10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(10):1021-1031. doi:10.7150/ijbs.6100 This issue Cite
Research Paper
Isolation and Quantification of MicroRNAs from Urinary Exosomes/Microvesicles for Biomarker Discovery
Institute of Nephrology, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, China.
Received 2013-2-20; Accepted 2013-9-24; Published 2013-10-12
Abstract
Recent studies indicate that microRNA (miRNA) is contained within exosome. Here we sought to optimize the methodologies for the isolation and quantification of urinary exosomal microRNA as a prelude to biomarker discovery studies. Exosomes were isolated through ultracentrifugation and characterized by immunoelectron microscopy. To determine the RNA was confined inside exosomes, the pellet was treated with RNase before RNA isolation. The minimum urine volume, storage conditions for exosomes and exosomal miRNA was evaluated. The presence of miRNAs in patients with various kidney diseases was validated with real-time PCR. The result shows that miRNAs extracted from the exosomal fraction were resistant to RNase digestion and with high quality confirmed by agarose electrophoresis. 16ml of urine was sufficient for miRNA isolation by absolute quantification with 4.15×105 copies/ul for miR-200c. Exosomes was stable at 4℃ 24h for shipping before stored at -80℃ and was stable in urine when stored at -80°C for 12months. Exosomal miRNA was detectable despite 5 repeat freeze-thaw cycles. The detection of miRNA by quantitative PCR showed high reproducibility (>94% for intra-assay and >76% for inter-assay), high sensitivity (positive call 100% for CKD patients), broad dynamic range (8-log wide) and good linearity for quantification (R2>0.99). miR-29c and miR-200c showed different expression in different types of kidney disease. In summary, the presence of urinary exosomal miRNA was confirmed for patients with a diversity of chronic kidney disease. The conditions of urine collection, storage and miRNA detection determined in this study may be useful for future biomarker discovery efforts.
Keywords: chronic kidney disease, exosome, miRNA, urine, biomarker.
Introduction
Micro(mi)RNAs are short (20-22 nucleotides in length), non-coding RNAs that control the translation of proteins of many genes[1]. Changes in the profile of cellular miRNAs have been shown to correlate with different pathophysiological conditions. Exosomes are small membrane vesicles with a size of 30-120 nm that are released by different cell types[2,3]. They can be isolated from various body fluids, including plasma, malignant ascites, urine, amniotic fluid, breast milk and saliva[4,5]. In addition to exosomes, microvesicles(larger in size) can also be included because of the overlap in size when they are isolated. In this study, the pellet isolated using ultracentrifugation (at 200,000 g) was collectively termed the exosomes because the majority of the pellet consisted of the exosomes[3].
More recently, Valadi and colleagues demonstrated that exosomes contain both mRNA and miRNA[6]. The findings triggered the hypothesis that extracellular miRNA from the exosomes may also play a role in cell-to-cell communication[7,8] and serve as a biomarker for disease. Taylor et al. demonstrated that miRNA profiling of circulating tumor exosomes could potentially be used as surrogate diagnostic markers for biopsy profiling.[9] More recently, Bala et al. showed that circulating miRNAs may serve as biomarkers to differentiate between hepatocyte injury and inflammation[10].Murakami et al. demonstrated that the miRNA expression pattern in the exosome-rich, fractionated serum has high potential as a biomarker for diagnosing the grade and stage of liver diseases.[11]
In the kidneys, miRNAs are indispensable for development and homeostasis[12]. Clearly, miRNAs play a critical role in the regulatory mechanisms involved in renal development, maintenance of renal function, and progression of kidney disease[13,14]. Exosomes are normally secreted into the urine from all nephron segments[15]; therefore, we hypothesize that the urinary exosomes may contain miRNAs that might be associated with various kidney diseases.
Before performing biomarker discovery studies, the optimal conditions for urinary exosomal isolation and detection need to be defined. Zhou et al. have studied effective methods for collecting, storing, and preserving urinary exosomal proteins[16]. Recently, Alvarez et al. sought to identify the best exosome isolation methods for proteomic analysis and RNA profiling[17]. However, to the best of our knowledge, there have been no in-depth studies investigating the isolation of urinary exosomal miRNAs as potential diagnostic biomarkers for kidney disease. Therefore, there is still a need to evaluate a methodology to obtain high-quality miRNA and reliable miRNA measurement for diagnostic applications. Here, we report for the first time the optimal conditions for the successful isolation and initial characterization of urinary exosomal miRNA from human urine. As a proof of principle, we also examined urine samples from individuals with nephrotic syndrome due to a variety of causes of chronic kidney disease (CKD). The method used in this study might be useful for future urinary exosomal miRNA detection for biomarker discovery in CKD.
Materials and Methods
Ethics Statement
All of the studies were approved by the Ethical Committee of Southeast University. Written informed consent was obtained from all of the subjects to use their urine for research purposes.
Sample collection and exosomes purification
A whole-stream, early-morning urine specimen was collected from patients with diabetic nephropathy (DN, n=4), focal segmental glomerulosclerosis (FSGS, n=5) or IgA nephropathy (IgA, n=7). The exosomes were isolated from the human urine samples using differential centrifugation. The urine was centrifuged at 2000 g for 20 min to eliminate the cells and debris and at 13,500g for 20 min, followed by ultracentrifugation at 200,000 g for 60 min. The pellet was stored at -80℃ for subsequent applications.
Immunoelectron microscopy
The exosomes suspensions were mixed 1:1 with 4% paraformaldehyde and were then applied to 200-mesh nickel grids. After blocking with 1% BSA (Bovine Serum Albumin) and washing, the grid was incubated with a primary antibody recognizing AQP-2 , CD9 and Alix (Santa Cruz, CA, USA) in 0.02% Triton X-100 for 1 hour at room temperature (AQP2, an aquaporin water channel protein; CD9, a cell surface glycoprotein, both were considered exosome markers). After washing, the grids were exposed to species-specific anti-IgG antibodies conjugated to colloidal gold particles (5 nm) (Boster, Wuhan, China). Then, the membranes were washed with PBS (Phosphate Buffered Saline) once and water twice. Control labeling was performed in an identical experiment, but the diluting solution was substituted for the primary antibody.
RNA isolation and miRNA measurement
RNA isolation
The total RNA was isolated using the RNeasy micro kit, and miRNA was isolated using the miRNeasy micro kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The exosomes were disrupted and homogenized in RLT buffer (Qiagen) or 700 µl QIAzol lysis reagent, and the rest of the procedure was performed according to the manufacturer's protocol. To confirm that the RNA was confined to the exosomes, urinary exosomes were treated with 0.1 μg μl-1RNase A (Biosharp) for 20 min at 37 °C. As a control, an equal amount of exosomes from the same patient was treated with PBS before RNA isolation. The test was repeated using 3 samples from three different patients. The RNA concentration and purity were confirmed by the spectrophotometric ratio using absorbance measurements at wavelengths of 260 nm and 280 nm on a Nanodrop 2000 (Thermo, Wilmington, USA). Isolated RNA was also analyzed on a RNA Pico 6000 chip (Agilent, Palo Alto, CA, USA) using an Agilent Bioanalyzer (Agilent) and on a 2% agarose electrophoresis.
Reverse-transcription quantitative real-time PCR
For mRNA quantification, cDNA was synthesized from isolated RNA using the Takara PrimeScript® RT reagent kit (Takara, Dalian, China) according to the manufacturer's instructions. Reverse-transcription quantitative real-time polymerase chain reaction (RT-qPCR) was performed using Takara SYBR® Premix Ex Taq™ (Takara, Dalian, China). CD24 and AQP2 detection was performed using the following primers:
CD24(NM_013230.2)-FWD,
5'CTCCTACCCACGCAGATTTATT3';
CD24(NM_013230.2)-REV,
5'GGTGGTGGCATTAGTTGGATTT3'; AQP2(NM_000486.5)-FWD,
5'GCTCCGCTCCATAGCCTTCTCC3';
AQP2(NM_000486.5)-REV,
5' GTGCCAATACCCAAGCCAAACG 3'.
Detection of the mature miRNAs was performed by reverse transcription using the Takara One Step PrimeScript® miRNA cDNA synthesis kit (product code: D350A,Takara, Dalian, China) according to the manufacturer's instructions, and the RT-qPCR reaction was performed using SYBR® Premix Ex Taq™ II (product code: DRR081, Takara, Dalian, China). The primer mix for RNU6B, hsa-miR-29c, and hsa-miR-200c was obtained from Takara Biotechnology (Takara, Dalian, China). miR-200c was synthesized by Invitrogen (Shanghai, China) as stardard for absolute quantification.
Statistical analysis
Statistical analysis was performed by SPSS for Windows software version 11.0. Briefly, raw threshold cycles (Ct) values from PCR assay were imported from ABI7300 SDS software. All the results were presented in mean ± SD (standard deviation) for data. We used Spearman's rank-order correlations to assess associations between different technical replicate and plots. A P value of below 0.05 was considered statistically significant. All probabilities were two-tailed.
Results
1. Identification and characterization of the exosomes
The exosomes were isolated from the urine of patients with kidney disease through a series of centrifugation and ultracentrifugation steps. To confirm that the structures studied were indeed exosomes, they were examined using electron microscopy and immunogold staining.
1.1 Size distribution of the urinary exosomes
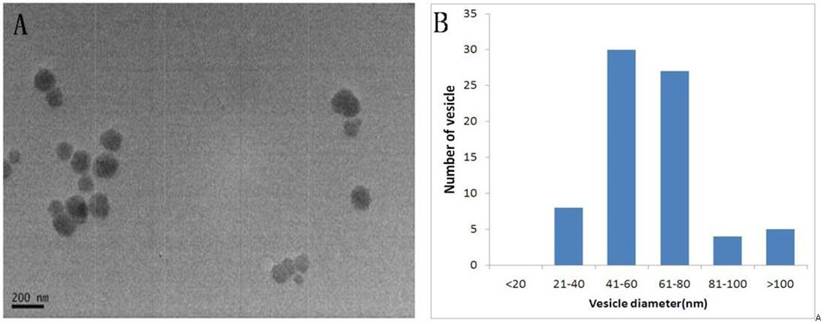
Transmission electron microscopy showed that the isolated vesicles were round with relative sizes ranging from 21-160nm. The images show that there was no contamination with cellular debris. Quantitative analysis of the electron micrographs for the urinary exosomes revealed the distribution of exosome sizes;92.7% of the exosomes were between 30-120nm, consistent with a small size for the exosomes[15](Fig.1).
1.2 Immunogold staining of the vesicles with exosome markers
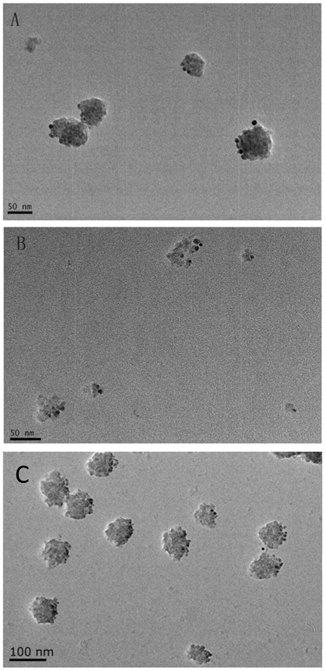
The identity of the studied vesicles was further confirmed to be exosomes using immunogold-staining analysis with antibodies against AQP2, CD9 and Alix, which are commonly used markers for urinary exosomes[18]. The results show that the majority of the vesicles were immunogold-labeled with anti-AQP2, anti-CD9 and anti-Alix (Fig. 2).
Shape and size distribution of the urinary exosomes. Exosomes were isolated using differential centrifugation. (A) Electron micrograph of a urinary exosome. (B) Histogram of the exosome diameter. Size analysis of 10 fields of view showing that 92.7% of the exosomes were between 30-120nm.
Immunoelectron microscopy of isolated urinary exosomes. The exosomes were immunogold labeled with anti-AQP2 (A) , anti-CD9 (B) and Anti-Alix (C). The scale bar represents 50 nm and 100nm.
2. Confirmation of the isolated RNA source
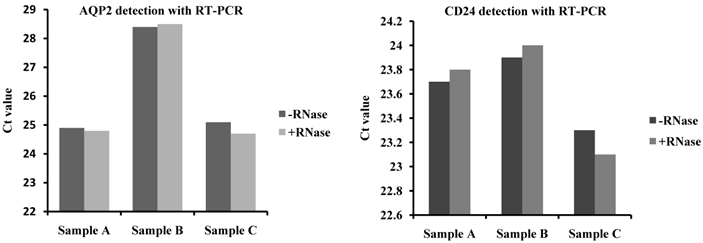
To confirm that the RNA was confined to the exosomes, RNA was isolated from RNase-treated and control urinary exosomes, and a real-time RT-PCR analysis was performed. The primer specificity of AQP2 and CD24 was confirmed by dissociation curve and agarose gel electrophoresis (Supplementary Fig.1). We observed that the exosomal RNA was indeed protected from RNase digestion because CD24- and AQP2-specific products could still be amplified. Moreover, there is no significant difference in the Ct values for AQP2 and CD24 between the RNase-treated and -untreated groups. The results indicate no difference in RNA degradation between the RNase-treated and control exosomes (Fig.3), indicating that RNA was not present on the exterior of the exosomes.
3. Evaluation of urinary exosomal miRNA
To confirm the presence of miRNAs within the urinary exosomes, we performed miRNA quantitative PCR amplification for two miRNAs (hsa-miR-29c, hsa-miR-200c) that have previously been identified as critical regulators in kidney disease development[13], and the minimum urine volume for miRNA isolation, stability of exosomal miRNA and the reproducibility, specificity, and dynamic range of RT-PCR detection were evaluated.
3.1 Minimum urine volume for miRNA isolation
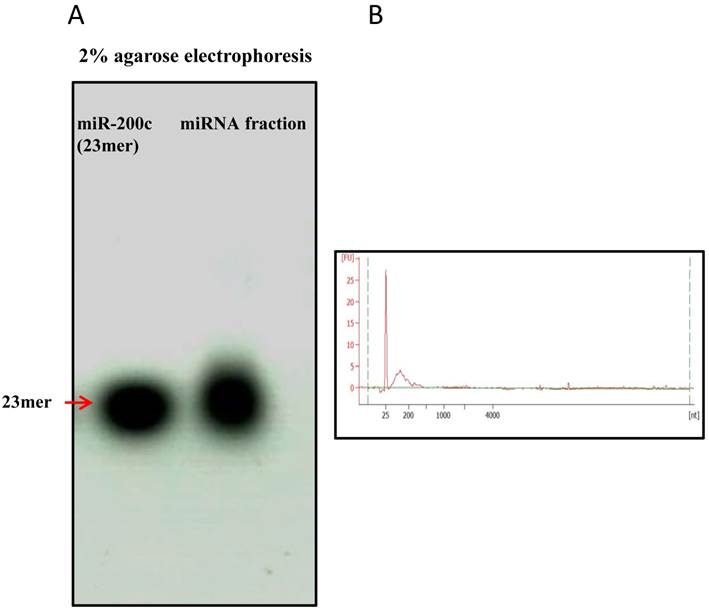
The miRNA fraction of was purified with removal of larger RNAs. The average concentration of the miRNA fraction was 22.8ng/ul. And the miRNA-enriched fraction was confirmed by running on 2% agarose gel and Agilent bioanlyzer. For comparison, synthetic miR-200c was also run on the gel. Fig.4 showed the size of miRNA fraction was approximately the same as miR-200c of 23mer. The plot showed a prominent 'small RNA' peak (between ∼25-200 nt) when miRNA was isolated separately.
RNA source confirmation by RNase treatment of the exosomes before RNA purification. To confirm that the RNA was confined to the exosomes, the exosomes were treated with RNase to remove other RNA and compared to control samples. The exosome markers CD24 and AQP2 were detected using RT-PCR. The results showed that CD24- and AQP2-specific products could still be amplified and that there are no significant differences in the Ct value for AQP2 and CD24 between the RNase-treated and untreated groups.
Enrichement of miRNA fraction run on agarose gel and Agilent bioanalyzer. miRNA fraction was purified and run on a 2% agarose gel and Agilent bioanalyzer 2100 . As a comparison, synthetic miR-200c (23mer) was also run on the gel. The gel revealed pure miRNA fraction approximately 23mer size. Figure4B showed plot with a prominent 'small RNA' peak (between ∼25-200 nt).
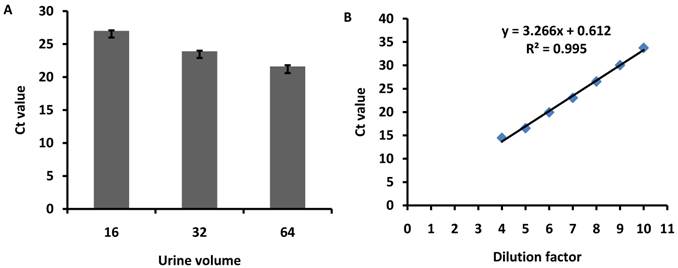
To determine the minimum urine volume for exosomal miRNA isolation, freshly obtained urine samples were pooled from three healthy volunteers and then subjected to exosomes purification with 16, 32 or 64ml of urine. The hsa-miR-200c was measured with same volume of RNA solution reverse transcribed and detected with real-time PCR. The results showed that the Ct values were 27±0.09, 23.9±0.11, and 21.6±0.2 for 16, 32 and 64ml of urine, respectively (Fig.5A).
Synthesized miRNA-200c was 10 fold serial diluted and standard curve was plotted with R2=0.995 (Fig.5B). The absolute abundance of miR-200c is determined as 4.15×105 copies/ul for miRNA fraction purified from 16ml urine. Besides, other members of miRNAs were also detected. The result showed that Ct value were 28.0, 28.2, 25.6 and 26.2 for miR-29a, 29b, 29c, and 200b with 10 fold dilution of reverse transcription solution with miRNA purified from 16ml urine.
3.2 Stability of exosomal miRNA
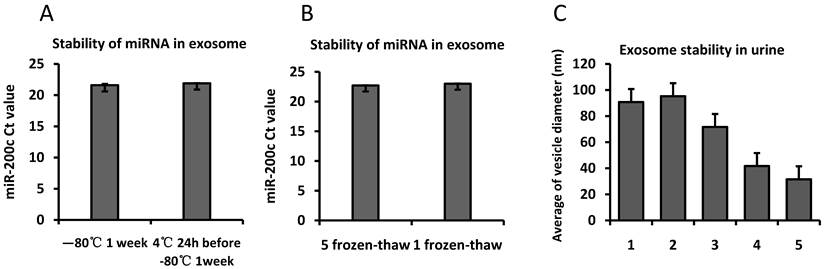
The stability of the miRNA oligonucleotides was further evaluated to determine the effect of different storage conditions. The isolated exosomes was stored at -80°C for 1 week or at 4°C for 24h before moving -80°C to mimic shipping on ice from a distant clinic to a central analysis laboratory. We examined the abundance of miR-200c isolated from the exosome fraction in the first morning urine from three volunteers. The Ct value was 21.9±0.03 for the exosomes stored at 4°C for 24h before storing at -80°C for 1 week, and the Ct value was 21.6±0.20 when the exososme was stored at -80°C for 1 week directly after purification. We found no difference in the miRNA amount from an equal volume of urine at these two conditions (Fig.6).
The effect of freeze-thaw cycles on the stability of miRNA containing RNase-free water was examined. The isolated RNA product was aliquoted into 2 separate RNase-free tubes. MiRNA from 1 tube was processed with 5 freeze-thaw cycles before reverse transcription, and the other miRNA was reverse transcribed directly. Hsa-miR-200c was detected by real-time PCR, and the results show that the Ct values were 22.7±0.03 and 23±0.04, respectively, for the two different conditions. Hsa-miR-200c remained detectable after 5 freeze-thaw cycles (Fig.6).
We also observe the exosome stability in urine samples by looking at the shape and size of the exosome structure with TEM. Urine was stored at different conditions to evaluate the exosome stability in urine samples: 1). 4°C, 24hours, 2). 37°C, 24hours, 3). -80°C, 24hours, 4). -80°C, 6months, 5). -80°C, 12months, 6). -80°C, 24months. Exosome was isolated and observed by TEM. The result indicated that in 24 months, the size of the exosome was significant reduced to 31.6nm which is the lower limit range of exosome diameter and the shape became irregular which indicated partial degradation after long time storage (Fig.6).
3.3 Specificity of miRNA detection
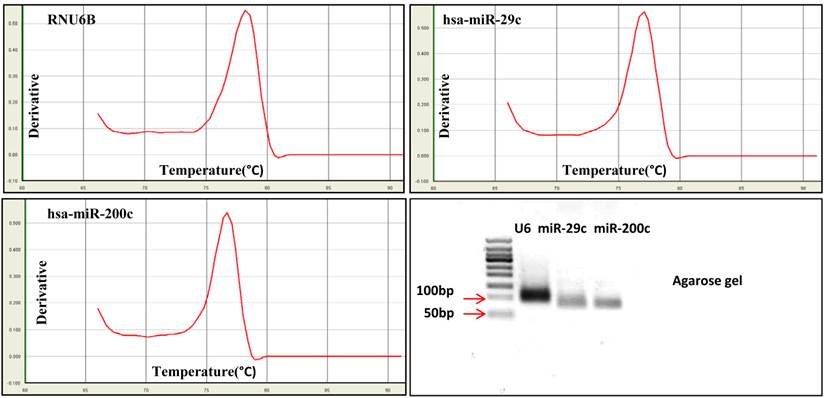
Exosomal miRNA was detected by RT real-time PCR for hsa-miR-29c, hsa-miR-200c and the internal control RNU6B followed by dissociation (melting) curve analysis. The melting curve analysis showed specific amplification, and no primer dimers or non-specific products were observed. The PCR products were run on 3% agarose gels to further detect the specificity. Electrophoresis showed single bands at the predicted sizes of the products for both miRNAs and the internal control (the predicted PCR product size was 82bp for has-miR-29c, 82bp for hsa-miR-200c and 104bp for RNU6B) (Fig.7).
Minimum urine volume for exosome miRNA isolation. Urine samples were pooled from 3 healthy volunteers and then subjected to exosome purification using 16, 32 or 64ml of urine. hsa-miR-200c was measured with same volume of RNA solution, reverse transcribed and detected using qPCR. The diagram shows the Ct values for miRNAs detected from different volumes of urine. The error bars represent the standard deviation of three replicate PCR test.
Stability of exosomal miRNA and urinary exosome. Exosomes isolated from the same volume of pooled urine was stored at 4℃ for 24h and moved to -80℃ for 1 week or stored at -80℃ for 1 week immediately. Fig.6A shows the Ct value for hsa-miR-200c using qPCR with the same volume of RNA solution. Equal amounts of miRNA in RNase-free water were aliquoted into two tubes. One tube was frozen and thawed 5 times, and the other was frozen once before being reverse transcribed. Fig.6B shows the Ct value for hsa-miR-200c using qPCR. Fig.6C. Urine was stored at different conditions to evaluate the exosome stability in urine samples: 1. 4°C, 24hours, 2, 37°C, 24hours, 3. -80°C, 24hours, 4. -80°C, 6months, 5. -80°C, 12months, 6. -80°C, 24months. Exosome was isolated and observed by TEM.The error bars represent the standard deviation of three replicate PCR test.
Specificity of miRNA detection using real-time PCR. MiRNA was detected using real-time PCR followed by dissociation (melting) curve analysis. The dissociation curve and agarose gel electrophoresis results show that RNU6B, hsa-miR-29c and hsa-miR-200c could be accurately detected.
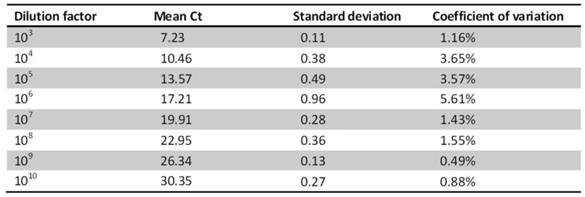
The intra-assay variability was evaluated by detecting hsa-miR-200c in ten-fold serially dilutions in triplicate.
3.4 Reproducibility of miRNA detection
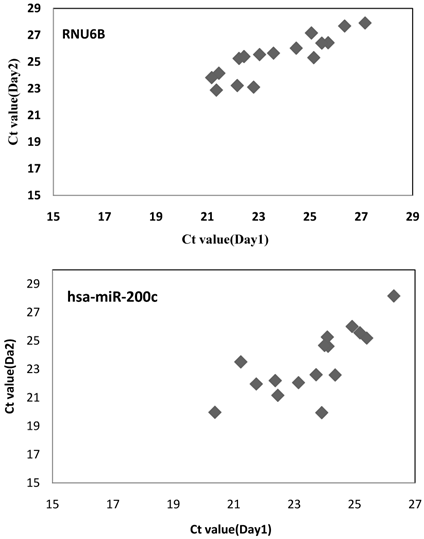
To determine the reproducibility of miRNA detection by RT-PCR, intra-assay variability was estimated by running the RT-PCR product for hsa-miR-200c in triplicate. The RT-PCR product was serially diluted by ten-fold. The results show that among the 8 tests for different dilutions, the coefficients of variation of 7 of the tests were below 5%, whereas that of the remaining test was 5.61% (Table 1). The inter-assay variability was measured by running tests on samples from CKD patients by different technicians on different days. The results show strong correlations across the technical replicates and lots (r=0.876, P <0.001 for RNU6B; r=0.762, P <0.001 for hsa-miR-200c) (Fig.8), ensuring reliable detection of differences in expression among the biological samples for biomarker discovery.
3.5 Dynamic range and sensitivity of miRNA detection
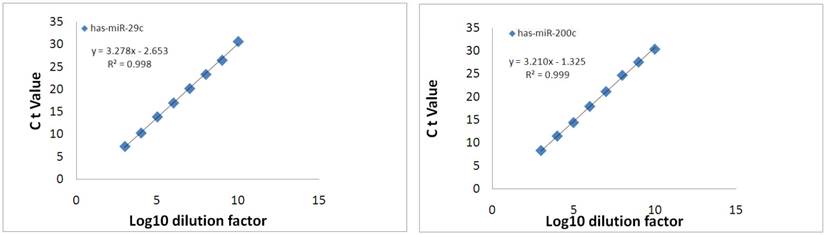
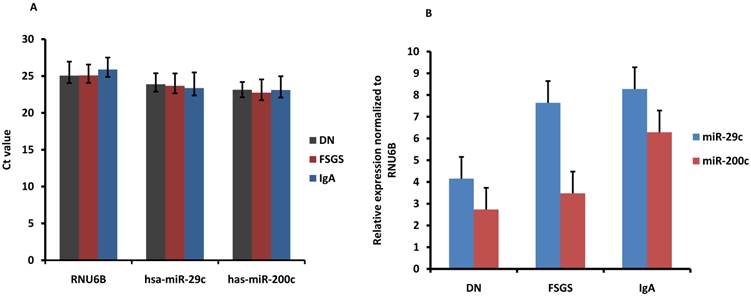
The PCR products for hsa-miR-29c and hsa-miRNA-200c were serially diluted by ten-fold and detected by real-time PCR. The results showed that both miRNAs could be detected in 8-log wide dynamic range, and the standard curve generated by plotting the log10 dilution factor and Ct values showed very good linearity, with R2=0.998 for hsa-miR-29c and R2=0.999 for hsa-miR-200c (Fig.9). To evaluate the sensitivity of miRNA detection, urinary exosomal miRNAs from 16 CKD patients with DN, FSGS or IgA were detected. The results showed that the positive call was 100% (Ct<30). For more than 90% of the samples, the Ct value was below 25 (Fig.10), and the average Ct values for hsa-miR-29c and hsa-miR-200c were 23.0 and 23.6, respectively.
Moreover, we also evaluate the difference of miRNA expression in different types of kidney disease by normalizing the expression to RNU6B. The result showed that miR-29c and miR-200c was 2.0 and 2.3 fold increased in IgA nephropathy patients compared with DN patients although no significant difference was found among the 3 types of kidney disease.
Discussion
Currently, miRNAs represent attractive candidate biomarkers for various diseases. Interestingly, recent studies have indicated that miRNAs are incorporated into exosomes and are more stable than their cellular counterparts. MiRNAs can resist degradation through protection in vesicles released from cultured cells[6] or during circulation in the body [19]. This apparent stability underscores the attractiveness of exosome miRNAs as disease biomarkers.
The inter-assay variability for miRNA detection. RNU6B and hsa-miR-200c were measured in 16 CKD patients by different technicians on different days. Spearman's rank-order correlation was applied to assess associations between the technical replicates and lots. The result show strong correlations across technical replicates and lots (r=0.876, p=0.000 for RNU6B; r=0.762, p=0.000 for hsa-miR-200c).
Dynamic range of exosomal miRNA detection. The specific PCR product was serially diluted ten-fold and analyzed using qPCR. The resulting threshold cycles for the assays are plotted versus the log10 of the dilution factor.
Sensitivity and difference of urinary exosomal miRNA detection for various kidney diseases. (A) RNU6B, hsa-miR-29c and hsa-miR-200c were detected in 16 CKD patients to assess the positive call of RT-PCR detection. Figure showed the average Ct values. (DN, diabetic nephropathy; FSGS, focal segmental glomerulosclerosis ; IgA, IgA nephropathy). (B) miR-29c and miR-200c was normalized to RNU6B. The relative expression level of miRNAs was showed (2.0, 2.3 fold increase in IgA compared with DN). The error bars represent the standard deviation of Ct values or normalized expression from different patients in each group.
Urinary exosomes are being studied intensively to identify potential biomarkers for renal disease[20]. Miranda et al. demonstrated that urinary exosomes have an RNA integrity profile similar to that of kidney tissue mRNA. They also detected mRNA-encoding proteins from all regions of the nephron and the collecting duct[19]. Eldh et al. compared different RNA extraction methods to analyze exosomal RNA from cultured cells and found that different isolation methods had extensive variation in exosomal RNA yield and patterns[21]. However, techniques for isolating and quantifying miRNA species from biological samples are relatively recent developments. The only report (Alvarez et al.) mainly focused on the isolation method for exosomal mRNA and miRNA, and the samples were only from healthy donors[17]. Therefore, the collection, storage, and preservation methods must be clarified before urinary exosomal miRNAs are used for further evaluation as candidate biomarkers.
In previous studies, different protocols have been presented for exosome purification from various biologic samples. The most common ones are differential centrifugation, combing ultracentrifugation[16] and nanomembrane ultrafiltration[22]. Recently, a precipitation method called Exo-Quick (System Biosciences, Mountain View, CA, U.S.A.) has become commercially available. It is relatively quick compared with the other two approaches. Yamada et al. showed that the highest yield of exosomes was achieved using ultracentrifugation with ExoQuick precipitation, while it was not the optimal way for persevering exosome structure[23]. Alvarez et al. demonstrated that, if an ultracentrifuge is available, ultracentrifugation methods are better options for exosomes isolation among the 6 single or combination approaches studied [17]. In this study, we chose to purify exosomes with the simple and low-cost way of ultracentrifugation. Electronmicroscopy verified a typical shape of urinary vesicles with a relatively uniform size. Although there is a very small fraction (7%) of vesicles larger than 100nm, the diameter of larger vesicles was less than 160nm; we believe these larger vesicles are exosome-like vesicles. The results indicate that the majority of microvesicles isolated from urine are exosomes, although a small amount of other microvesicles do co-isolate during ultracentrifugation and ultrafiltration isolation techniques. However, the microvesicles were formed by budding off the cell's plasma membrane and may also contain a snapshot of the parent cell's RNA[24,25]. The microvesicles could also serve as extracellular miRNA markers. The exosomes were further verified by detecting exosomal markers CD9, AQP2 and Alix[15]. The results showed that almost all of the isolated exosomes were positively stained with gold particles, which further verified the identity and purity of the isolated exosomes. Thus, we concluded that the exosomes mainly contributed to the findings in this report.
To confirm that the isolated RNA was confined to the exosomes, the exosome pellets were subjected to RNase before RNA isolation. The results indicated that there is no difference in the amount of RNA isolated between the RNase-treated and control groups. Thus, the majority of RNA appears to be contained within the exosomes and is thereby protected from RNases by the surrounding membrane. The absence of extraneous RNA contamination may be because urine contains ribonucleases[26]. The fact that microvesicles can resist RNase digestion adds further support to the previously reported stable nature of nucleic acids within urinary exosomes[19,27]. However, a recent study by Arroyo et al. showed that Argonaute2-miRNA complexes and vesicle-associated miRNA are the two modes for stabilizing circulating miRNAs. The author hypothesized that the two models of miRNAs originated from different cell types and might reflect cell type-specific miRNAs[28]. Previous studies have shown that miRNAs are stable in serum[29] and urine sediment[30]. However, there is little evidence regarding miRNA stability in the urine exosomes. Although the true mechanism for miRNA stability in the urine is not clear, in this study, at least the miRNA that could be detected was protected from RNases by the exosomes.
Considering the protective effect of exosomes, methods to preserve the exosomes before RNA isolation should be evaluated. There is still a critical need for developing effective protocols for collecting and storing urinary exosomes[31]. In this study, we determined the minimum urine volume needed and the influence of the storage condition on the isolation of the exosome miRNA.
Zhou et al. processed 10 ml of fresh first morning urine samples from three individuals to verify whether the exosome fraction could be isolated from much smaller volumes compared to typical clinical samples, and the results showed that 10 ml of urine was sufficient to detect urinary exosome-associated proteins by western blot analysis[16]. Miranda et al. used 200ml of urine for exosomal mRNA detection[19]. However, the minimum urine volume for miRNA isolation and quantification is not clear. This study showed that 16 ml of urine was sufficient to detect miRNA. All 5 miRNAs were readily detected with miRNA fraction purified from 16ml urine. Moreover, gel electrophoresis and absolute quantification showed high abundance and high quality of miRNA was purified. The minimum urine volume for miRNA detection in this study indicated high levels of miRNA inside the urinary exosomes, demonstrating the value of this organelle in biomarker discovery.
Moreover, the results indicate that the exosomes and exosome miRNA were stable despite storage at 4°C for 24h before cooling to -80°C and 5 freeze-thaw cycles. Moreover, exosome was stable in urine when stored at -80°C for 12months, but become degraded at 24months. Cheruvanky et al. demonstrated that urinary exosomes could be used for protein isolation after 24 h when shipped on ice[22]. The findings in this study also suggest that an exosomes pellet can be shipped from a distant clinical site and stored at -80°C before miRNA isolation and that the isolated miRNA in RNase-free water could be used for repeat measurements for biomarker tests.
Before using urinary miRNAs for biomarker discovery, the techniques used to quantify these molecules must be reliable. In this study, we concluded that for miRNA quantification, the method was highly precise over a wide range of concentrations with low inter-assay and intra-assay variability. Moreover, the high linearity of the quantification standard curve further indicated the concise measurement of the miRNA. The average Ct values for hsa-miR-29c and hsa-miR-200c were23.0 and 23.6, respectively, which indicates high levels of miRNA within the urinary exosomes. In the study from Alvarez et al., high levels of exosomal miR-192 and miR-1207-5p were obtained only using modified exosomes precipitation methods with a commercial kit compared to the 6 methods studied[17]. The high recovery of miRNA using ultracentrifugation in this study supports this simple and fast strategy for exosomal miRNA isolation. Purifying exosome from nephrotic urine with abundant protein is a very important problem to be addressed for CKD patients. A group has reported that ultrafiltration cannot be used to isolate exosomes from nephrotic urine because the proteins clog the filter[32]. Rood et al. showed that exosomes could be further purified using a method based on ultracentrifugation and an extra step of size-exclusion chromatography[32]. In this study, the successful isolation of miRNA from nephrotic urine suggests that protein does not pose a problem for miRNA profiling when isolating exosomes using ultracentrifugation[17].
Moreover, miR-29c and miR-200c was found 2.0 and 2.3 fold increased in IgA nephropathy patients compared with DN patients although no significant difference was found among the 3 types of kidney disease. However, the number of patients is small and a larger number of patients are needed in the future to confirm this primary result.
In summary, this current investigation has shown that high levels of miRNA were confined to the urinary exosomes for patients with a variety of kidney diseases. The collection, storage, and processing conditions of the urinary samples established in this study may be useful for future biomarker discovery efforts. Moreover, the stability of the urinary exosomes and exosomal miRNA and their accurate detection by qPCR further support their potential as non-invasive biomarkers for kidney disease.
Supplementary Material
Supplementary Figure 1Abbreviations
miRNA: microRNA; CKD: chronic kidney disease; PCR: Polymerase Chain Reaction; AQP2: aquaporin2.
Acknowledgements
We thank Juan Chen for help with the electron microscopy measurements. This study was funded by the National Natural Science Foundation of China (81100544, 81130010), Natural Science Foundation of Jiangsu Province (BK2011602, BK2011061), major State Basic Research Development Program (2012CB517706) and the Research Fund for the Doctoral Program of Ministry of Education (20110092120059).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215-33
2. Vlassov AV, Magdaleno S, Setterquist R. et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940-8
3. Camussi G, Deregibus MC, Bruno S. et al. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78(9):838-48
4. Keller S, Ridinger J, Rupp AK. et al. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86
5. Gu Y, Li M, Wang T. et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS One. 2012;7(8):e43691
6. Valadi H, Ekstrom K, Bossios A. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654-9
7. Montecalvo A, Larregina AT, Shufesky WJ. et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119(3):756-66
8. Wahlgren J, Karlson Tde L, Brisslert M. et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40(17):e130
9. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13-21
10. Bala S, Petrasek J, Mundkur S. et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012
11. Murakami Y, Toyoda H, Tanahashi T. et al. Comprehensive miRNA Expression Analysis in Peripheral Blood Can Diagnose Liver Disease. PLoS One. 2012;7(10):e48366
12. Chandrasekaran K, Karolina DS, Sepramaniam S. et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81(7):617-27
13. Patel V, Noureddine L. MicroRNAs and fibrosis. Curr Opin Nephrol Hypertens. 2012;21(4):410-6
14. Lorenzen JM, Haller H, Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol. 2011;7(5):286-94
15. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101(36):13368-73
16. Zhou H, Yuen PS, Pisitkun T. et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69(8):1471-6
17. Alvarez ML, Khosroheidari M, Kanchi Ravi R. et al. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82(9):1024-32
18. Fernandez-Llama P, Khositseth S, Gonzales PA. et al. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77(8):736-42
19. Miranda KC, Bond DT, McKee M. et al. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78(2):191-9
20. van Balkom BW, Pisitkun T, Verhaar MC. et al. Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int. 2011;80(11):1138-45
21. Eldh M, Lotvall J, Malmhall C. et al. Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods. Mol Immunol. 2012;50(4):278-86
22. Cheruvanky A, Zhou H, Pisitkun T. et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol. 2007;292(5):F1657-61
23. Yamada T, Inoshima Y, Matsuda T. et al. Comparison of methods for isolating exosomes from bovine milk. J Vet Med Sci. 2012;74(11):1523-5
24. Marzesco AM, Janich P, Wilsch-Brauninger M. et al. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci. 2005;118(Pt 13):2849-58
25. Hong BS, Cho JH, Kim H. et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10:556
26. Sugiyama RH, Blank A, Dekker CA. Multiple ribonucleases of human urine. Biochemistry. 1981;20(8):2268-74
27. Skog J, Wurdinger T, van Rijn S. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470-6
28. Arroyo JD, Chevillet JR, Kroh EM. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003-8
29. Mitchell PS, Parkin RK, Kroh EM. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513-8
30. Sharkey JW, Antoine DJ, Park BK. Validation of the isolation and quantification of kidney enriched miRNAs for use as biomarkers. Biomarkers. 2012;17(3):231-9
31. Dimov I, Jankovic Velickovic L, Stefanovic V. Urinary exosomes. ScientificWorldJournal. 2009;9:1107-18
32. Rood IM, Deegens JK, Merchant ML. et al. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78(8):810-6
Author contact
![]() Corresponding author: Lin-Li Lv, M.D. Ph.D. Email: lvlinliedu.cn. Or Bi-Cheng Liu, M.D. Ph.D. Email: liubc64com. Institute of Nephrology, Zhongda Hospital, School of Medicine, Southeast University. No.87 Ding JiaQiao Road, Nanjing,China 210009. Phone number: +86 2583272512.
Corresponding author: Lin-Li Lv, M.D. Ph.D. Email: lvlinliedu.cn. Or Bi-Cheng Liu, M.D. Ph.D. Email: liubc64com. Institute of Nephrology, Zhongda Hospital, School of Medicine, Southeast University. No.87 Ding JiaQiao Road, Nanjing,China 210009. Phone number: +86 2583272512.

 Global reach, higher impact
Global reach, higher impact