hand, C/EBPδ can also contribute to genomic instability by directly inducing the expression of Aurora C kinase, which leads to centrosome abnormalities in cervical carcinoma cells that have been treated with TNFα [114]. This last study highlights how C/EBPδ may switch functions specifically under inflammatory conditions.
Role of C/EBPδ in inflammatory responses
One of the earliest discoveries about C/EBPδ was that its expression is induced by LPS from gram-negative bacteria and by pro-inflammatory cytokines in many cell types and tissues [1]. Studies of C/EBPδ in inflammatory signaling have focused on cells of the myeloid lineage [1]. Collectively, the reports suggest a role for C/EBPδ in the activation of the innate immune response and in the generation of pro-inflammatory conditions. However, a few reports also point to possible functions of C/EBPδ in the inhibition of inflammation. In addition, other C/EBP family members play critical roles in the innate immune response and inflammation (e.g.[115,116,5]), and it remains to be determined how mutual regulation or distinct functions are played out in the overall control of the inflammatory response.
C/EBPδ amplifies LPS signaling in macrophages, supports the expression of many LPS-induced genes, and is necessary to clear persistent gram-negative bacterial infections [21]. Innate immune responses are largely mediated by toll-like receptors (TLRs), which act as a primary line of defense against pathogens, and activate the NFκB and IRF3 transcription program [117]. Mechanistically, C/EBPδ promotes LPS responses in part by inducing the expression of the LPS-receptor TLR4 and by inhibiting FBXW7 expression. In turn, FBXW7α downregulates C/EBPδ expression and is an attenuator of inflammatory signaling [34, 57]. This negative feedback loop reveals an important role of the C/EBPδ/FBXW7α pathway in inflammatory signaling and opens up new avenues for investigation of their functions in immune cells and inflammation-associated diseases.
C/EBPδ also binds directly and activates the promoters of many LPS-induced genes [21]. However, because C/EBPδ also induces expression of the LPS receptor subunit TLR4 [57], some of the reported defects in LPS responses due to Cebpd-null mutations or siRNA could also be due to reduced levels of the LPS receptor itself. For example, TLR4 reconstitution partially rescues the LPS-induced IL-6 expression in Cebpd null cells [57]. Interestingly, a low dose of LPS specifically induces C/EBPδ expression rather than NFκB [118], and the suppressor of cytokine signaling 3 (SOCS3) attenuates LPS-induced activation of C/EBPδ but not of NFκB [119]. Together, these findings suggest that C/EBPδ is critical in sensitizing cells to LPS and prepares them for activation of the innate immune response. Accordingly, C/EBPδ is necessary for lung infiltration by macrophages and to clear pulmonary infection by Klebsiella pneumoniae [75]. On the other hand, the loss of C/EBPδ protects against infection by S. pneumoniae [120]. While this result appears at odds with the aforementioned reports, this particular phenotype is explained by the role of C/EBPδ in lung epithelial expression of PAFR, the receptor used by S. pneumoniae to enter the organism [120]. Therefore, the relative resistance of Cebpd-null mutant mice to S. pneumoniae is unrelated to C/EBPδ's role in the immune system.
Although inflammatory responses are essential for activation of the innate immune response and to enhance adaptive immunity, excessive inflammation is detrimental and can lead to autoimmune disorders and cancer (or sepsis in the case of bacterial triggers) [121]. Indeed, a Cebpd null mutation partly protects mice from sepsis, i.e. disseminated intravascular coagulation (DIC)-induced mortality in response to LPS [122]. Furthermore, C/EBPδ deficiency protects against LPS-induced lung injury [82]. In contrast, C/EBPδ deficiency does not protect against immune-complex-induced lung injury [83], although it participates in Fcγ receptor-mediated activation of cultured macrophages [79]. These data indicate that the role of C/EBPδ in inflammation depends in part on the inflammatory stimulant. Because most studies so far have used LPS, C/EBPδ's role in the response to other specific triggers of the innate immune response still needs to be determined. Interestingly, a Cebpd null mutation has profound effects on the expression of several TLR genes in peritoneal macrophages [57], raising the possibility that C/EBPδ has a much broader role in the innate immune response beyond LPS.
So-called sterile inflammation in the absence of foreign antigens also contributes to pathological conditions. CEBPD expression is induced by inflammatory cytokines, including IL-6, and in turn C/EBPδ directly activates IL-6 gene expression [123], providing the potential for an autocrine pro-inflammatory feedback loop. C/EBPδ expression in astrocytes is implicated in the pathology of Alzheimer disease through the activation of inflammatory cytokines and pentraxin-3 (PTX3), a secreted molecule shown to inhibit the phagocytosis of damaged neurons by macrophages [124, 125]. In CNS dendritic cells, C/EBPδ suppresses the expression of IL-10, consequently altering the ratio of T helper17 cells (Th17) and T regulatory cells (Treg), and thereby contributing to the severity of experimental autoimmune encephalomyelitis [126]. C/EBPδ is also implicated in the promotion of amyotrophic lateral sclerosis, because of its expression in the activated microglia of patients and in a mouse model of this disease, and because it promotes the expression of inflammatory cytokines in the brain in response to systemic LPS [127]. Indeed, the anti-inflammatory flavanoid chrysin inhibits microglial C/EBPδ expression and the concomitant neural cell death in culture [128].
In another inflammatory pathology, C/EBPδ promotes the pathogenesis of collagen-induced rheumatoid arthritis in mice, most likely by contributing to the expression of chemokines and cytokines in macrophages that promote the formation of endothelial cell tubes and the migration and proliferation of synoviocytes [129]. In addition, C/EBPδ mediates the IL-1β-induced expression of secreted type IIA phospholipase A(2) in chondrocytes, which can also contribute to rheumatoid arthritis [130]. Taken together, these reports strongly support a role for C/EBPδ in the pathology of chronic inflammation and autoimmune disorders.
A few reports point to additional anti-inflammatory functions for C/EBPδ. For example, C/EBPδ expression can be induced by glucocorticoids and PGE2, which are potent anti-inflammatory and immunosuppressant agents [54, 131, 132]. Most strikingly, C/EBPδ attenuates the Il-1β+IFN-γ-induced chemokine production in rat insulinoma cells by promoting the expression of IRF-1 [112]. The role of C/EBPδ in supporting or inhibiting inflammatory signaling may depend on the context or cell type. Indeed, C/EBPδ activates the expression of anti-inflammatory IL-10 in response to LPS or PGE2 in macrophages [54, 133]. On the other hand, a Cebpd null mutation leads to increased IL-10 expression in dendritic cells, which is associated with the progression of CNS autoimmune inflammatory disease [126]. Elevated IL-10 mRNA levels have also been observed in Cebpd null mammary tumor tissue [57]; however, it remains to be determined if this is due to a direct or indirect role of C/EBPδ.
Negative feedback regulation of C/EBPδ expression appears to play an important role in the resolution of inflammation and can be achieved through ATF-3 [21], Myc, and the Myc-interacting protein Miz1 [134, 135]. It will be very interesting to learn if and how C/EBPδ, ATF3, and the Myc/Miz1 complex engage in mutual regulatory feedback loops to modulate the magnitude and duration of inflammatory signaling.
Interestingly, C/EBPδ can activate an indirect negative feedback mechanism by inducing the expression of the peroxisome proliferator-activated receptor-gamma (PPARγ), a member of the nuclear receptor superfamily, in VSMCs. PPARγ plays a key role in the inhibition of inflammation, in part by inactivating the STAT3 transcription factor, thereby downregulating CEBPD expression [136]. PPARγ agonists are currently in clinical trials for the treatment of many inflammatory disorders [137]. Thus, by inducing PPARγ expression, C/EBPδ contributes to the attenuation of inflammatory responses in the early stages of atherosclerosis [136]. On the other hand, sumoylated C/EBPδ is reported to inhibit PPARγ gene transcription in hepatocytes [36]. Therefore, the regulation of C/EBPδ's sumoylation may provide a mechanism for increasing the inflammatory response. The potential for another feedback loop was raised in a report on the role of IKKi/IKKε (IKBKE) in the LPS induction of C/EBPδ expression and activity, in which C/EBPδ was found to bind to the IKBKE promoter [61]. However, it is not known if IKBKE expression is reduced or activated by C/EBPδ.
As mentioned earlier, C/EBPδ also inhibits its own suppressor. C/EBPδ protein is targeted for ubiquitination and degradation by FBXW7α, and FBXW7a gene expression is in turn inhibited by C/EBPδ. This negative feedback loop depends on GSK-3β's phosphorylation of C/EBPδ, which is inhibited by LPS. Therefore, LPS leads to the stabilization of C/EBPδ [57]. Interestingly, GSK3β is mostly known as a driver of inflammation, and its inhibitors are under investigation as therapeutics for a variety of diseases [138]. The discovery that GSK-3β can promote the degradation of C/EBPδ therefore opens up new avenues for investigation into the cross-talk between signaling pathways and the regulation of the innate immune response and inflammation.
C/EBPδ expression in human cancers
In accordance with the diversity of C/EBPδ functions described above, analyses of C/EBPδ expression in human cancers paint a similarly complex picture, indicating that it functions both as a tumor suppressor and tumor promoter. In breast cancer, CEBPD mRNA is downregulated with malignant progression, i.e. it is significantly reduced or undetectable in invasive/metastatic disease [108, 139, 140], and CEBPD is part of a 70-gene signature that correlates with longer survival of breast cancer patients [141]. These data suggest that C/EBPδ has a tumor-suppressor function. On the other hand, C/EBPδ protein expression is positively correlated with cyclin D1 and cyclin E in human breast cancer tissue [72], in contrast to several reports showing that C/EBPδ downregulates these cyclins in cultured cell models [85, 89, 91, 93]. While this study [72] did not analyze the correlation of C/EBPδ expression with survival/prognosis, proliferation markers are usually correlated with poor prognosis. Thus, it will be very interesting to follow up on this initial report with a more extensive analysis of C/EBPδ protein expression in breast cancer, especially given that findings from a mouse model suggest that C/EBPδ has a dual activity as tumor suppressor and a promoter of metastasis [34].
In support of a tumor suppressor function, C/EBPδ protein expression correlates with longer disease-free survival in meningioma patients [73]. CEBPD gene methylation was identified in breast, liver, and cervical carcinomas [142, 143]. A recent report suggests that metastatic lesions in breast cancer patients exhibit a higher incidence of methylation within a CpG island of CEBPD, which was associated with increased risk of relapse [144]. Consistent with the observation that C/EBPδ promotes myeloid differentiation in cell culture models (see above), the CEBPD promoter was found to be silenced in 35% of 260 human acute myeloid leukemia cases [8] and its expression is downregulated in the blast crisis phases of CML [100]. Interestingly, in the U937 leukemia cell line, eicosapentaenoic acid causes the demethylation of a specific CpG in the CEBPD promoter, increased C/EBPδ expression, and cell differentiation [145]. Furthermore, the CEBPD gene resides within a common fragile site that is frequently lost in certain cancer cell lines. However, it is not known if this finding has any functional bearing on the behavior of tumor cells or is a byproduct of CEBPD's vicinity to the KIAA0146 gene [146]. While these data are consistent with a tumor-suppressor function of C/EBPδ, mutations in CEBPD are not common in breast cancer [147]. In addition, analyses of the CEBPD gene sequence in leukemias, lymphomas, and a variety of solid tumors indicate that mutations of CEBPD may not contribute to the development of neoplastic disease [148]. Interestingly, single nucleotide polymorphisms in the CEBPD gene are infrequent, perhaps indicative of a stringent need for gene conservation, and none has so far been associated with disease(s) [149, 150]
As in breast cancer, CEBPD mRNA is downregulated in human and rat prolactinomas, and C/EBPδ inhibits prolactinoma cell proliferation in culture through the inhibition of prolactin expression [18, 151]. In contrast, CEBPD mRNA levels are upregulated in mesothelioma [152], gliomas [7], and in Anaplastic Large Cell Lymphoma but not in Hodgkin's Lymphoma [153]. However, correlation studies of expression and cancer biology will be more informative if performed at the protein level, because the C/EBPδ level can be regulated by its protein stability [56, 57], and the CEBPD mRNA expression level may not therefore correlate with the protein level and hence function. Furthermore, analysis in the context of tissue histology will be necessary in light of C/EBPδ's potential expression in tumor-infiltrating host cells.
A strong case for C/EBPδ as a tumor promoter can be made in brain cancer. CEBPD mRNA is overexpressed in mesenchymal glioblastoma cells and associated with poor prognosis [7, 154]. C/EBPδ protein expression is detected near necrotic glioblastoma tissue [7]. Based on the observation that C/EBPδ promotes HIF-1α expression and survival under hypoxic conditions [34], we speculate that this function is responsible for the association of C/EBPδ expression with poor prognosis in glioblastoma (see also section below). Furthermore, C/EBPδ inhibits the expression of FBXW7α in glioblastoma cell lines [34]. FBXW7α is a substrate-binding subunit of the SCF E3 ubiquitin ligase complex and targets many oncoproteins for degradation, including mTOR, NOTCH, c-myc, and cyclin E [59] and is a bona fide tumor suppressor in glioblastoma [155]. Therefore, we suggest that C/EBPδ's inhibition of FBXW7α contributes to glioblastoma progression.
Taken together, expression studies in human neoplasias suggest that C/EBPδ can be associated with and contribute to either a better or worse prognosis, depending on the tumor type or cell of origin. Given the role of C/EBPδ in inflammation and the cross-talk between tumor cells and the immune system, care must be taken to determine if the expression of C/EBPδ is associated with a cell differentiation state, hypoxic or inflammatory induction, or is a result of immune cell infiltration.
Role of C/EBPδ in cancer: a special case for hypoxia and inflammation?
Accumulating evidence supports a role for hypoxia and inflammation in the progression of tumors [156, 157]. As outlined above, C/EBPδ expression is induced by inflammatory effectors and hypoxia, and it promotes pro-inflammatory signaling and adaptation to hypoxia. Thus, C/EBPδ expression and functions are strongly associated with conditions that correlate with cancer progression, which is in contrast to its initial designation as a tumor suppressor. Mechanistically, one C/EBPδ pathway stands out as highly relevant for both conditions. Inhibition of the tumor suppressor FBXW7α by C/EBPδ leads to the upregulation of many oncoproteins and indirectly to the upregulation of HIF-1α [34, 57], which has a prominent role in hypoxia adaptation, inflammation, and cancer [59, 158]. Consistent with this notion, MMTV-Neu-induced mammary tumors arising in Cebpd null mice are less metastatic than those arising in control mice [34], and the growth of experimental metastases of a lung cancer cell line is impaired in Cebpd null host mice [48]. Furthermore, C/EBPδ is associated with a mesenchymal phenotype and poor prognosis in glioblastoma [7, 154].
The association of C/EBPδ with hypoxia and inflammation raises additional questions. For example, although HIF-1α has many pro-oncogenic functions, it too can act as a tumor suppressor or promote therapeutic responses [159]. Indeed, HIF-1α can promote cell survival or cell death depending on the degree of hypoxia [160]. However, it is unclear if C/EBPδ supports HIF-1α expression and function under all of these conditions. Furthermore, it remains to be determined how HIF-1-independent functions of C/EBPδ affect cells under hypoxic conditions and whether HIF-1 induces CEBPD expression directly.
Monocyte and dendritic cell infiltrations are seen in the hypoxic regions of solid tumors, particularly associated with necrotic tissue, and HIF-1α is required for the recruitment of myeloid cells [161]. Does C/EBPδ support the recruitment of monocytes to the tumors and/or the function of tumor-associated macrophages (TAMs)? Altered cytokine/chemokine expression patterns in Cebpd null mammary tumors suggest that the answer is yes [57]. The immune system's role is to eliminate tumor cells, and subversion of the immune system by the tumor leads to escape from immune surveillance and the promotion of chronic inflammatory signaling. Interestingly, C/EBPδ promotes the differentiation of bone-marrow macrophages to the classical M1 type, while Cebpd silencing promotes the alternatively activated M2-type, which is considered immunosuppressive [55]. This finding suggests a potential tumor-suppressor function of C/EBPδ in the immune system. Indeed, it is conceivable that C/EBPδ functions in the innate immune response may contribute to the reduced tumor burden of Cebpd null mice in the mammary tumor model [34]. On the other hand, a very recent study indicates that C/EBPδ expression in TAMs correlates by several measures with the malignant progression of nasopharyngeal carcinoma [54]. Thus, an important question to resolve is the role of C/EBPδ in tumor cells as opposed to in the tumor microenvironment along tumor progression. It needs to be determined what types of immune cells express C/EBPδ in vivo and how C/EBPδ's functions in these cells affect the metastatic phenotype. The dual role of C/EBPδ is reminiscent of the “TGF-β paradox” [11, 12]. While TGF-β has been shown to inhibit C/EBPδ expression and activity in VSMCs and pre-adipocytes respectively [40, 162] an interesting open question is the extent of cross-talk between TGF-β and C/EBPδ signaling in tumor and immune cells. A conditional null allele of C/EBPδ, which is being developed in our laboratory, will be useful for addressing some of the above questions in mouse models.
Summary and Perspectives
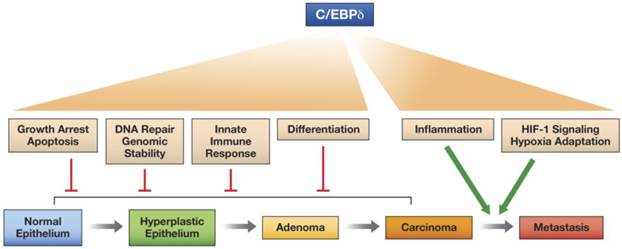
In this review we have highlighted the versatile nature of the C/EBPδ protein, a transcription factor that can activate or repress target genes and promotes many different, and sometimes contrasting, cellular functions, depending on context, that can affect intracellular functions and/or the microenvironment. C/EBPδ even exhibits at least one documented non-transcriptional function. We propose that C/EBPδ has potential as a tumor suppressor when it inhibits proliferation, promotes differentiation, contributes to genomic integrity, and supports the innate immune response. On the other hand, C/EBPδ promotes tumor progression through its functions in hypoxia adaptation and pro-inflammatory signaling, which may also operate in the tumor parenchyma and the tumor microenvironment (Figure 2).
Far too little is known about physiologically relevant C/EBPδ protein modifications or how specific protein interactions are regulated and affect the regulation of C/EBPδ target genes and C/EBPδ's cell-type-specific activities. Because C/EBPδ is linked to so many cellular functions, insights into its roles and regulation will lead to a better understanding of the complexity and diversity of cellular processes in the normal development and progression of diseases.
Model for the differential effects of C/EBPδ on epithelial tumor progression. C/EBPδ may function as a tumor suppressor at the early stages of tumor development, because of its role in promoting growth arrest, cell death, differentiation, DNA repair, genomic stability, and the innate immune response. However, by supporting HIF-1 activity and hypoxia adaptation and by amplifying inflammatory signaling, C/EBPδ promotes tumor progression.
Acknowledgements
The authors are supported by the Intramural Research Program of the NIH, National Cancer Institute. We are grateful to Drs. Peter Johnson, Howard Young, and Ju-Ming Wang for helpful discussion and critical comments on the manuscript, and Allen Kane for preparation of the figures. We apologize for not citing all the publications in the field and welcome feedback.
The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Competing Interests
The authors have declared that no conflict of interest exists.
References
1. Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561-75 doi:10.1042/BJ20020508
2. Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6-19 doi:10.1016/j.cyto.2010.12.019
3. Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318-24 doi:10.1016/j.tcb.2007.07.004
4. Sebastian T, Johnson PF. Stop and go: anti-proliferative and mitogenic functions of the transcription factor C/EBPbeta. Cell Cycle. 2006;5:953-7
5. Roos AB, Nord M. The emerging role of C/EBPs in glucocorticoid signaling: lessons from the lung. J Endocrinol. 2012;212:291-305 doi:10.1530/JOE-11-0369
6. Takiguchi M. The C/EBP family of transcription factors in the liver and other organs. Int J Exp Pathol. 1998;79:369-91
7. Cooper LA, Gutman DA, Chisolm C, Appin C, Kong J, Rong Y. et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. The American journal of pathology. 2012;180:2108-19 doi:10.1016/j.ajpath.2012.01.040
8. Agrawal S, Hofmann WK, Tidow N, Ehrich M, van den Boom D, Koschmieder S. et al. The C/EBPdelta tumor suppressor is silenced by hypermethylation in acute myeloid leukemia. Blood. 2007;109:3895-905 doi:10.1182/blood-2006-08-040147
9. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74 doi:10.1016/j.cell.2011.02.013
10. Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18:884-901 doi:10.1016/j.devcel.2010.05.012
11. Tian M, Schiemann WP. The TGF-beta paradox in human cancer: an update. Future Oncol. 2009;5:259-71 doi:10.2217/14796694.5.2.259
12. Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100:8621-3 doi:10.1073/pnas.1633291100
13. Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859-64 doi:10.1038/nature07885
14. Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K. et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. The Journal of clinical investigation. 2010;120:127-41 doi:10.1172/JCI40027
15. Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem. 1996;271:3891-6
16. Zhou J, Gurates B, Yang S, Sebastian S, Bulun SE. Malignant breast epithelial cells stimulate aromatase expression via promoter II in human adipose fibroblasts: an epithelial-stromal interaction in breast tumors mediated by CCAAT/enhancer binding protein beta. Cancer Res. 2001;61:2328-34
17. Enwright JF 3rd, Kawecki-Crook MA, Voss TC, Schaufele F, Day RN. A PIT-1 homeodomain mutant blocks the intranuclear recruitment of the CCAAT/enhancer binding protein alpha required for prolactin gene transcription. Mol Endocrinol. 2003;17:209-22
18. Tong Y, Zhou J, Mizutani J, Fukuoka H, Ren SG, Gutierrez-Hartmann A. et al. CEBPD suppresses prolactin expression and prolactinoma cell proliferation. Molecular endocrinology. 2011;25:1880-91 doi:10.1210/me.2011-1075
19. Zhang Y, Liu T, Yan P, Huang T, Dewille J. Identification and characterization of CCAAT/enhancer binding protein delta (C/EBPdelta) target genes in G0 growth arrested mammary epithelial cells. BMC Mol Biol. 2008;9:83. doi:10.1186/1471-2199-9-83
20. Borrelli S, Fanoni D, Dolfini D, Alotto D, Ravo M, Grober OM. et al. C/EBPdelta Gene Targets in Human Keratinocytes. PLoS One. 2010;5:e13789. doi:10.1371/journal.pone.0013789
21. Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE. et al. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437-43 doi:10.1038/ni.1721
22. Kovacs KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J Biol Chem. 2003;278:36959-65 doi:10.1074/jbc.M303147200
23. Wang JM, Ko CY, Chen LC, Wang WL, Chang WC. Functional role of NF-IL6beta and its sumoylation and acetylation modifications in promoter activation of cyclooxygenase 2 gene. Nucleic Acids Res. 2006;34:217-31 doi:10.1093/nar/gkj422
24. Chamberlain W, Gonnella P, Alamdari N, Aversa Z, Hasselgren PO. Multiple muscle wasting-related transcription factors are acetylated in dexamethasone-treated muscle cells. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2012;90:200-8 doi:10.1139/o11-082
25. Chiang BT, Liu YW, Chen BK, Wang JM, Chang WC. Direct interaction of C/EBPdelta and Sp1 at the GC-enriched promoter region synergizes the IL-10 gene transcription in mouse macrophage. J Biomed Sci. 2006;13:621-35 doi:10.1007/s11373-006-9101-y
26. Meruvu S, Hugendubler L, Mueller E. Regulation of adipocyte differentiation by the zinc finger protein ZNF638. The Journal of biological chemistry. 2011;286:26516-23 doi:10.1074/jbc.M110.212506
27. Zhang J, Chang L, Chen C, Zhang M, Luo Y, Hamblin M. et al. Rad GTPase inhibits cardiac fibrosis through connective tissue growth factor. Cardiovasc Res. 2011;91:90-8 doi:10.1093/cvr/cvr068
28. Bezy O, Elabd C, Cochet O, Petersen RK, Kristiansen K, Dani C. et al. Delta-interacting protein A, a new inhibitory partner of CCAAT/enhancer-binding protein beta, implicated in adipocyte differentiation. J Biol Chem. 2005;280:11432-8 doi:10.1074/jbc.M411741200
29. Delany AM, Durant D, Canalis E. Glucocorticoid suppression of IGF I transcription in osteoblasts. Mol Endocrinol. 2001;15:1781-9
30. Okazaki K, Li J, Yu H, Fukui N, Sandell LJ. CCAAT/enhancer-binding proteins beta and delta mediate the repression of gene transcription of cartilage-derived retinoic acid-sensitive protein induced by interleukin-1 beta. J Biol Chem. 2002;277:31526-33 doi:10.1074/jbc.M202815200. M202815200 [pii]
31. Lacorte JM, Beigneux A, Parant M, Chambaz J. Repression of apoC-III gene expression by TNFalpha involves C/EBPdelta/NF-IL6beta via an IL-1 independent pathway. FEBS Lett. 1997;415:217-20
32. Lacorte JM, Ktistaki E, Beigneux A, Zannis VI, Chambaz J, Talianidis I. Activation of CAAT enhancer-binding protein delta (C/EBPdelta) by interleukin-1 negatively influences apolipoprotein C-III expression. J Biol Chem. 1997;272:23578-84
33. Qiu X, Aiken KJ, Chokas AL, Beachy DE, Nick HS. Distinct functions of CCAAT enhancer-binding protein isoforms in the regulation of manganese superoxide dismutase during interleukin-1beta stimulation. J Biol Chem. 2008;283:25774-85 doi:10.1074/jbc.M801178200
34. Balamurugan K, Wang JM, Tsai HH, Sharan S, Anver M, Leighty R. et al. The tumour suppressor C/EBPdelta inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J. 2010;29:4106-17 doi:emboj2010280 [pii] 10.1038/emboj.2010.280
35. Turgeon N, Valiquette C, Blais M, Routhier S, Seidman EG, Asselin C. Regulation of C/EBPdelta-dependent transactivation by histone deacetylases in intestinal epithelial cells. J Cell Biochem. 2008;103:1573-83 doi:10.1002/jcb.21544
36. Lai PH, Wang WL, Ko CY, Lee YC, Yang WM, Shen TW. et al. HDAC1/HDAC3 modulates PPARG2 transcription through the sumoylated CEBPD in hepatic lipogenesis. Biochim Biophys Acta. 2008;1783:1803-14 doi:S0167-4889(08)00225-5 [pii] 10.1016/j.bbamcr.2008.06.008
37. Xu L, Panel V, Ma X, Du C, Hugendubler L, Gavrilova O. et al. The winged helix transcription factor foxa3 regulates adipocyte differentiation and depot-selective fat tissue expansion. Mol Cell Biol. 2013;33:3392-9 doi:10.1128/MCB.00244-13
38. Turgeon N, Rousseau D, Roy E, Asselin C. GATA-4 modulates C/EBP-dependent transcriptional activation of acute phase protein genes. Biochem Biophys Res Commun. 2008;370:371-5 doi:S0006-291X(08)00603-7 [pii] 10.1016/j.bbrc.2008.03.107
39. McCarthy TL, Ji CH, Chen Y, Kim KK, Imagawa M, Ito Y. et al. Runt domain factor (Runx)-dependent effects on CCAAT/enhancer-binding protein delta expression and activity in osteoblasts. Journal of Biological Chemistry. 2000;275:21746-53 doi:Doi 10.1074/Jbc.M002291200
40. Choy L, Derynck R. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J Biol Chem. 2003;278:9609-19 doi:10.1074/jbc.M212259200
41. Wang J, Sarkar TR, Zhou M, Sharan S, Ritt DA, Veenstra TD. et al. CCAAT/enhancer binding protein delta (C/EBP{delta}, CEBPD)-mediated nuclear import of FANCD2 by IPO4 augments cellular response to DNA damage. Proc Natl Acad Sci U S A. 2010;107:16131-6 doi:1002603107 [pii] 10.1073/pnas.1002603107
42. Matsushita N, Endo Y, Sato K, Kurumizaka H, Yamashita T, Takata M. et al. Direct inhibition of TNF-alpha promoter activity by Fanconi anemia protein FANCD2. PLoS One. 2011;6:e23324. doi:10.1371/journal.pone.0023324
43. Park E, Kim H, Kim JM, Primack B, Vidal-Cardenas S, Xu Y. et al. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol Cell. 2013;50:908-18 doi:10.1016/j.molcel.2013.05.017
44. Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. Embo J. 1997;16:7432-43 doi:10.1093/emboj/16.24.7432
45. Sterneck E, Paylor R, Jackson-Lewis V, Libbey M, Przedborski S, Tessarollo L. et al. Selectively enhanced contextual fear conditioning in mice lacking the transcriptional regulator CCAAT/enhancer binding protein delta. Proc Natl Acad Sci U S A. 1998;95:10908-13
46. Takiguchi M, Mori M. In vitro analysis of the rat liver-type arginase promoter. J Biol Chem. 1991;266:9186-93
47. Tang Y, Pacary E, Freret T, Divoux D, Petit E, Schumann-Bard P. et al. Effect of hypoxic preconditioning on brain genomic response before and following ischemia in the adult mouse: identification of potential neuroprotective candidates for stroke. Neurobiol Dis. 2006;21:18-28 doi:10.1016/j.nbd.2005.06.002
48. Min Y, Ghose S, Boelte K, Li J, Yang L, Lin PC. C/EBP-delta regulates VEGF-C autocrine signaling in lymphangiogenesis and metastasis of lung cancer through HIF-1alpha. Oncogene. 2011;30:4901-9 doi:10.1038/onc.2011.187
49. Wotton S, Terry A, Kilbey A, Jenkins A, Herzyk P, Cameron E. et al. Gene array analysis reveals a common Runx transcriptional programme controlling cell adhesion and survival. Oncogene. 2008;27:5856-66 doi:onc2008195 [pii] 10.1038/onc.2008.195
50. Glaros T, Fu Y, Xing J, Li L. Molecular mechanism underlying persistent induction of LCN2 by lipopolysaccharide in kidney fibroblasts. PLoS One. 2012;7:e34633. doi:10.1371/journal.pone.0034633
51. Liu YW, Wang SA, Hsu TY, Chen TA, Chang WC, Hung JJ. Inhibition of LPS-induced C/EBP delta by trichostatin A has a positive effect on LPS-induced cyclooxygenase 2 expression in RAW264.7 cells. J Cell Biochem. 2010;110:1430-8 doi:10.1002/jcb.22682
52. Borrelli S, Testoni B, Callari M, Alotto D, Castagnoli C, Romano RA. et al. Reciprocal regulation of p63 by C/EBP delta in human keratinocytes. BMC Mol Biol. 2007;8:85. doi:10.1186/1471-2199-8-85
53. Li B, Si J, DeWille JW. Ultraviolet radiation (UVR) activates p38 MAP kinase and induces post-transcriptional stabilization of the C/EBPdelta mRNA in G0 growth arrested mammary epithelial cells. J Cell Biochem. 2008;103:1657-69 doi:10.1002/jcb.21554
54. Hsiao YW, Li CF, Chi JY, Tseng JT, Chang Y, Hsu LJ. et al. CCAAT/Enhancer Binding Protein delta in Macrophages Contributes to Immunosuppression and Inhibits Phagocytosis in Nasopharyngeal Carcinoma. Sci Signal. 2013;6:ra59. doi:10.1126/scisignal.2003648
55. Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M. et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190:6542-9 doi:10.4049/jimmunol.1202496
56. Sarkar TR, Sharan S, Wang J, Pawar SA, Cantwell CA, Johnson PF. et al. Identification of a Src tyrosine kinase/SIAH2 E3 ubiquitin ligase pathway that regulates C/EBPdelta expression and contributes to transformation of breast tumor cells. Mol Cell Biol. 2012;32:320-32 doi:10.1128/mcb.05790-11
57. Balamurugan K, Sharan S, Klarmann KD, Zhang Y, Coppola V, Summers GH. et al. FBXW7alpha attenuates inflammatory signalling by downregulating C/EBPdelta and its target gene Tlr4. Nature communications. 2013;4:1662. doi:10.1038/ncomms2677
58. House CM, Moller A, Bowtell DD. Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res. 2009;69:8835-8 doi:10.1158/0008-5472.can-09-1676
59. Cheng Y, Li G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 2012;31:75-87 doi:10.1007/s10555-011-9330-z
60. Ji C, Chang W, Centrella M, McCarthy TL. Activation domains of CCAAT enhancer binding protein delta: regions required for native activity and prostaglandin E2-dependent transactivation of insulin-like growth factor I gene expression in rat osteoblasts. Mol Endocrinol. 2003;17:1834-43 doi:10.1210/me.2002-0235
61. Kravchenko VV, Mathison JC, Schwamborn K, Mercurio F, Ulevitch RJ. IKKi/IKKepsilon plays a key role in integrating signals induced by pro-inflammatory stimuli. J Biol Chem. 2003;278:26612-9 doi:10.1074/jbc.M303001200
62. Ray A, Ray BK. Serum amyloid A gene expression under acute-phase conditions involves participation of inducible C/EBP-beta and C/EBP-delta and their activation by phosphorylation. Mol Cell Biol. 1994;14:4324-32
63. Yin M, Yang SQ, Lin HZ, Lane MD, Chatterjee S, Diehl AM. Tumor necrosis factor alpha promotes nuclear localization of cytokine-inducible CCAAT/enhancer binding protein isoforms in hepatocytes. J Biol Chem. 1996;271:17974-8
64. Billiard J, Umayahara Y, Wiren K, Centrella M, McCarthy TL, Rotwein P. Regulated nuclear-cytoplasmic localization of CCAAT/enhancer-binding protein delta in osteoblasts. J Biol Chem. 2001;276:15354-61 doi:10.1074/jbc.M009973200
65. Kitami Y, Fukuoka T, Hiwada K, Inagami T. A high level of CCAAT-enhancer binding protein-delta expression is a major determinant for markedly elevated differential gene expression of the platelet-derived growth factor-alpha receptor in vascular smooth muscle cells of genetically hypertensive rats. Circ Res. 1999;84:64-73
66. Fukuoka T, Kitami Y, Okura T, Hiwada K. Transcriptional regulation of the platelet-derived growth factor alpha receptor gene via CCAAT/enhancer-binding protein-delta in vascular smooth muscle cells. J Biol Chem. 1999;274:25576-82
67. Zhou S, Si J, Liu T, DeWille JW. PIASy represses CCAAT/enhancer-binding protein delta (C/EBPdelta) transcriptional activity by sequestering C/EBPdelta to the nuclear periphery. J Biol Chem. 2008;283:20137-48 doi:10.1074/jbc.M801307200
68. Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999;13:2231-41
69. Billiard J, Grewal SS, Lukaesko L, Stork PJ, Rotwein P. Hormonal control of insulin-like growth factor I gene transcription in human osteoblasts: dual actions of cAMP-dependent protein kinase on CCAAT/enhancer-binding protein delta. J Biol Chem. 2001;276:31238-46 doi:10.1074/jbc.M103634200
70. Thangaraju M, Rudelius M, Bierie B, Raffeld M, Sharan S, Hennighausen L. et al. C/EBPdelta is a crucial regulator of pro-apoptotic gene expression during mammary gland involution. Development. 2005;132:4675-85 doi:10.1242/dev.02050
71. Huang AM, Rudelius M, Sharan S, McAllister JM, Raffeld M, Christenson LK. et al. The Cebpd (C/EBPdelta) gene is induced by luteinizing hormones in ovarian theca and interstitial cells but is not essential for mouse ovary function. PLoS One. 2007;2:e1334. doi:10.1371/journal.pone.0001334
72. Milde-Langosch K, Loning T, Bamberger AM. Expression of the CCAAT/enhancer-binding proteins C/EBPalpha, C/EBPbeta and C/EBPdelta in breast cancer: correlations with clinicopathologic parameters and cell-cycle regulatory proteins. Breast Cancer Res Treat. 2003;79:175-85
73. Barresi V, Vitarelli E, Cerasoli S, Barresi G. The cell growth inhibitory transcription factor C/EBPdelta is expressed in human meningiomas in association with low histological grade and proliferation index. J Neurooncol. 2010;97:233-40 doi:10.1007/s11060-009-0024-0
74. Sugahara K, Sadohara T, Sugita M, Iyama K, Takiguchi M. Differential expression of CCAAT enhancer binding protein family in rat alveolar epithelial cell proliferation and in acute lung injury. Cell Tissue Res. 1999;297:261-70
75. Duitman J, Hoogendijk AJ, Groot AP, Ruela de Sousa RR, van der Poll T, Florquin S. et al. CCAAT-enhancer binding protein delta (C/EBPdelta) protects against Klebsiella pneumoniae-induced pulmonary infection: potential role for macrophage migration. The Journal of infectious diseases. 2012;206:1826-35 doi:10.1093/infdis/jis615
76. Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. Journal of cell science. 2005;118:2545-55 doi:10.1242/jcs.02459
77. Jones LC, Lin ML, Chen SS, Krug U, Hofmann WK, Lee S. et al. Expression of C/EBPbeta from the C/ebpalpha gene locus is sufficient for normal hematopoiesis in vivo. Blood. 2002;99:2032-6
78. Chiu CH, Lin WD, Huang SY, Lee YH. Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 2004;18:1970-5 doi:10.1101/gad.1213104
79. Yan C, Zhu M, Staiger J, Johnson PF, Gao H. C5a-regulated CCAAT/enhancer binding protein beta and -delta are essential in Fcgamma receptor- mediated inflammatory cytokine and chemokine production in macrophages. J Biol Chem. 2012;287:3217-30 doi:10.1074/jbc.M111.280834
80. Lu YC, Kim I, Lye E, Shen F, Suzuki N, Suzuki S. et al. Differential role for c-Rel and C/EBPbeta/delta in TLR-mediated induction of proinflammatory cytokines. J Immunol. 2009;182:7212-21 doi:10.4049/jimmunol.0802971
81. Staiger J, Lueben MJ, Berrigan D, Malik R, Perkins SN, Hursting SD. et al. C/EBPbeta regulates body composition, energy balance-related hormones and tumor growth. Carcinogenesis. 2009;30:832-40 doi:10.1093/carcin/bgn273
82. Yan C, Johnson PF, Tang H, Ye Y, Wu M, Gao H. CCAAT/Enhancer-Binding Protein delta Is a Critical Mediator of Lipopolysaccharide-Induced Acute Lung Injury. The American journal of pathology. 2012 doi:10.1016/j.ajpath.2012.10.013
83. Yan C, Wu M, Cao J, Tang H, Zhu M, Johnson PF. et al. Critical role for CCAAT/enhancer-binding protein beta in immune complex-induced acute lung injury. J Immunol. 2012;189:1480-90 doi:10.4049/jimmunol.1200877
84. Yang L, Jiang Y, Wu SF, Zhou MY, Wu YL, Chen GQ. CCAAT/enhancer-binding protein alpha antagonizes transcriptional activity of hypoxia-inducible factor 1 alpha with direct protein-protein interaction. Carcinogenesis. 2008;29:291-8 doi:10.1093/carcin/bgm262
85. O'Rourke JP, Newbound GC, Hutt JA, DeWille J. CCAAT/enhancer-binding protein delta regulates mammary epithelial cell G0 growth arrest and apoptosis. J Biol Chem. 1999;274:16582-9
86. Dearth LR, DeWille J. Posttranscriptional and posttranslational regulation of C/EBP delta in G0 growth-arrested mammary epithelial cells. J Biol Chem. 2003;278:11246-55 doi:10.1074/jbc.M207930200
87. Huang AM, Montagna C, Sharan S, Ni Y, Ried T, Sterneck E. Loss of CCAAT/enhancer binding protein delta promotes chromosomal instability. Oncogene. 2004;23:1549-57 doi:10.1038/sj.onc.1207285
88. Hutt JA, DeWille JW. Oncostatin M induces growth arrest of mammary epithelium via a CCAAT/enhancer-binding protein delta-dependent pathway. Mol Cancer Ther. 2002;1:601-10
89. Pawar SA, Roy Sarkar T, Balamurugan K, Sharan S, Wang J, Zhang Y. et al. C/EBP{delta} targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression. Proc Natl Acad Sci U S A. 2010;107:9210-5 doi:0913813107 [pii] 10.1073/pnas.0913813107
90. Gigliotti AP, Johnson PF, Sterneck E, DeWille JW. Nulliparous CCAAT/enhancer binding proteindelta (C/EBPdelta) knockout mice exhibit mammary gland ductal hyperlasia. Exp Biol Med (Maywood). 2003;228:278-85
91. Gery S, Tanosaki S, Hofmann WK, Koppel A, Koeffler HP. C/EBPdelta expression in a BCR-ABL-positive cell line induces growth arrest and myeloid differentiation. Oncogene. 2005;24:1589-97 doi:1208393 [pii] 10.1038/sj.onc.1208393
92. Sanford DC, DeWille JW. C/EBPdelta is a downstream mediator of IL-6 induced growth inhibition of prostate cancer cells. Prostate. 2005;63:143-54 doi:10.1002/pros.20159
93. Ikezoe T, Gery S, Yin D, O'Kelly J, Binderup L, Lemp N. et al. CCAAT/enhancer-binding protein delta: a molecular target of 1,25-dihydroxyvitamin D3 in androgen-responsive prostate cancer LNCaP cells. Cancer Res. 2005;65:4762-8 doi:10.1158/0008-5472.CAN-03-3619
94. Yang G, Gregory CW, Shang Q, O'Brien DA, Zhang YL. Differential expression of CCAAT/enhancer-binding protein-delta (c/EBPdelta) in rat androgen-dependent tissues and human prostate cancer. J Androl. 2001;22:471-80
95. Umayahara Y, Ji C, Centrella M, Rotwein P, McCarthy TL. CCAAT/enhancer-binding protein delta activates insulin-like growth factor-I gene transcription in osteoblasts. Identification of a novel cyclic AMP signaling pathway in bone. J Biol Chem. 1997;272:31793-800
96. Hishida T, Nishizuka M, Osada S, Imagawa M. The role of C/EBPdelta in the early stages of adipogenesis. Biochimie. 2009;91:654-7 doi:10.1016/j.biochi.2009.02.002
97. Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A. et al. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. Embo J. 2011;30:1459-72 doi:10.1038/emboj.2011.65
98. Chen PL, Riley DJ, Chen YM, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes & Development. 1996;10:2794-804 doi:Doi 10.1101/Gad.10.21.2794
99. Pan YC, Li CF, Ko CY, Pan MH, Chen PJ, Tseng JT. et al. CEBPD reverses RB/E2F1-mediated gene repression and participates in HMDB-induced apoptosis of cancer cells. Clin Cancer Res. 2010;16:5770-80 doi:10.1158/1078-0432.CCR-10-1025
100. Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B. et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:2794-9 doi:10.1073/pnas.0510423103
101. Wang QF, Friedman AD. CCAAT/enhancer-binding proteins are required for granulopoiesis independent of their induction of the granulocyte colony-stimulating factor receptor. Blood. 2002;99:2776-85
102. Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725-35
103. Barbaro V, Testa A, Di Iorio E, Mavilio F, Pellegrini G, De Luca M. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J Cell Biol. 2007;177:1037-49 doi:jcb.200703003 [pii] 10.1083/jcb.200703003
104. Wright LS, Li J, Caldwell MA, Wallace K, Johnson JA, Svendsen CN. Gene expression in human neural stem cells: effects of leukemia inhibitory factor. Journal of neurochemistry. 2003;86:179-95
105. Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K. et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783-98 doi:10.1016/j.stem.2012.09.011
106. Wang Z, Inuzuka H, Fukushima H, Wan L, Gao D, Shaik S. et al. Emerging roles of the FBW7 tumour suppressor in stem cell differentiation. EMBO Rep. 2012;13:36-43 doi:10.1038/embor.2011.231
107. Takeishi S, Matsumoto A, Onoyama I, Naka K, Hirao A, Nakayama KI. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer cell. 2013;23:347-61 doi:10.1016/j.ccr.2013.01.026
108. Porter DA, Krop IE, Nasser S, Sgroi D, Kaelin CM, Marks JR. et al. A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res. 2001;61:5697-702
109. Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K. et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604-16
110. Resemann HK, Watson CJ, Lloyd-Lewis B. The Stat3 paradox: A killer and an oncogene. Molecular and cellular endocrinology. 2013 doi:10.1016/j.mce.2013.06.029
111. Ranger JJ, Levy DE, Shahalizadeh S, Hallett M, Muller WJ. Identification of a Stat3-dependent transcription regulatory network involved in metastatic progression. Cancer Res. 2009;69:6823-30 doi:10.1158/0008-5472.CAN-09-1684
112. Moore F, Santin I, Nogueira TC, Gurzov EN, Marselli L, Marchetti P. et al. The transcription factor C/EBP delta has anti-apoptotic and anti-inflammatory roles in pancreatic beta cells. PLoS One. 2012;7:e31062. doi:10.1371/journal.pone.0031062
113. Hour TC, Lai YL, Kuan CI, Chou CK, Wang JM, Tu HY. et al. Transcriptional up-regulation of SOD1 by CEBPD: A potential target for cisplatin resistant human urothelial carcinoma cells. Biochem Pharmacol. 2010;80:325-34 doi:S0006-2952(10)00252-2 [pii] 10.1016/j.bcp.2010.04.007
114. Wu SR, Li CF, Hung LY, Huang AM, Tseng JT, Tsou JH. et al. CCAAT/enhancer-binding protein delta mediates tumor necrosis factor alpha-induced Aurora kinase C transcription and promotes genomic instability. J Biol Chem. 2011;286:28662-70 doi:10.1074/jbc.M111.270710
115. Huber R, Pietsch D, Panterodt T, Brand K. Regulation of C/EBPbeta and resulting functions in cells of the monocytic lineage. Cellular signalling. 2012;24:1287-96 doi:10.1016/j.cellsig.2012.02.007
116. Akagi T, Thoennissen NH, George A, Crooks G, Song JH, Okamoto R. et al. In vivo deficiency of both C/EBPbeta and C/EBPepsilon results in highly defective myeloid differentiation and lack of cytokine response. PLoS One. 2010;5:e15419. doi:10.1371/journal.pone.0015419
117. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637-50 doi:10.1016/j.immuni.2011.05.006
118. Maitra U, Gan L, Chang S, Li L. Low-Dose Endotoxin Induces Inflammation by Selectively Removing Nuclear Receptors and Activating CCAAT/Enhancer-Binding Protein {delta}. J Immunol. 2011;186:4467-73 doi:10.4049/jimmunol.1003300
119. Yan C, Ward PA, Wang X, Gao H. Myeloid depletion of SOCS3 enhances LPS-induced acute lung injury through CCAAT/enhancer binding protein delta pathway. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27:2967-76 doi:10.1096/fj.12-225797
120. Duitman J, Schouten M, Groot AP, Daalhuisen JB, Florquin S, van der Poll T. et al. CCAAT/enhancer-binding protein delta facilitates bacterial dissemination during pneumococcal pneumonia in a platelet-activating factor receptor-dependent manner. Proc Natl Acad Sci U S A. 2012;109:9113-8 doi:10.1073/pnas.1202641109
121. Goldszmid RS, Trinchieri G. The price of immunity. Nat Immunol. 2012;13:932-8 doi:10.1038/ni.2422
122. Slofstra SH, Groot AP, Obdeijn MH, Reitsma PH, ten Cate H, Spek CA. Gene expression profiling identifies C/EBPdelta as a candidate regulator of endotoxin-induced disseminated intravascular coagulation. Am J Respir Crit Care Med. 2007;176:602-9 doi:10.1164/rccm.200609-1250OC
123. Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279-82
124. Li R, Strohmeyer R, Liang Z, Lue LF, Rogers J. CCAAT/enhancer binding protein delta (C/EBPdelta) expression and elevation in Alzheimer's disease. Neurobiology of aging. 2004;25:991-9 doi:10.1016/j.neurobiolaging.2003.10.016
125. Ko CY, Chang LH, Lee YC, Sterneck E, Cheng CP, Chen SH. et al. CCAAT/enhancer binding protein delta (CEBPD) elevating PTX3 expression inhibits macrophage-mediated phagocytosis of dying neuron cells. Neurobiology of aging. 2012;33:422.e11-25 doi:10.1016/j.neurobiolaging.2010.09.017
126. Tsai VW, Mohammad MG, Tolhurst O, Breit SN, Sawchenko PE, Brown DA. CCAAT/enhancer binding protein-delta expression by dendritic cells regulates CNS autoimmune inflammatory disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:17612-21 doi:10.1523/JNEUROSCI.3449-11.2011
127. Valente T, Straccia M, Gresa-Arribas N, Dentesano G, Tusell JM, Serratosa J. et al. CCAAT/enhancer binding protein delta regulates glial proinflammatory gene expression. Neurobiology of aging. 2013;34:2110-24 doi:10.1016/j.neurobiolaging.2013.02.007
128. Gresa-Arribas N, Serratosa J, Saura J, Sola C. Inhibition of CCAAT/enhancer binding protein delta expression by chrysin in microglial cells results in anti-inflammatory and neuroprotective effects. Journal of neurochemistry. 2010;115:526-36 doi:10.1111/j.1471-4159.2010.06952.x
129. Chang LH, Huang HS, Wu PT, Jou IM, Pan MH, Chang WC. et al. Role of macrophage CCAAT/enhancer binding protein delta in the pathogenesis of rheumatoid arthritis in collagen-induced arthritic mice. PLoS One. 2012;7:e45378. doi:10.1371/journal.pone.0045378
130. Massaad C, Paradon M, Jacques C, Salvat C, Bereziat G, Berenbaum F. et al. Induction of secreted type IIA phospholipase A2 gene transcription by interleukin-1beta. Role of C/EBP factors. J Biol Chem. 2000;275:22686-94 doi:10.1074/jbc.M001250200
131. Sai S, Esteves CL, Kelly V, Michailidou Z, Anderson K, Coll AP. et al. Glucocorticoid regulation of the promoter of 11beta-hydroxysteroid dehydrogenase type 1 is indirect and requires CCAAT/enhancer-binding protein-beta. Mol Endocrinol. 2008;22:2049-60 doi:10.1210/me.2007-0489
132. Allen DL, Cleary AS, Hanson AM, Lindsay SF, Reed JM. CCAAT/enhancer binding protein-delta expression is increased in fast skeletal muscle by food deprivation and regulates myostatin transcription in vitro. American journal of physiology Regulatory, integrative and comparative physiology. 2010;299:R1592-601 doi:10.1152/ajpregu.00247.2010
133. Liu YW, Tseng HP, Chen LC, Chen BK, Chang WC. Functional cooperation of simian virus 40 promoter factor 1 and CCAAT/enhancer-binding protein beta and delta in lipopolysaccharide-induced gene activation of IL-10 in mouse macrophages. J Immunol. 2003;171:821-8
134. Si J, Yu X, Zhang Y, DeWille JW. Myc interacts with Max and Miz1 to repress C/EBPdelta promoter activity and gene expression. Mol Cancer. 2010;9:92. doi:10.1186/1476-4598-9-92
135. Do-Umehara HC, Chen C, Urich D, Zhou L, Qiu J, Jang S. et al. Suppression of inflammation and acute lung injury by Miz1 via repression of C/EBP-delta. Nat Immunol. 2013;14:461-9 doi:10.1038/ni.2566
136. Takata Y, Kitami Y, Yang ZH, Nakamura M, Okura T, Hiwada K. Vascular inflammation is negatively autoregulated by interaction between CCAAT/enhancer-binding protein-delta and peroxisome proliferator-activated receptor-gamma. Circ Res. 2002;91:427-33
137. Kaundal RK, Sharma SS. Peroxisome proliferator-activated receptor gamma agonists as neuroprotective agents. Drug news & perspectives. 2010;23:241-56 doi:10.1358/dnp.2010.23.4.1437710
138. Klamer G, Song E, Ko KH, O'Brien TA, Dolnikov A. Using small molecule GSK3beta inhibitors to treat inflammation. Current medicinal chemistry. 2010;17:2873-81
139. Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J. et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362-75
140. Zucchi I, Mento E, Kuznetsov VA, Scotti M, Valsecchi V, Simionati B. et al. Gene expression profiles of epithelial cells microscopically isolated from a breast-invasive ductal carcinoma and a nodal metastasis. Proc Natl Acad Sci U S A. 2004;101:18147-52 doi:10.1073/pnas.0408260101
141. Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG. et al. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene. 2007;26:1507-16 doi:10.1038/sj.onc.1209920
142. Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J Biol Chem. 2008;283:30919-32 doi:10.1074/jbc.M804029200
143. Tang D, Sivko GS, DeWille JW. Promoter methylation reduces C/EBPdelta (CEBPD) gene expression in the SUM-52PE human breast cancer cell line and in primary breast tumors. Breast Cancer Res Treat. 2005;95:161-70 doi:10.1007/s10549-005-9061-3
144. Palmieri C, Monteverde M, Lattanzio L, Gojis O, Rudraraju B, Fortunato M. et al. Site-specific CpG methylation in the CCAAT/enhancer binding protein delta (CEBPdelta) CpG island in breast cancer is associated with metastatic relapse. Br J Cancer. 2012;107:732-8 doi:10.1038/bjc.2012.308
145. Ceccarelli V, Racanicchi S, Martelli MP, Nocentini G, Fettucciari K, Riccardi C. et al. Eicosapentaenoic acid demethylates a single CpG that mediates expression of tumor suppressor CCAAT/enhancer-binding protein delta in U937 leukemia cells. J Biol Chem. 2011;286:27092-102 doi:10.1074/jbc.M111.253609
146. Brueckner LM, Hess EM, Schwab M, Savelyeva L. Instability at the FRA8I common fragile site disrupts the genomic integrity of the KIAA0146, CEBPD and PRKDC genes in colorectal cancer. Cancer Lett. 2013;336:85-95 doi:10.1016/j.canlet.2013.04.007
147. Tang D, DeWille J. Detection of base sequence changes in the CEBPD gene in human breast cancer cell lines and primary breast cancer isolates. Mol Cell Probes. 2003;17:11-4
148. Vegesna V, Takeuchi S, Hofmann WK, Ikezoe T, Tavor S, Krug U. et al. C/EBP-beta, C/EBP-delta, PU.1, AML1 genes: mutational analysis in 381 samples of hematopoietic and solid malignancies. Leuk Res. 2002;26:451-7
149. Bennett CE, Nsengimana J, Bostock JA, Cymbalista C, Futers TS, Knight BL. et al. CCAAT/enhancer binding protein alpha, beta and delta gene variants: associations with obesity related phenotypes in the Leeds Family Study. Diab Vasc Dis Res. 2010;7:195-203 doi:10.1177/1479164110366274
150. Angeloni D, Lee JD, Johnson BE, Teh BT, Dean M, Lerman MI. et al. C306A single nucleotide polymorphism in the human CEBPD gene that maps at 8p11.1-p11.2. Molecular and Cellular Probes. 2001;15:395-7 doi:Doi 10.1006/Mcpr.2001.0377
151. Tong Y, Zheng Y, Zhou J, Oyesiku NM, Koeffler HP, Melmed S. Genomic characterization of human and rat prolactinomas. Endocrinology. 2012;153:3679-91 doi:10.1210/en.2012-1056
152. Covell DG, Wallqvist A, Rabow AA, Thanki N. Molecular classification of cancer: unsupervised self-organizing map analysis of gene expression microarray data. Mol Cancer Ther. 2003;2:317-32
153. Willenbrock K, Kuppers R, Renne C, Brune V, Eckerle S, Weidmann E. et al. Common features and differences in the transcriptome of large cell anaplastic lymphoma and classical Hodgkin's lymphoma. Haematologica. 2006;91:596-604
154. Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY. et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318-25 doi:10.1038/nature08712
155. Hagedorn M, Delugin M, Abraldes I, Allain N, Belaud-Rotureau MA, Turmo M. et al. FBXW7/hCDC4 controls glioma cell proliferation in vitro and is a prognostic marker for survival in glioblastoma patients. Cell Div. 2007;2:9. doi:10.1186/1747-1028-2-9
156. Shay JE, Celeste Simon M. Hypoxia-inducible factors: crosstalk between inflammation and metabolism. Seminars in cell & developmental biology. 2012;23:389-94 doi:10.1016/j.semcdb.2012.04.004
157. Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annual review of immunology. 2012;30:677-706 doi:10.1146/annurev-immunol-020711-075008
158. Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends in pharmacological sciences. 2012;33:207-14 doi:10.1016/j.tips.2012.01.005
159. Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721-32 doi:10.1038/nrc1187
160. Greijer AE, van der Wall E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J Clin Pathol. 2004;57:1009-14 doi:10.1136/jcp.2003.015032
161. Bosco MC, Puppo M, Blengio F, Fraone T, Cappello P, Giovarelli M. et al. Monocytes and dendritic cells in a hypoxic environment: Spotlights on chemotaxis and migration. Immunobiology. 2008;213:733-49 doi:10.1016/j.imbio.2008.07.031
162. Feinberg MW, Watanabe M, Lebedeva MA, Depina AS, Hanai J, Mammoto T. et al. Transforming growth factor-beta1 inhibition of vascular smooth muscle cell activation is mediated via Smad3. J Biol Chem. 2004;279:16388-93 doi:10.1074/jbc.M309664200
163. de Heredia LL, Magoulas C. Lack of the transcription factor C/EBPdelta impairs the intrinsic capacity of peripheral neurons for regeneration. Exp Neurol. 2012 doi:10.1016/j.expneurol.2012.10.012
164. Chang LH, Huang HS, Wu PT, Jou IM, Pan MH, Chang WC. et al. Role of Macrophage CCAAT/Enhancer Binding Protein Delta in the Pathogenesis of Rheumatoid Arthritis in Collagen-Induced Arthritic Mice. PLoS ONE. 2012;7:e45378
165. Thangaraju M, Ananth S, Martin PM, Roon P, Smith SB, Sterneck E. et al. c/ebpdelta Null mouse as a model for the double knock-out of slc5a8 and slc5a12 in kidney. J Biol Chem. 2006;281:26769-73 doi:C600189200 [pii] 10.1074/jbc.C600189200
166. Takeji M, Kawada N, Moriyama T, Nagatoya K, Oseto S, Akira S. et al. CCAAT/Enhancer-binding protein delta contributes to myofibroblast transdifferentiation and renal disease progression. J Am Soc Nephrol. 2004;15:2383-90 doi:10.1097/01.ASN.0000136426.01160.2F
167. Wang JM, Tseng JT, Chang WC. Induction of human NF-IL6beta by epidermal growth factor is mediated through the p38 signaling pathway and cAMP response element-binding protein activation in A431 cells. Mol Biol Cell. 2005;16:3365-76 doi:10.1091/mbc.E05-02-0105
168. Chen H, Libertini SJ, George M, Dandekar S, Tepper CG, Al-Bataina B. et al. Genome-wide analysis of androgen receptor binding and gene regulation in two CWR22-derived prostate cancer cell lines. Endocrine-related cancer. 2010;17:857-73 doi:10.1677/ERC-10-0081
169. Khan JA, Bellance C, Guiochon-Mantel A, Lombes M, Loosfelt H. Differential regulation of breast cancer-associated genes by progesterone receptor isoforms PRA and PRB in a new bi-inducible breast cancer cell line. PLoS One. 2012;7:e45993. doi:10.1371/journal.pone.0045993
170. Ali S, Singh NN, Yildirim H, Ramji DP. Requirement for nuclear factor kappa B signalling in the interleukin-1-induced expression of the CCAAT/enhancer binding protein-delta gene in hepatocytes. The international journal of biochemistry & cell biology. 2010;42:113-9 doi:10.1016/j.biocel.2009.09.018
171. Cantwell CA, Sterneck E, Johnson PF. Interleukin-6-specific activation of the C/EBPdelta gene in hepatocytes is mediated by Stat3 and Sp1. Mol Cell Biol. 1998;18:2108-17
172. Sekine O, Nishio Y, Egawa K, Nakamura T, Maegawa H, Kashiwagi A. Insulin activates CCAAT/enhancer binding proteins and proinflammatory gene expression through the phosphatidylinositol 3-kinase pathway in vascular smooth muscle cells. J Biol Chem. 2002;277:36631-9 doi:10.1074/jbc.M206266200
173. Hallenborg P, Feddersen S, Francoz S, Murano I, Sundekilde U, Petersen RK. et al. Mdm2 controls CREB-dependent transactivation and initiation of adipocyte differentiation. Cell death and differentiation. 2012;19:1381-9 doi:10.1038/cdd.2012.15
174. Cardinaux JR, Magistretti PJ. Vasoactive intestinal peptide, pituitary adenylate cyclase-activating peptide, and noradrenaline induce the transcription factors CCAAT/enhancer binding protein (C/EBP)-beta and C/EBP delta in mouse cortical astrocytes: involvement in cAMP-regulated glycogen metabolism. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1996;16:919-29
175. Borges TJ, Lopes RL, Pinho NG, Machado FD, Souza AP, Bonorino C. Extracellular Hsp70 inhibits pro-inflammatory cytokine production by IL-10 driven down-regulation of C/EBPbeta and C/EBPdelta. Int J Hyperthermia. 2013;29:455-63 doi:10.3109/02656736.2013.798037
176. Wang Y, Sul HS. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell metabolism. 2009;9:287-302 doi:10.1016/j.cmet.2009.01.013
177. Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G. et al. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. Embo J. 2011;30:1357-75 doi:10.1038/emboj.2011.52
178. Svotelis A, Doyon G, Bernatchez G, Desilets A, Rivard N, Asselin C. IL-1 beta-dependent regulation of C/EBP delta transcriptional activity. Biochem Biophys Res Commun. 2005;328:461-70 doi:10.1016/j.bbrc.2005.01.002
Author contact
![]() Corresponding author: Email: sternecgnih.gov Phone: +1-301-846-1471; Fax: +1-301-846-1666.
Corresponding author: Email: sternecgnih.gov Phone: +1-301-846-1471; Fax: +1-301-846-1666.

 Global reach, higher impact
Global reach, higher impact