10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(4):426-437. doi:10.7150/ijbs.8754 This issue Cite
Research Paper
The Blunt Trichoid Sensillum of Female Mosquitoes, Anopheles gambiae: Odorant Binding Protein and Receptor Types
University of Hohenheim, Institute of Physiology, Stuttgart, Germany.
Received 2014-2-4; Accepted 2014-3-1; Published 2014-3-22
Abstract
In order to find a blood host and to select appropriate oviposition sites female Anopheles gambiae mosquitoes rely on olfactory cues which are sensed by olfactory sensory neurons (OSNs) located within morphologically different sensilla hairs. While the sharp type trichoid sensilla are most abundant and intensely studied, the striking blunt type trichoid sensilla exist only in small numbers and their specific function is unknown. It has been suggested that they may play a role in the detection of chemical cues indicating oviposition sites. With the aim of identifying molecular elements in blunt type trichoid sensilla, which may be relevant for chemosensory function of this sensillum type, experiments were performed which include whole mount fluorescence in situ hybridization (WM-FISH) and fluorescence immunohistochemistry (WM-FIHC). The studies were concentrated on odorant binding proteins (AgOBPs) and odorant receptors (AgORs). WM-FISH approaches using a probe for the plus-C class AgOBP47 led to the labeling of cells, which resembled in number and antennal distribution pattern the blunt type trichoid sensilla. Moreover, WM-FIHC with an antiserum for AgOBP47 allowed to assign the AgOBP47-expressing cells to blunt type trichoid sensilla and to allocate the protein within the sensillum hair shafts. The result of double WM-FISH-experiments and combined WM-FIHC/FISH approaches indicated that the AgOBP47-expressing cells are co-localized with cells, which express AgOR11, AgOR13 and AgOR55. In addition, it turned out that the two receptor types AgOR13 and AgOR55 are co-expressed in the same cells. Together, the results indicate that the blunt type trichoid sensilla contain a characteristic binding protein, plus-C AgOBP47, in the sensillum lymph and two sensory neurons, one cell which express the odorant receptor AgOR11 and a second cell which express the receptor types AgOR13 and AgOR55. The expression of characteristic chemosensory elements in blunt type trichoid sensilla supports the notion that this sensillum type is involved in sensing distinct odorous compounds.
Keywords: olfaction, odorant receptor proteins, odorant binding proteins, mRNA expression.
Introduction
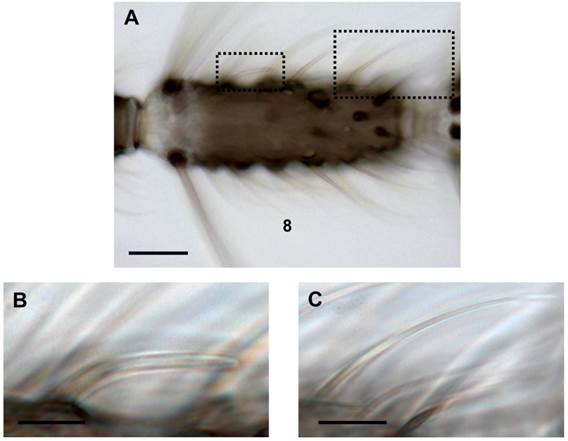
Females of the malaria mosquito Anopheles gambiae, the major vector of the life-threatening parasite Plasmodium falciparum, strongly rely on their sense of smell to find appropriate human blood hosts, nectar sources and oviposition sites [1-3]. The detection and discrimination of behavioral relevant odorants in the environment of females is mediated by olfactory sensory neurons (OSNs) housed in hair-like sensilla on different appendices of the body [4-6]. The major olfactory appendices, the two antennae, each carry about 740 sensilla hairs while lower numbers of the chemosensory units are present on the maxillary palps and the proboscis [5, 7, 8]. Among the three morphologically different olfactory hair types on the female antenna, sensilla trichodea (~630) are most numerous, while only ~80 sensilla basiconica (grooved pegs) and few peg-in-pit sensilla coeloconica (~30) have been counted [9]. Based on their external morphology sensilla trichodea are further divided into three subtypes [7, 9]: The most frequent sharp-tipped trichoid hairs are found on all 13 antennal segments and significantly increase in number towards more distal flagellomeres. A second subtype is the short-sharp trichoid sensillum, which is less numerous and mainly differs by the length of its hair shaft. Finally, the most striking sensillum trichodeum subtype is blunt-tipped, rather short and has a nearly consistent diameter from the base to the tip. Moreover, the sensillum shaft often parallels the flagellum surface (Fig.1). It is found in very low numbers almost exclusively on each of the segments 2-9 [6, 9].
Single sensillum recordings from A. gambiae antenna have shown that the sharp sensilla trichodea respond to distinct but partly overlapping spectra of volatile compounds [6] whereas odorants which activate blunt type trichoid sensilla are still unknown. However, it has been reported that short blunt trichoid sensilla (SBT II) of the culicine mosquito species, Culex quinquefasciatus and Aedes aegypti [10, 11] respond to a panel of behavioral relevant odorants [12-14]. Remarkably, for Aedes aegypti it was demonstrated that the sensitivity to oviposition cues (primarily indole and phenolic compounds) of SBT II sensilla increased both at 24h and 72h after a blood meal, while the sensitivity of other trichoid sensilla types remained unchanged [15]. Based on this findings it has been speculated that blunt type trichoid sensilla may be particular important for the detection of oviposition sites by female mosquitoes.
Several studies have indicated that odorant detection in insects requires distinct combinations of small globular odorant binding proteins (OBPs) in the sensillum lymph [16-20] as well as of olfactory receptor proteins (ORs), localized in the dendritic membrane of OSNs [21-25]. In the genome of A. gambiae 76 genes encoding ORs (AgORs) [26, 27] and ~60 genes encoding OBPs (AgOBPs) have been annotated [28-30]. It was subsequently shown that at least 52 AgORs and 21 AgOBPs are expressed in female antenna [31]. The response spectra for 50 different AgORs was assessed and found to range from broad to rather narrow [32, 33]. Similarly, for the AgOBPs different but partially overlapping binding spectra were found for five classical AgOBPs and the plus-C AgOBP47 [34]. Plus-C class AgOBPs differ from classical OBPs by (i) their overall length, (ii) at least 12 compared to 6 cysteins (forming three additional disulphide bridges) and (iii) by the proposed structure of their ligand binding pocket [35-38]. The majority of AgOBPs expressed in female antenna are classical AgOBPs while only four plus-C class AgOBPs have been found. However, the plus-C class binding proteins AgOBP47 and AgOBP48 show very high transcript levels and are among the most abundantly transcribed AgOBPs [29, 31].
Different trichoid sensilla types on the female antenna. (A) Brightfield image of an 8th antennal segment carrying various sensilla hairs. (B) Higher magnification of the area boxed in A (left) showing a blunt type trichoid sensillum. (C) Long sharp trichoid sensillum of the same segment (right box) shown at higher magnification. Scale bars: 20 µm in A, 5 µm in B, 10 µm in C.
Recently we have determined the number and the antennal topography of cells expressing AgORs [39] and cells expressing AgOBPs, including AgOBP48 [40, 41]. For the various AgOBP types as well as AgOR types considerable differences in the number of cells were found, which indicates expression in distinct subpopulations of sensilla. Furthermore, a combinatorial expression pattern of distinct AgOBPs and AgORs in long sharp trichoid sensilla was detected [41], suggesting an interplay of the two functional proteins in this sensillum type. In order to explore the molecular basis for odorant detection in blunt type trichoid sensilla of female malaria mosquitoes, in this study we set out to identify AgOBP and AgOR subtypes that are expressed in the sensillum type.
Materials and Methods
Animal rearing
Larvae of the Anopheles gambiae s.s. strain “RSPH” (MR4-number: MRA-334) were kindly provided by Bayer CropScience AG, Monheim, Germany. This laboratory strain was originally derived from the region of Kisumu in Kenya. The animals were reared to adults in a 28°C incubator with a day/night cycle of 12:12 hours. After emergence, the adult mosquitoes had access to 10% sucrose ad libitum. For the experiments antennae were used from females 2 to 4 days after emergence.
Preparation of labeled probes for fluorescence in situ hybridization
For in situ hybridization Digoxigenin (DIG)-labeled and biotin-labeled antisense RNA probes were made from linearized pGem-T Easy and pBluescript plasmids containing coding regions of AgOBP and AgOR genes (Supplementary Material: Table S1). The RNA sequences were generated by performing in vitro transcription using the SP6/T3/T7 RNA transcription system (Roche) and recommended protocols. To improve hybridization signals the riboprobes were fragmented to an average length of about 800 bp for AgORs and 200 bp for some of the AgOBPs by incubation with carbonate buffer (80 mM NaHCO3, 120 mM Na2CO3, pH 10.2) following the protocol of [42].
Whole mount fluorescence in situ hybridization (WM-FISH)
Whole mount fluorescence in situ hybridization using complete female antennae and either a single DIG labeled probe or two differently labeled (DIG or biotin) antisense RNA probes (double WM-FISH) were performed as described previously [39-41] with few modifications. Unless specified otherwise, all incubation-steps and washes were done in thin walled PCR tubes (0.25 ml; Kisker, Germany) with slow rotation or moderate shaking of tubes at room temperature. Antennae were dissected from cold anesthetized animals under control of a stereo microscope and directly transferred to fixation solution (4% paraformaldehyde in 0.1 M NaCO3, pH 9.5, 0.03% Triton X-100). After incubation for 20 - 24 hours at 6°C, antennae were washed for 1 min in PBS (phosphate-buffered saline = 0.85% NaCl, 1.4 mM KH2PO4, 8 mM Na2HPO4, pH 7.1), 0.03% Triton X-100, incubated for 10 min in 0.2 M HCl, 0.03% Triton X-100 and transferred to PBS, 1% Triton X-100. In this solution antennae were carefully squeezed few times with a fine forceps to improve the penetration of solutions into tissue. Subsequently, antennae were prehybridized with in situ hybridization solution (50% formamide, 5x SSC, 1x Denhardt's reagent, 50 µg/ml yeast RNA, 1% Tween 20, 0.1% Chaps, 5 mM EDTA, pH 8.0) for 6-72 hours at 55°C. Then the solution was displaced by hybridization solution containing the respective labeled antisense RNA probes (1:100) and incubated at 55°C for at least 48 hours.
Antennae were then washed 4 x 15 min each in 0.1x SSC, 0.03% Triton X-100 at 60°C and incubated in 1% blocking reagent (Roche) in TBS (100 mM Tris, pH 7.5, 150 mM NaCl), 0.03% Triton X-100 for 5 hours at 6°C. In case of single in situ hybridization DIG-labeled antisense RNA probes were detected by using an anti-DIG AP-conjugated antibody (Roche) diluted 1:500 in TBS, 0.03% Triton X-100, 1% blocking reagent. In case of double in situ hybridizations biotin-labeled probes were simultaneously detected by adding a streptavidin horse radish peroxidase-conjugate (1:100, TSA kit, Perkin Elmer). After incubation at 6°C for at least 48 hours, antennae were washed five times in TBS with 0.05% Tween 20 for 10 min each. For visualization of cells labeled by DIG-probes, tissues were incubated first with HNPP/Fast Red TR (Roche) diluted 1:100 in DAP-buffer (100 mM Tris, pH 8.0, 100 mM NaCl, 50 mM MgCl2) for 5 - 6 hours at 6°C and washed 3 x 5 min in TBS with 0.05% Tween 20. In case of double in situ hybridizations employing a biotin-labeled riboprobe, after HNPP development, the antennae were treated with the components of the TSA Fluorescein System (Perkin Elmer) for 17 - 18 hours at 6°C, following the protocol of the supplier. Finally, antennae were washed 3 x 5 min each in TBS with 0.05% Tween 20 and briefly rinsed in PBS, before they were mounted in mowiol (10% polyvinylalcohol 4-88, 20% glycerol in PBS).
Two color WM-FISH experiments with antennae and differently labeled probes specific for two AgORs were performed as described above with following modifications: After fixation in 4% paraformaldehyde in 0.1 M NaCO3, pH 9.5, 0.03% Triton X-100 for 24 h at 6°C, antennae were washed for 1 min in PBS, 0.03% Triton X-100 and incubated for 10 min in 0.2 M HCl, 0.03% Triton X-100 both at room temperature. Subsequently the antennae were washed two times in PBS for 2 min, carefully squeezed few times with a fine forceps and incubated for 10 min in acetylation solution (25% acetic anhydride freshly added to a 0.1 M triethanolamine solution) at room temperature. Then antennae were washed 3 x 3 min each in PBS, 0.03% Triton X-100 and prehybridized in hybridization solution for 4-72 hours at 6°C.
Whole mount fluorescence immunohistochemistry (WM-FIHC)
For immunolocalization of the AgOBP47 protein in female antenna we used an antiserum against AgOBP47 [34] and adapted a whole mount fluorescence immunohistochemistry protocol previously used with antisera against AgOBP1 and AgOBP4 [41]. Briefly, antennae were dissected from cold anesthetized animals and directly transferred to ZnFA solution (0.25% ZnCl2, 1% formaldehyde, 135 mM NaCl, 1.2% sucrose, 0.03% Triton X-100). After fixation over night at room temperature, antennae were washed 2 x 15 min with HBS buffer (150 mM, NaCl, 5 mM KCl, 25 mM sucrose, 10 mM Hepes, 5 mM CaCl2, 0.03% Triton X-100) and carefully squeezed few times with a fine forceps in HBS buffer under control of a stereo microscope followed by incubation for 1 hour in 80% Methanol/20% DMSO. Antennae were washed for 5 min in 0.1 M Tris pH 7.4, 0.03% Triton X-100 and incubated in blocking solution (PBS, 5% NGS (Dianova, Hamburg, Germany), 1% DMSO, 0.03% Triton X-100) for at least 3 hours. The solution was replaced by blocking solution containing the primary antibody (dilution 1:200) and antennae were subsequently placed in a water bath sonifier (Bansonic 1200, Branson, Danbury, CT) for 30 sec followed by the incubation for 4 days at 6°C. Then antennae were washed 3 x 15 min in PBS, 1% DMSO, 0.03% Triton X-100 and incubated in blocking solution containing an anti-guinea pig Alexa488 coupled secondary antibody (Invitrogen, Karlsruhe, Germany, Dilution 1:500). After 30 sec sonification-step, antennae were incubated for 3 days at 6°C and washed 3 x 15 min each in PBS with 1% DMSO, 0.03% Triton X-100. Finally antennae were shortly rinsed in PBS and mounted in mowiol solution.
Combined whole mount fluorescence in situ hybridization and fluorescence immunohistochemistry
Combined WM-FISH and WM-FIHC was performed as described previously [41]. Briefly, female antennae were dissected and incubated for 24 h in ZnFA fixation solution at room temperature. After fixation, antennae were washed 2 x 15 min in HBS-buffer and squeezed few times with a fine forceps in this solution. Subsequently, antennae were washed for 1 min with PBS at room temperature, transferred to hybridization solution and stored up to 72 h at 6°C. Afterwards the tubes were incubated for 5-6 h at 55°C and then the solution was replaced by hybridization solution containing the specific DIG-labeled antisense RNA probe (1:100). Antennae were subsequently incubated for 72 h at 55°C, washed 4 x 15 min in 0.1 x SSC, 0.03% Triton X-100 at 60°C and incubated 5-6 h in blocking solution (TBS, 5% NGS, 0.03% Triton X-100). Before antennae were incubated 72-96 h in blocking solution containing the primary antibody (1:100 for OR55/OBP47 and 1:200 for the OR11/OBP47 and OBP13/OBP47) and the anti-DIG AP-conjugated antibody (Roche) (1:500) at 6°C, tubes were placed in a waterbath sonifier (Bansonic 1200, Branson, Danbury, CT) for 30 sec. Antennae were washed 3 x 15 min with TBS, 0.05% Tween 20, before blocking solution containing a goat anti-rabbit Alexa488 coupled secondary antibody (Invitrogen, Karlsruhe, Germany, Dilution 1:500) was added. Before antennae were incubated for 72-96 h in the dark at 6°C, tubes were placed again in the waterbath sonifier and treated for 30 sec. Subsequently, antennae were washed 3 x 15 min with TBS, 0.05% Tween 20 and shortly rinsed in DAP-buffer, pH 8.0, followed by the incubation in HNPP/Fast Red TR (Roche) diluted 1:100 in DAP-buffer for 5-6 h at 6°C in the dark. Finally, antennae were washed 3 x 5 min in TBS, 0.05% Tween 20, briefly rinsed in PBS and mounted in mowiol solution.
Analysis of antennae by microscopy
Antennae from WM-FISH and WM-FIHC experiments were analyzed with a Zeiss LSM510 Meta laser scanning microscope (Zeiss, Oberkochen, Germany). Confocal image stacks of the red and green fluorescence channel as well as the transmitted-light channel were taken from single antennal segments. Pictures represent projections of selected optical planes, with the fluorescence channels and the transmitted-light channel overlaid or shown separately. Longer antennal stretches were arranged from single pictures representing projections of selected planes of confocal image stacks.
Brightfield images illustrating different sensilla types were taken using an Axioskop 2 Mot (Zeiss, Jena, Germany) equipped with an AxioCam MRc5 and the AxioVision LE 4.3 software.
Results
Expression of AgOBPs in blunt type trichoid sensilla
Towards a molecular phenotyping of the blunt type trichoid sensillum on the antenna of female Anopheles gambiae (Figure 1) we first set out to screen for AgOBPs which show an expression pattern similar to the topographic distribution of this sensillum type. Based on the assumption that blunt type trichoid sensilla may play an important role in the detection of odorants which trigger female specific behaviors, AgOBPs were assessed, for which abundant and predominant expression levels were found in female antennae compared to males by RT-PCR experiments, microarray studies and whole transcriptome RNA sequencing [29, 31, 43]. Employing whole mount approaches with labeled antisense RNA-probes for the classical odorant binding proteins AgOBP1, 3, 4, 5, 7, 19 and 20 as well as for the plus-C class AgOBP48 we obtained hybridization signals (not shown) indicating expression of OBPs in distinct and divergent populations of supporting cells, thus confirming previous results [39-41]. However, for none of these AgOBPs we found a distribution pattern of the hybridization signals that resembles the spatial arrangement of blunt type trichoid sensilla along the female antenna [9].
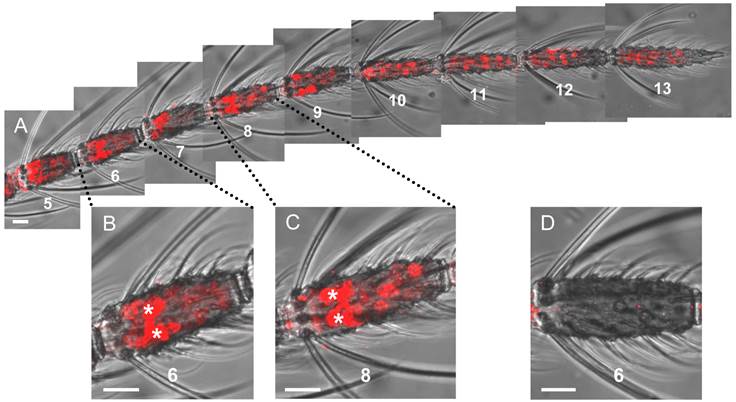
Using a riboprobe for the plus-C class AgOBP47 resulted in a picture different from the labeling pattern obtained for the other AgOBPs tested (Figure 2). We identified two populations of labeled cells, which can be clearly distinguished by the intensity and size of the WM-FISH signals as well as by their distribution along the antenna (Figure 2A). One large population of AgOBP47-positive cells is characterized by only weak labeling and was found on segments (flagellomeres) 1 - 13 with increasing number of positive cells towards the distal part of the antenna; only in rare cases weakly labeled cells were seen on segment number 1. Interestingly, a second rather small population of AgOBP47-positive cells showed very intense and extraordinary large WM-FISH signals (Figure 2B and Figure 2C). A few of these strong signals were regularly found on each of the segments 2 - 9 thus resembling the number and distribution of blunt type trichoid sensilla on the female antenna [9]. No labeling was obtained in control experiments using a sense riboprobe, confirming the specificity of the weak and strong WM-FISH signals obtained with the antisense probe (Figure 2D).
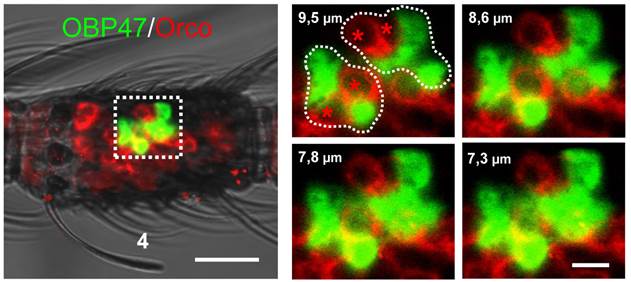
Within insect sensilla the OBPs are generally expressed and secreted by supporting cells [44-47]. However, due to the large size of the WM-FISH signals it could not be excluded that AgOBP47 may also be expressed in olfactory sensory neurons (ORNs). To scrutinize this notion, we performed two-color WM-FISH with the insect olfactory co-receptor (AgOrco). These double staining experiments revealed that red labeled ORNs and green labeled AgOBP47-expressing cells are located in close proximity (Figure 3), however, no double-labeling of cells was found. This result is in accordance with a selective expression of AgOBP47 only in supporting cells.
Gene expression of the plus-C class AgOBP47 in the antenna of a female mosquito. Cells having the transcript for AgOBP47 were detected and subsequently visualized using a specific DIG-labeled antisense RNA probe and detection systems leading to red fluorescence. (A) Antennal stretch (segments 5 to 13) with two populations of labeled cells which differ in WM-FISH signal intensities. Segments are numbered from proximal (1) to distal (13). (B and C) Higher magnification of segments 6 and 8 showing very strong (asterisks) and weak labeled OBP47-expressing cells. (D) Segment 6 of a female antenna hybridized with a DIG-labeled sense RNA probe for AgOBP47. Scale bars: 20 µm.
Two-color WM-FISH visualizing AgOrco-expression in neurons (red) and AgOBP47-expressing supporting cells (green) in a proximal segment of a female antenna. The left image represents an overlay of optical planes from a confocal image stack. The right image represents single optical planes showing the region boxed in the left image. The optical depth of each plane is indicated by the numbers in µm. AgOrco-positive cells (asterisks) are localized in direct proximity to AgOBP47-positive cells but do not co-express the OBP. Scale bars: 20 µm left image; 5 µm right image.
Immunolocalization of AgOBP47
The marked similarities between the distribution pattern of the AgOBP47 labeled cells and the antennal topography of blunt type trichoid sensilla suggested that AgOBP47 protein is expressed in this trichoid sensillum type.
To support this hypothesis we performed whole mount immunohistochemical (WM-FIHC) experiments applying an antiserum generated against AgOBP47 [34]. Immunoreactivity in the antenna was visualized using an Alexa488-coupled secondary antibody and confocal laser scanning microscopy. We found intensive green staining under individual sensilla hairs which can be morphologically classified as blunt type trichoid sensilla (Figure 4). At higher magnification green labeled cells could be assigned to individual sensilla hair shafts with additional staining within the hair structures. Morphological analysis have identified about 15 blunt type trichoid sensilla on a complete antenna of female Anopheles gambiae with a rare appearance of sensilla on the four most distal segments [9]. While the WM-FIHC experiments did not allow for a reliable determination of the number of blunt type trichoid sensilla expressing AgOBP47, in our WM-FISH analysis we regularly found 1-3 strong signals on segment 2-9, with rare cases of single strong labeling also on segment 10. Counting all strong signals on six complete antennae revealed an average of 14.3 (+/- 1.8) FISH-signals per antenna.
Co-expression of AgOBPs and AgORs in blunt type trichoid sensilla
The expression patterns which were obtained for other AgOBPs did not exclude a co-expression with AgOBP47 in blunt type trichoid sensilla. To elucidate this aspect we conducted double WM-FISH experiments with all combinations of the various AgOBPs and AgOBP47. We did not find any indication for a co-expression of AgOBP47 with another AgOBP as indicated by the clearly separated red and green signals (Figure 5). In addition, we did not find AgOBP47-positive cells in close vicinity to supporting cells expressing one of the other AgOBPs, indicating that the blunt trichoid sensillum type houses neither any of the classical AgOBPs tested nor AgOBP48.
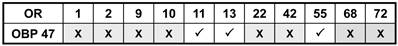
In order to further illuminate the molecular phenotype of blunt type trichoid sensilla, we next explored which odorant receptor genes are expressed in ORNs of this sensillum type. Toward this goal, 11 AgORs were chosen which were found to be highly expressed in female antenna [31, 48], including seven with a known ligand spectrum [32, 33]. Double WM-FISH was performed using AgOR-specific RNA-probes in combination with the probe for AgOBP47 (as marker for blunt type trichoid sensilla). The results are shown in Figure 6. As exemplarily shown for the combinations AgOR1/AgOBP47 and AgOR10/AgOBP47 in Figure 6A there was a clear separation of AgOR (red) and AgOBP47 (green) signals within a segment, indicating that cells are located in different sensilla (Table 1). However, for the combinations AgOR11/AgOBP47, AgOR13/AgOBP47 and AgOR55/AgOBP47, it was found that AgOR- (red) and AgOBP47- (green) expressing cells are in very close vicinity, indicating different cells which are located within the same sensillum (Figure 6B). Interestingly, for all three combinations we also observed AgOR-expressing cells which were not associated with the AgOBP47-cells, indicating that AgOR11, AgOR13 and AgOR55 are not only expressed in ORNs of blunt type trichoid sensilla. To further verify AgOBP47/AgOR co-localizations to the same blunt trichoid sensillum, we performed combined WM-FISH/WM-FIHC-experiments (Figure 7). This allowed visualization of single AgOR11-, AgOR13- or AgOR55-expressing cells in close association with AgOBP47-expressing cells. Furthermore, immunolocalization of AgOBP47-protein in the hair shafts made it also possible to clearly assign the co-localized cells to a distinct blunt type trichoid sensillum (Figure 7).
Combinations of AgORs and AgOBP47 tested for co-localization in double WM-FISH. X = No co-localization found, √ = Sensilla co-localization.
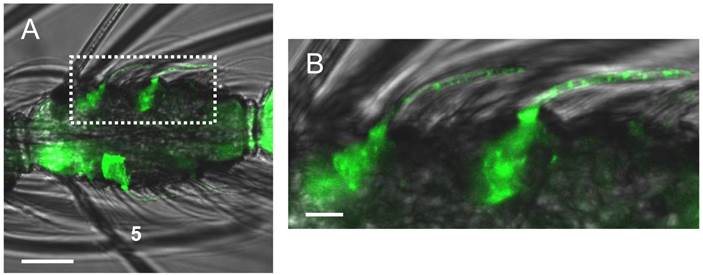
Whole mount immunolocalization of AgOBP47 protein using an OBP-specific antiserum and an Alexa488-coupled secondary antibody. (A) WM-FIHC with green labeled cells positive for anti-AgOBP47 in a 5th segment of a female antenna. (B) Higher magnification of the area boxed in A. Immunostaining is visible in supporting cells below single sensilla as well as within these hair structures that can be identified as blunt type trichoid sensilla. Scale bars: 20 µm in A; 5 µm in B.
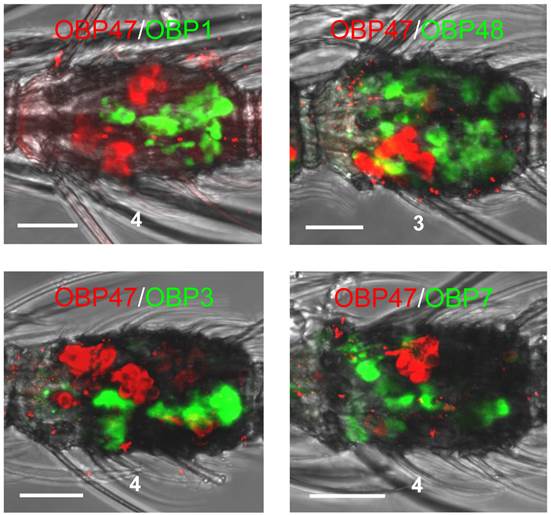
Two-color WM-FISH on proximal segments of female antennae using a biotin-labeled antisense RNA-probe specific for the AgOBPs indicated in combination with a DIG-labeled riboprobe for AgOBP47. Cells carrying OBP-transcripts are visualized by red (DIG) and green (biotin) fluorescence, respectively. No co-labeled cells are visible, indicating expression of the four AgOBPs and AgOBP47 in different cell populations. Scale bars: 20 µm.
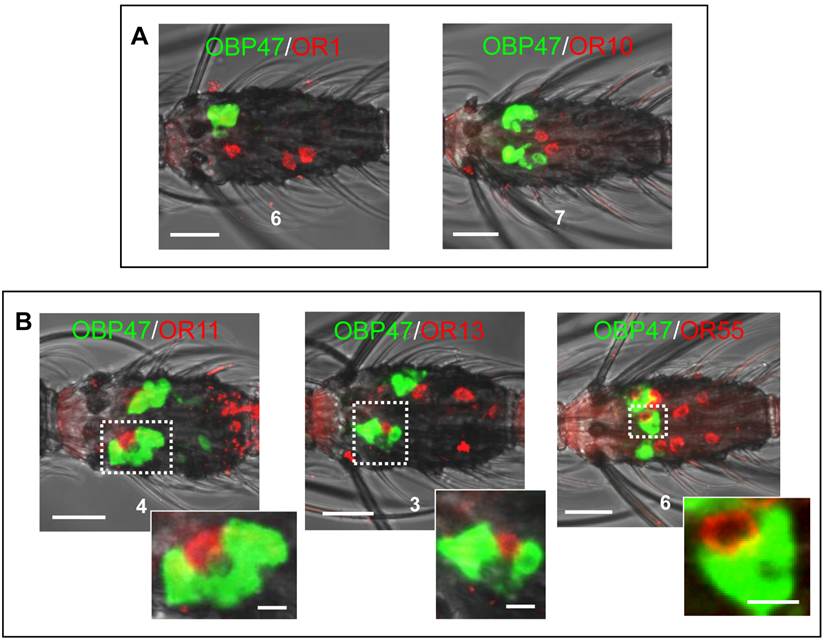
Relative localization of cells bearing transcripts for different Anopheles gambiae olfactory receptors and AgOBP47 in proximal segments of female antennae. WM-FISH with DIG (ORs) and biotin (AgOBP47) labeled riboprobes; the tested combinations are indicated. (A) No co-localization of AgOBP47- and AgOR1- or AgOR10-expressing cells indicated by the clear spatial separation of red and green WM-FISH signals. (B) WM-FISH using combinations of AgOBP47 with AgOR11, AgOR13 and AgOR55, respectively giving evidences for co-localization in the same sensillum. As depicted by the higher magnification of the boxed areas AgOR-cells (red) are located in close proximity to the AgOBP47-cells (green). In addition, AgOR-positive cells which are not associated with green AgOBP47 signals are found. Scale bars: 20 µm in A, B; 5 µm in enlarged boxes.
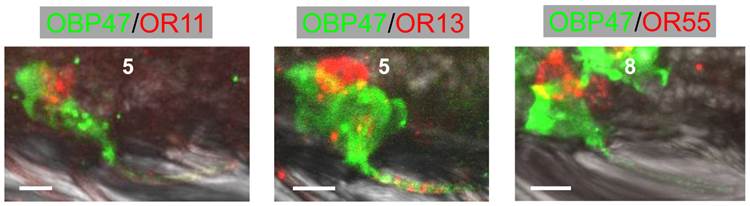
Combined WM-FIHC using the antiserum against AgOBP47 and WM-FISH employing antisense RNA probes specific for AgOR11, AgOR13 and AgOR55, respectively. Images represent single optical planes of hybridized antennae showing AgOR-expressing cells in direct association with AgOBP47-immunoreactivity for the three different combinations. Immunostaining is visible also within the sensilla hairshafts and allows to assign the expressing cells to single trichoid sensilla. Scale bars: 5 µm
Two-color WM-FISH using three different combinations of DIG- or biotin-labeled AgOR11-, AgOR13- and AgOR55-antisense RNA probes. (A and B, left) Localization of the AgOR11/AgOR13 and AgOR55/AgOR11 gene expression in the 7th and the 6th segment of two single antennae. Clearly separated red- and green-labeled cells (white arrow heads) are found. In addition, positive cells are visible for the AgOR-pairs in direct proximity (boxed) indicating sensilla co-localization. Pictures on the right side show adjacent red and green cells in enlarged size with the red and green channel separated and overlaid. (C) Localization of the transcripts for AgOR13 and AgOR55. Cells are co-labeled in all cases, visible as yellow color in the overlaid channels. Co-expression of AgOR13 and AgOR55 is also indicated by the identical morphology of the cell visualized in red and green fluorescence separately on the right side. Scale bars: 20 µm left side; 5 µm right side.
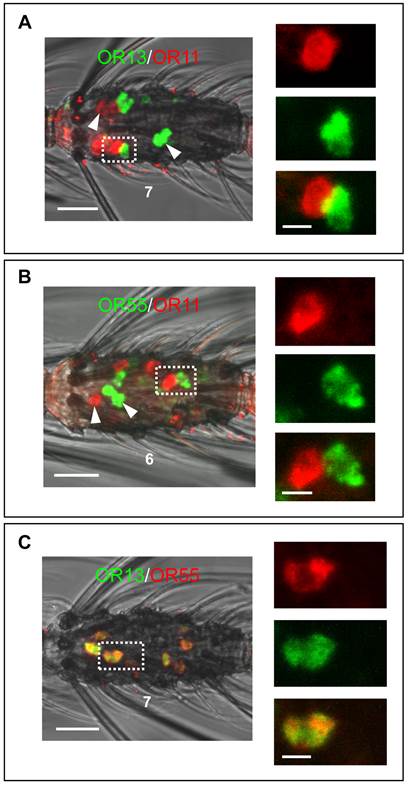
Generally, trichoid sensilla of A. gambiae contain two ORNs (Qiu et al., 2006) and single sensilla recordings from short blunt trichoid sensilla (SBT II) of the mosquito species Aedes aegypti and Culex quinquefasciatus have demonstrated the presence of two ORNs in this sensillum type [10, 11]. The finding that three different AgORs are expressed in cells which are co-localized with AgOBP47-cells implies that blunt type trichoid sensilla may house three (ORNs). Alternatively, ORNs in blunt type sensilla of A. gambiae may express more than one OR. In order to test these possibilities we have employed combinations of antisense RNA-probes for AgOR11, AgOR13 and AgOR55 in double WM-FISH. The combination AgOR11/AgOR13 (Figure 8A) resulted in clearly distinguishable red and green labeled AgOR11- and AgOR13-cells, respectively. Similar results were obtained for the combination AgOR11/AgOR55 (Figure 8B), indicating the expression of AgOR11 in a different population of cells than AgOR13 and AgOR55. For both combinations we found also cases of an AgOR11-cell paired with individual AgOR13- and AgOR55-cells, respectively (Figure 8A and B); these results are in line with a co-localization of the ORNs in the same blunt type trichoid sensillum. In addition, cells located spatially separated from each other indicate that AgOR11/AgOR13- and AgOR11/AgOR55- expressing cells are not strictly paired in a sensillum. In contrast, the WM-FISH with the combination AgOR13/AgOR55, showed cells which are positive for both receptors (Figure 8C) indicating expression of the two AgORs in the same cells. The complete overlap of AgOR13- and AgOR55 labeling indicates that Anopheles blunt type trichoid sensilla house two ORNs, one which expresses both AgOR13 and AgOR55 and the second which expresses AgOR11.
Discussion
In this study, attempts were made to elucidate the molecular phenotype of the relatively scarce blunt type trichoid sensilla on the antennae of female Anopheles gambiae mosquitoes in view of their potential role in odor detection. The WM-FISH experiments with OBP-specific probes revealed that the binding protein plus-C AgOBP47 is expressed in this sensillum type. No evidence was found for an expression of plus-C AgOBP48 or any classical odorant binding protein in blunt type trichoid sensilla. However, due to the limited number of OBP-subtypes analyzed in the study which were selected based on the abundance of their transcripts in the antenna [28, 29, 31, 49], it cannot be ruled out that additional AgOBP types are expressed in the blunt type trichoid sensilla. Based on the very strong hybridization signals under blunt trichoid sensilla it is assumed that AgOBP47 is highly expressed in this sensillum type. In recent studies the number of blunt type trichoid sensilla on the female antenna was determined as 14.6 (+/-1.8) [9]; interestingly, the total number of strong AgOBP47 signals on the antenna (14.3 (+/- 1.8)) closely matches this number. It is therefore conceivable that all blunt type trichoid sensilla of female Anopheles antenna abundantly express the binding protein AgOBP47. In addition, the AgOBP47 probe visualized weakly labeled cells, mainly in distal parts of the antenna (segments 9 - 13). Since the very strong and the weak AgOBP47-signals were obtained under the same experimental conditions, the finding suggests that two populations of supporting cells exist, which significantly differ in the level of AgOBP47 transcripts and may indicate that high levels of AgOBP47 protein exist in blunt type trichoid sensilla. In accordance with this notion, experiments with the anti-OBP47 antibody resulted in labeled cells and hair shafts only in the blunt type trichoid sensilla.
Any functional implication of the plus-C class OBPs, including AgOBP47, in insect olfaction are unknown. However, the characteristic expression pattern of AgOBP47 (this study) as well as for AgOBP48 [40] suggests a decisive role of these proteins. High transcript levels for plus-C OBPs have been found in the antennae and maxillary palps of various mosquitoes [29, 31]. The notion that plus-C OBPs are involved in odorant detection is supported by the results of 1-NPN-based competitive fluorescence binding assays; it was found that several alcoholic and aromatic compounds bind to AgOBP47 [34]. Moreover, the 3D structures and ligand binding modelling of AgOBP47 [37] and AgOBP48 [38] revealed the existence of putative binding pockets for odorants in the plus-C OBPs; interestingly these binding sites differ from the binding pocket of classical OBPs.
The in situ hybridization experiments in combination with immunohistochemical studies have shown that strongly labeled AgOBP47-cells are closely associated with cells expressing the receptor types AgOR11, AgOR13 or AgOR55 (Figures 6 and 7). Furthermore, it was found that the receptor types AgOR13 and AgOR55 are expressed in the same cells. Together, the data indicate that blunt trichoid sensilla comprise molecular elements which render this sensillum type capable for responding to odorants: the binding protein AgOBP47 in the sensillum lymph and two types of sensory neurons, one expressing the receptor type AgOR11 and a second neuron expressing the two receptor types AgOR13 and AgOR55. Although it was originally considered as a general rule that insect olfactory neurons express only one odorant detecting OR-type in addition to Orco [21, 23-25], meanwhile it has been reported for a number of Drosophila OR genes to be co-expressed in antennal neurons [50-52]. It has turned out that such co-expressed OR genes are often conserved and closely linked in the genome, for example OR22a and OR22b [53]. Interestingly, although AgOR13 and AgOR55 share only rather low sequence identity (43%) the encoding genes are located less than 500 bp apart in the genome. Thus, Anopheles OR genes in close genomic vicinity may likely be under control of the same regulatory elements leading to co-expression.
The results of double WM-FISH experiments indicate that there is no strict pairing of the AgOR11-cells with the AgOR13/AgOR55-expressing ORNs in the same sensillum. Moreover, AgOR11-cells as well as AgOR13/AgOR55-cells were also found in olfactory hairs other than blunt type trichoid sensilla, most likely of the sharp trichoid type paired with different receptor-specific neurons. These data indicate that in different sensilla types neurons which express these three receptor types are combined with neurons which express receptor of different identity. Previously, it was generally accepted that in sensilla on the antennae or the maxillary palps, olfactory neurons with ORs of defined identity are strictly paired and housed together in a morphological distinct sensillum type [5, 23, 50, 54]. Our results represent a deviation from this rule, however, they are in line with findings in the fruitfly, where neurons expressing the receptor OR33b are located in large (ab2) and thin (ab5) type basiconic sensilla and are combined with cells expressing OR59b and OR82a, respectively [51, 54]. It is worth mentioning, that in Drosophila neurons which express OR33b also express a second OR type, however, this is different in neurons of the ab2 and ab5 sensilla [51]. The functional implications for the expression of the same receptor type in cells of morphologically different sensilla types and their co-existence with neurons which express ORs of different identity is unknown. Such a combinatorial arrangement of receptor specific ORNs may serve to extend the detection and coding capacity of the olfactory system. In this regard it has to be taken into consideration that cells with the same ORs in different sensilla types may be involved in the detection of a different odorant spectrum. This could be based on sensillum-specific OBP types, which are supposed to transfer different subsets of odorants to the chemosensory cells. In fact, recent studies in Drosophila [18] as well as in Anopheles gambiae [41] have shown that specific subsets of OBP-types are present in distinct populations of trichoid sensilla and contribute to their specific responsive spectrum [18].
Whether and which odorants are detected by the blunt type trichoid sensilla of female Anopheles is unknown, however, it has been reported that in Culex quinquefasciatus and Aedes aegypti short blunt trichoid sensilla are responsive to a panel of behavioral relevant odorants [10, 11, 13]. In addition, in A. aegypti the sensitivity to oviposition cues (indole and phenolic compounds) was increased after a blood meal in short blunt trichoid sensilla, while the sensitivity of other trichoid sensilla types remained unchanged [15]. In this respect it is interesting to note that functional expression experiments in Xenopus oocytes revealed that indole and phenolic compounds (2-methylphenol, 3-methylindole and indole) strongly activate AgOR11 and AgOR13 receptors [33]. In addition, competitive binding assays have shown that indole interacts with binding protein AgOBP47, characteristic for blunt type trichoid sensilla [34]. In order to investigate, if AgOR13 and AgOR55 recognize a similar panel of odorants, in future studies it would be interesting to test the ligands for AgOR55. Together, the emerging picture suggests that the receptor types AgOR11 and AgOR13 as well as the binding protein AgOBP47 may be involved in the detection of odorants emitted from oviposition sites and that the neurons of blunt type trichoid sensilla of A. gambiae have the molecular elements for sensing the olfactory cues of appropriate breeding sites.
Supplementary Material
Table S1.
Acknowledgements
We gratefully acknowledge Prof. P. Pelosi, University of Pisa, Italy, for providing us the AgOBP47 antibody and the AgOBP19 cDNA clone. We thank and acknowledge Prof. K. Iatrou from National Centre for Scientific Research ”Demokritos”, Athens, Greece, for providing us with cDNA clones for A. gambiae OBPs, OR1 and OR2. We are grateful to Dr. Günther Nentwig, Dr. Maike Hink and Jörg Egger from Bayer CropScience for collaboration and providing Anopheles gambiae eggs and larvae.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Takken W, Knols BG. Odor-mediated behavior of Afrotropical malaria mosquitoes. Annu Rev Entomol. 1999;44:131-157
2. Zwiebel LJ, Takken W. Olfactory regulation of mosquito-host interactions. Insect Biochem Mol Biol. 2004;34:645-652
3. Himeidan YE, Temu EA, El rayah EA. et al. Chemical cues for malaria vectors oviposition site selection: Challenges and opportunities. Journal of Insects. 2013 Article ID685182: 1-9
4. Meijerink J, Braks MA, van Loon JJ. Olfactory receptors on the antennae of the malaria mosquito Anopheles gambiae are sensitive to ammonia and other sweat-borne components. J Insect Physiol. 2001;47:455-464
5. Lu T, Qiu YT, Wang G. et al. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr Biol. 2007;17:1533-1544
6. Qiu YT, van Loon JJ, Takken W. et al. Olfactory coding in antennal neurons of the malaria mosquito, Anopheles gambiae. Chem Senses. 2006;31:845-863
7. McIver SB. Sensilla of mosquitoes (Diptera: Culicidae). J Med Entomol. 1982;19:489-535
8. Kwon HW, Lu T, Rutzler M. et al. Olfactory responses in a gustatory organ of the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2006;103:13526-13531
9. Pitts RJ, Zwiebel LJ. Antennal sensilla of two female anopheline sibling species with differing host ranges. Malaria J. 2006;5:26
10. Hill SR, Hansson BS, Ignell R. Characterization of antennal trichoid sensilla from female southern house mosquito, Culex quinquefasciatus Say. Chem Senses. 2009;34:231-252
11. Ghaninia M, Larsson M, Hansson BS. et al. Natural odor ligands for olfactory receptor neurons of the female mosquito Aedes aegypti: use of gas chromatography-linked single sensillum recordings. J exp Biol. 2008;211:3020-3027
12. Puri SN, Mendki MJ, Sukumaran D. et al. Electroantennogram and behavioral responses of Culex quinquefasciatus (Diptera: Culicidae) females to chemicals found in human skin emanations. J Med Entomol. 2006;43:207-213
13. Davis EE. A receptor sensitive to oviposition site attractants on the antennae of the mosquito, Aedis agyptyi. J Insect Physiol. 1976;22:1371-1376
14. Bowen MF. Terpene-sensitive receptors in female Culex pipiens mosquitoes: Electrophysiology and behaviour. J Insect Physiol. 1992;38:759-764
15. Siju KP, Hill SR, Hansson BS. et al. Influence of blood meal on the responsiveness of olfactory receptor neurons in antennal sensilla trichodea of the yellow fever mosquito, Aedes aegypti. J Insect Physiol. 2010;56:659-665
16. Vogt RG, Rybczynski R, Lerner MR. Molecular cloning and sequencing of general odorant-binding proteins GOBP1 and GOBP2 from the tobacco hawk moth Manduca sexta: comparisons with other insect OBPs and their signal peptides. J Neurosci. 1991;11:2972-2984
17. Hekmat-Scafe DS, Steinbrecht RA, Carlson JR. Coexpression of two odorant-binding protein homologs in Drosophila: implications for olfactory coding. J Neurosci. 1997;17:1616-1624
18. Swarup S, Williams TI, Anholt RR. Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 2011;10:648-657
19. Biessmann H, Andronopoulou E, Biessmann MR. et al. The Anopheles gambiae Odorant Binding Protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS ONE. 2010;5:e9471
20. Xu PX, Atkinson R, Jones DNM. et al. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193-200
21. Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143-160
22. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373-391
23. Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147-159
24. Vosshall LB, Amrein H, Morozov PS. et al. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725-736
25. Clyne PJ, Warr CG, Freeman MR. et al. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327-338
26. Fox AN, Pitts RJ, Zwiebel LJ. A cluster of candidate odorant receptors from the malaria vector mosquito, Anopheles gambiae. Chem Senses. 2002;27:453-459
27. Hill CA, Fox AN, Pitts RJ. et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176-178
28. Xu PX, Zwiebel LJ, Smith DP. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol Biol. 2003;12:549-560
29. Biessmann H, Nguyen QK, Le D. et al. Microarray-based survey of a subset of putative olfactory genes in the mosquito Anopheles gambiae. Insect Mol Biol. 2005;14:575-589
30. Justice RW, Dimitratos S, Walter MF. et al. Sexual dimorphic expression of putative antennal carrier protein genes in the malaria vector Anopheles gambiae. Insect Mol Biol. 2003;12:581-594
31. Pitts RJ, Rinker DC, Jones PL. et al. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics. 2011;12:271
32. Carey AF, Wang G, Su CY. et al. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66-71
33. Wang G, Carey AF, Carlson JR. et al. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:4418-4423
34. Qiao H, He X, Schymura D. et al. Cooperative interactions between odorant-binding proteins of Anopheles gambiae. Cell Mol Life Sci. 2010;68:1799-1813
35. Hekmat-Scafe DS, Scafe CR, McKinney AJ. et al. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357-1369
36. Zhou JJ, Huang W, Zhang GA. et al. "Plus-C" odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene. 2004;327:117-129
37. Lagarde A, Spinelli S, Qiao H. et al. Crystal structure of a novel type of odorant binding protein from Anopheles gambiae, belonging to the C+ class. Biochem J. 2011;427:423-430
38. Tsitsanou KE, Drakou CE, Thireou T. et al. Crystal and solution studies of the "Plus-C" odorant-binding protein 48 from Anopheles gambiae: control of binding specificity through three-dimensional domain swapping. J Biol Chem. 2013;288:33427-33438
39. Schymura D, Forstner M, Schultze A. et al. Antennal expression pattern of two olfactory receptors and an odorant binding protein implicated in host odor detection by the malaria vector Anopheles gambiae. Int J Biol Sci. 2010;6:614-626
40. Schultze A, Schymura D, Forstner M. et al. Expression pattern of a "Plus-C" class odorant binding protein in the antenna of the malaria vector Anopheles gambiae. Insect Mol Biol. 2012;21:187-195
41. Schultze A, Pregitzer P, Walter MF. et al. The co-expression pattern of odorant binding proteins and olfactory receptors identify distinct trichoid sensilla on the antenna of the malaria mosquito Anopheles gambiae. PLoS ONE. 2013;8:e69412
42. Angerer LM, Angerer RC. In situ hybridization to cellular RNA with radiolabelled RNA probes. Wilkinson DG (ed), In situ hybridization. Oxford: IRL Press. 1992:15-32
43. Li ZX, Pickett JA, Field LM. et al. Identification and expression of odorant-binding proteins of the malaria-carrying mosquitoes Anopheles gambiae and Anopheles arabiensis. Arch Insect Biochem Physiol. 2005;58:175-189
44. Vogt RG, Kohne AC, Dubnau JT. et al. Expression of pheromone binding proteins during antennal development in the gypsy moth Lymantria dispar. J Neurosci. 1989;9:3332-3346
45. Steinbrecht RA, Laue M, Ziegelberger G. Immunolocalization of pheromone-binding protein and general odorant-binding protein in olfactory sensilla of the silk moths Antheraea and Bombyx. Cell Tiss Res. 1995;282:203-217
46. Steinbrecht RA, Ozaki M, Ziegelberger G. Immunocytochemical localization of pheromone-binding protein in moth antennae. Cell Tissue Res. 1992;270:287-302
47. Shanbhag SR, Hekmat-Scafe D, Kim MS. et al. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc Res Tech. 2001;55:297-306
48. Iatrou K, Biessmann H. Sex-biased expression of odorant receptors in antennae and palps of the African malaria vector Anopheles gambiae. Insect Biochem Mol Biol. 2008;38:268-274
49. Vieira FG, Rozas J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol Evol. 2011;3:476-490
50. Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535-1547
51. Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548-1553
52. Goldman AL, van Naters WV, Lessing D. et al. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45:661-666
53. Dobritsa AA, van der Goes van Naters, Warr CG. et al. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827-841
54. Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965-979
Author contact
![]() Corresponding author: Dr. Jürgen Krieger, University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany. e-mail: juergen.kriegerde.
Corresponding author: Dr. Jürgen Krieger, University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany. e-mail: juergen.kriegerde.

 Global reach, higher impact
Global reach, higher impact