10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(8):913-922. doi:10.7150/ijbs.11585 This issue Cite
Research Paper
Transmissible Gastroenteritis Virus (TGEV) Infection Alters the Expression of Cellular MicroRNA Species That Affect Transcription of TGEV Gene 7
College of Veterinary Medicine, Northwest A&F University, Yangling, Shaanxi 712100, P.R. China
* These authors contributed equally to this work.
Received 2015-1-13; Accepted 2015-5-14; Published 2015-6-7
Abstract
Transmissible gastroenteritis virus (TGEV) is a member of Coronaviridae family. TGEV infection has emerged as a major cause of severe gastroenteritis and leads to alterations of many cellular processes. Meanwhile, the pathogenic mechanism of TGEV is still unclear. microRNAs (miRNAs) are a novel class of small non-coding RNAs which are involved in the regulation of numerous biological processes such as viral infection and cell apoptosis. Accumulating data show that miRNAs are involved in the process of coronavirus infection such as replication of severe acute respiratory syndrome coronavirus (SARS-CoV). However, the link between miRNAs and TGEV infection is unknown. In this study, we performed microRNA microarray assay and predicted targets of altered miRNAs. The results showed TGEV infection caused the change of miRNAs profile. Then we selected miR-4331 for further analysis and subsequently identified cell division cycle-associated protein 7 (CDCA7) as the target of miR-4331. Moreover, miR-4331 showed the ability to inhibit transcription of TGEV gene 7 (a non-structure gene) via directly targeting CDCA7. In conclusion, differentially expressed miR-4331 that is caused by TGEV infection can suppress transcription of TGEV gene 7 via targeting cellular CDCA7. Our key finding is that TGEV selectively manipulates the expression of some cellular miRNAs to regulate its subgenomic transcription.
Keywords: TGEV, microRNAs (miRNAs), TGEV, CDCA7, Gene 7, viral transcription
Introduction
The miRNAs are small RNA species (containing about 22 nucleotides) which are found in animals, plants, and some viruses. It has been shown that miRNAs play a variety of roles in many cellular processes including development, differentiation, apoptosis, cell cycle (reviewed in reference [1]). Generally, miRNAs are derived from introns or intergenes of eukaryotic organisms or viruses and serve as regulators of gene expression [2]. The miRNA-mediated RNA interference includes two patterns. One is perfectly base-paring with 3' UTR of target mRNA leading to degradation of mRNA. The other is imperfectly binding to target 3' UTR of mRNA resulting in the inhibition of mRNA translation. In mammalians, miRNAs perform RNAi through translation inhibition mediated through imperfect complementarity [3]. As a result, a miRNA can directly interact with multiple targets and a gene can be targeted by many miRNAs [4].
During the interactions between virus and host, miRNAs are also playing important regulatory roles [5, 6]. Increasing evidence indicates that viral infection results in the changes of cellular miRNAs species. In turn, the altered miRNAs also affect viral infection [7, 8]. For example, SARS-CoV infection up-regulates miR-17*, -574-5p, and -214, and down-regulates miR-98 and miR-223. Of these miRNAs, miR-17*, -574-5p, and -214 suppress SARS-CoV replication and contribute to immune evasion in bronchoalveolar stem cells (BASC) [9]. TGEV is an enveloped virus with a positive-sense single-stranded RNA genome and causes a highly contagious, fatal gastroenteritis primarily in less than 2-week age piglets around the world. The mortality rate of seronegative suckling piglets infected with TGEV can reach to as high as 100% [10, 11]. The TGEV genome contains a leader sequence at the 5' end and a poly (A) tail at the 3' end. The TGEV genes are arranged in the order 5'-Rep-S-3a-3b-E-M-N-7-3' [12]. The TGEV gene 7 locates at the 3'end of the genome. During TGEV infection, protein 7 attenuates host antiviral response by interacting with protein phosphatase 1 catalytic subunit (PP1c), a key regulator of host antiviral defense [13], and reduces apoptosis, immune response, interferon response, and inflammation [14].
The mammalian cellular miRNAs are profoundly affected by viral infection and, in turn the miRNAs also regulate viral gene expression via targeting viral genes or cellular genes. The miR-122 promotes hepatitis C virus (HCV) RNA translation through hybridizing with the 5' end of viral transcript [15, 16]. The cellular miR-28, miR-125b, miR-150, miR-223, and miR-382 play important roles in the regulation of human immunodeficiency virus (HIV) gene expression through targeting the viral nef gene in CD4+ T-cells [17, 18]. The miR-146a binds to cellular ring finger (RNF) mRNA to increase hendra virus replication [19].
To date, complex regulatory networks associated with miRNAs have not been comprehensively assessed in TGEV infection. Overall, we observed that TGEV infection caused the change of miRNA profile and miR-4331 suppressed transcription of TGEV gene 7 via directly targeting CDCA7.
Methods
Antibodies, Cells and virus
CDCA7 polyclonal antibody (15249-1-AP) was obtained from ProteinTech Group (ProteinTech Group, Chicago, IL, US). Monoclonal β-actin (sc-69879) was purchased from SantaCruz Biotechnology (Santa Cruz, Inc., CA, US). Horseradish peroxidase (HRP)-conjugated secondary antibody was purchased from Pierce (Pierce, Rockford, IL, US). PK-15 cells were obtained from American Type Culture Collection (ATCC) (CCL-33) and grown in Dulbecco's Minimal Essential Medium (DMEM) supplemented with 10% fetal bovine serum (Gibco BRL, Gaithersburg, MD, US), 100 IU of penicillin and 100 mg of streptomycin per ml, at 37 ℃ in a 5% CO2 atmosphere incubator. The TGEV Shaanxi strain was isolated from TGEV-infected piglets by Ding L et al [20].
The miRNAs microarray and target prediction of differentially expressed miRNAs
Confluent PK-15 cells in 100-mm cell culture dish were infected with TGEV for 24 h at an MOI of 1.0. Meanwhile, the mock infection was carried out. At 24 hpi, total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, US). Microarray assay was performed as described previously [21] using an updated version of the chip (swine miRNA version 18, http://www.mirbase.org/). Targets of differentially expressed miRNAs were predicted by TargetScan and miRanda.
The quantification of miRNAs by real-time PCR
The total RNA was obtained using Trizol reagent (Invitrogen, Carlsbad, CA, US) from PK-15 cells infected with TGEV at 1.0 MOI for 24 h for the microarray analysis. Reverse transcription reactions were performed as described previously [22]. Briefly, 2 μg of total RNA initially treated with DNase I (Fermentas, St. Leon-Rot, Germany), 50 nM stem-loop RT primers, 1×first strand buffer, 0.25 U/μL RNase inhibitor, 10 U/μL M-MLV, and 10 mM DTT (Invitrogen, Carlsbad, CA, US), were incubated at 16 ℃ for 30 min, 42 ℃ for 30 min, and 85 ℃ for 5 min. Real-time PCR was carried out using the AccuPower 2×Greenstar qPCR Master mix (Bioneer, Daejeon, Korea) in a 25 μL reaction volume including 12.5 μL 2×Greenstar Master mix, 0.5 μL 50×ROX dye, 0.5 μL RT product, 1 mM forward primer, and 1 mM reverse primer. Reactions were incubated at 95 ℃ for 10 min, followed by 40 cycles of 95 ℃ for 15 sec, and 60 ℃ for 1 min on Bio-Rad iQ5 Real-Time PCR System (Bio-Rad, USA). The relative quantification of miRNAs was normalized to U6 using the ∆∆Ct method [23].
The quantification of subgenomic mRNAs by real-time PCR
Total RNA was obtained using Trizol reagent (Invitrogen, Carlsbad, CA, US) according to manufacturer's instructions. The primers for genomic RNA (gRNA) and subgenomic mRNAs (sgmRNA) of TGEV were described previously [24]. A total of 2 μg of RNA was treated with DNase I (Fermentas, St. Leon-Rot, Germany) for 30 min at 37 ℃. The treated total RNA was reversely transcribed with the First-strand cDNA synthesis kit (Invitrogen, Carlsbad, CA, US). Real-time PCR was performed using the AccuPower 2×Greenstar qPCR Master mix (Bioneer, Daejeon, Korea) in a 25 μL reaction volume on Bio-Rad iQ5 Real-Time PCR System (Bio-Rad, USA). Fold variations of the sgmRNAs were calculated (normalized to gRNA).
Luciferase reporter experiments
3' UTRs of 16 candidate target genes containing the binding site of miR-4331 were respectively amplified by PCR using primers containing sequences of Xho I and Not I cloning sites and were cloned into the vector psiCHECK-2 (Promega, Madison, WI, USA). To obtain mutation of miR-4331 complementary sites within the 3' UTR of CDCA7, seed region was mutated following a mutagenesis protocol [25]. The miR-4331 mimics (sense strand 5'-UGUGGCUGUGGUGUAGGCCAGC-3'; antisense strand 5'-GCUGGCCUACACCACAGCCAC A-3'), a negative control for mimics (an unrelated mimic, sense strand 5'-UCACAACCUCCUAGAAAGAGUAGA-3'; antisense strand 5'- UCUACUCUUUCUAGGAGGUUGUGA-3'), an inhibitor for miR-4331 (5'-GCUGGCCUACACCACAGCCACA-3'), and a control RNA inhibitor (a random sequence, 5'-UCUACUCUUUCUAGGAGGUUGUGA-3') were designed and synthesized by Ribo Biotech (RiboBio, Guangzhou, China). The miRNA inhibitors were modified with 2'-O-methyl. For the luciferase reporter assay, PK-15 cells were seeded in 24-well plates and then co-transfected with 100 ng plasmid and 100 nM of miR-4331 mimics, miR-4331 inhibitors, or negative control, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, US). At 48 h post transfection (hpt), the luciferase activities were measured using Dual-Glo Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer's manual.
RNA interference
Three siRNAs (siCDCA7-1, siCDCA7-2, siCDCA7-3) silencing CDCA7 gene and irrelevant siRNA were synthesized by Ribo Biotech (RiboBio, Guangzhou, China). The most effective siRNA (si-CDCA7-2), identified by western blot, was applied for the experiments. The sequence of si-CDCA7-2 is: sense sequence 5'-GAAGUUGAUUUCCAUGGAAdTdT-3' and antisense sequence 5'-dTdT CUUCAACUAAAGGUACCUU-3'. The negative control siRNA is an irrelevant siRNA. PK-15 cells were transfected with 50 nM CDCA7-specific siCDCA7-2 or irrelevant siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, US) according to the manufacturer's guidelines. Cells were grown at 37 ℃ for 48 h and then infected with the TGEV at MOI of 1.0. The total RNA was isolated at 12 and 24 hpi.
Cell viability assay
Cell viability assay was carried out using Cell Counting Kit-8 (CCK-8) reagent (Vazyme, Piscataway, NJ, USA). PK-15 cells were plated in 96-well dishes at a density of 1×104 cells/well. Cells were transfected with 50 nM siCDCA7-2 or irrelevant siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, US) according to the manufacturer's guidelines. After 48 h of incubation, the cells were treated with CCK-8 reagent for 1h. Absorbance was recorded with the microplate reader at 450 nm.
Vector construction
To construct the CDCA7 expression plasmid, Full-length CDCA7 clone was produced by PCR amplification (forward 5'-CTAGCTAGCTAGCTGGCCGTCAGCATGGATG-3'; reverse 5'-CTAGTCTAGACTAGCAAGTGGAAGGCAGGTAGTGG-3'). PCR products were digested and ligated into pCI-neo (Promega, Madison, WI, USA) at Nhe I and Xho I sites. The correct constructed plasmid was named pCI-neo-CDCA7. PK-15 cells were transfected with pCI-neo vector (control) or pCI-neo-CDCA7. At 48 hours post-infection (hpt), cells were infected with TGEV at MOI of 1.0. Total RNAs were extracted by Trizol reagent (Invitrogen, Carlsbad, CA, US) at 12 and 24 hpi according to the manufacturer's protocol. Real-time PCR was performed to detect the sgmRNAs level.
Western blot analysis
Cells were treated with RIPA lysis buffer with phenylmethyl sulfonylfluoride (PMSF). Protein concentration was determined with BCA Protein Assay Reagent (Pierce, Rockford, IL, US) and separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and subsequently transferred to a polyvinylidene difluoride (PVDF) membranes (Millipore Corp, Atlanta, GA, US). The PVDF membrane was blocked with 5% non-fat dry milk for 2 h at room temperature, and then incubated with primary antibodies overnight at 4 ℃ and subsequently with HRP-conjugated secondary antibody at room temperature for 1 h. The signal was detected with enhanced chemiluminescence (ECL).
TGEV subgenomic transcription in the presence of miR-4331 mimics and inhibitors
PK-15 cells were seeded in 6-well plate (5×105 cells/well) in growth medium and transfected using Lipofectamine 2000 with 100 nM miR-4331 mimics, miR-4331 inhibitors, or negative control respectively. At 24 hpt, the cells were infected with TGEV at MOI of 1.0. The transcription of virus was determined by real-time PCR analysis after 12 and 24 hpi.
Results
Cellular microRNAs profiling in response to TGEV infection
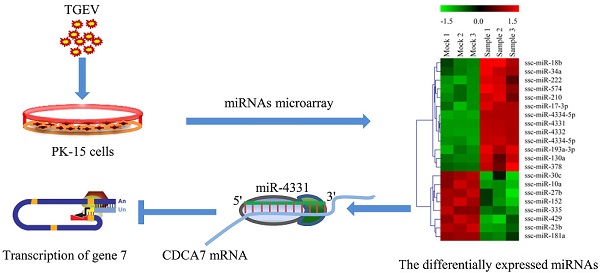
To gain the overview of the impact of TGEV infection on host cellular miRNAs expression, PK-15 cells were infected with TGEV Shaanxi strain at a multiplicity of infection (MOI) of 1.0 for microarray. The microarray was performed with the total RNA extracted from PK-15 cells infected with TGEV at 24 hpi. The differential expression of multiple miRNAs in TGEV-infected cells in comparison with mock-infected cells was observed in Fig. 1(A). The expression levels of 21 miRNAs were changed remarkably (fold change > 1.5, and p < 0.01). Among them 13 miRNAs were up-regulated and 8 miRNAs were down-regulated. To validate the microarray results of differentially expressed miRNAs, we tested the expression levels of them using real-time PCR. The fold changes of 21 miRNAs in TGEV-infected cells were referred to that in mock-infected cells. The miRNAs levels were normalized to U6. The results were correlated with microarray (Fig. 1B). A total of 21 differentially expressed miRNAs were chosen for target prediction. The potential targets of 21 differentially expressed miRNAs were predicted with TargetScan and miRanda and 7,844 potential target genes were gained (Supplementary Table S1).
The regulatory function of miR-4331 on TGEV subgenomic transcription
To investigate the impact of differentially expressed miRNAs on TGEV transcription, we selected miR-4331 as a representative based on p value (p < 0.01), fold change of miRNA expression (fold change > 1.5), and mean fluorescence intensities (MFI) (MFI > 400). PK-15 cells were transfected with miR-4331 mimics, mimics control, inhibitors, or inhibitors control. Subsequently all the samples and controls were infected with TGEV. The miR-4331 level of the samples was compared with the controls and normalized to U6. The miR-4331 level increased after transfection with the miRNA mimics and decreased after transfection of the inhibitors in comparison with the control (Fig. 2A). The transcription of TGEV gene 7 was decreased by overexpression of miR-4331 and increased by inhibition of miR-4331 at 12 and 24 hpi (Fig. 2B). Except for gene 7, the replication of TGEV genome and transcription of other subgenomes were not affected by miR-4331 (data are not shown).
miRNAs expression profile in TGEV-infected PK-15 cells. (A) Heat map analysis showing differentially expressed miRNAs in PK-15 cells infected with TGEV at 24 h. Red indicates higher expression and green indicates lower expression. Each colored block represents the expression of one miRNA (labeled on the right) in the indicated sample. The map showed all significantly differential expressed miRNAs in three independent samples (p < 0.01). (B) Verification of miRNAs microarray by real-time PCR. The fold change was determined using the 2-ΔΔCt method and all miRNAs expression values were normalized to endogenous U6. Data from real-time PCR were shown as mean ±standard deviation (S.D.) of three independent experiments. Similar results were obtained in three independent experiments.
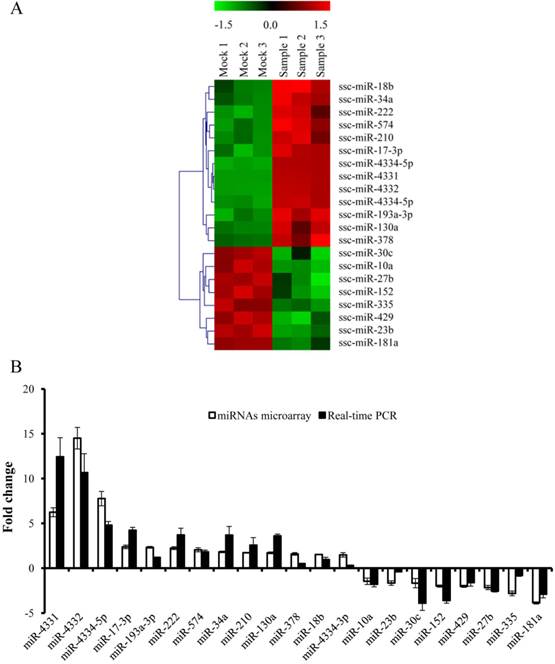
The effect of miR-4331 on transcription of TGEV gene 7. (A) miR-4331 levels in PK-15 cells transfected with mimics or inhibitors and infected with TGEV at 12 and 24 hpi. The expression of miR-4331 was measured by real-time PCR (Normalized to U6 and in reference to the level of the control). (B) The effect of miR-4331 on transcription of TGEV gene 7 at 12 and 24 hpi. The effect of miR-4331 on TGEV transcription of gene 7 was assessed by real-time PCR analysis (Normalized to gRNA and in reference to the level of the control). Data represent mean ±S.D. of three independent experiments. * P < 0.05 in comparison with the control. ** P < 0.01 in comparison with the control.
CDCA7 is the target of miR-4331
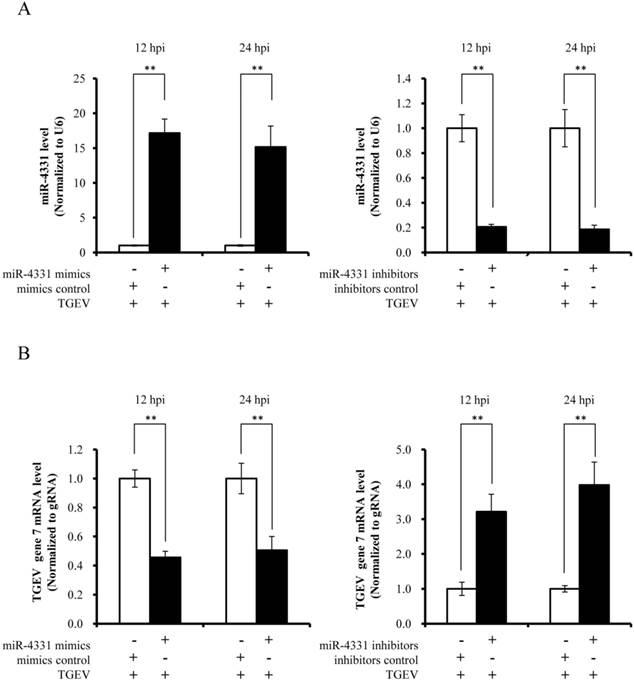
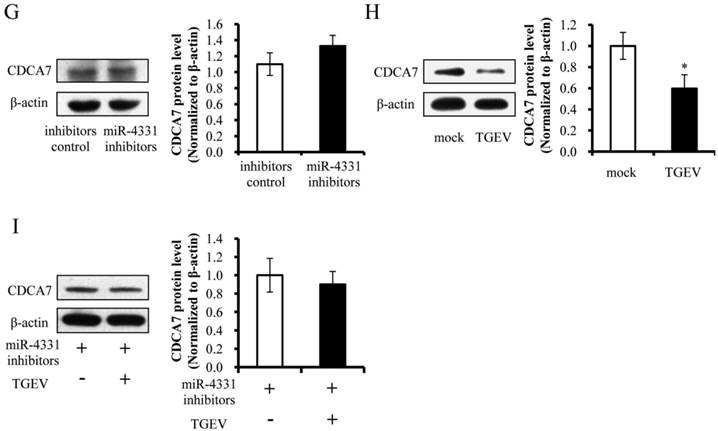
To identify the targets that affect the transcription of TGEV gene 7, targets of miR-4331 were predicted by TargetScan and miRanda (Supplementary Table S1), and TGEV gene 7 does not contain miR-4331 binding site. Therefore, we hypothesized that miR-4331 should target cellular gene to inhibit transcription of gene 7. In order to screen miR-4331 target gene, 16 recombinant dual-luciferase reporter plasmids that contain the binding site of miR-4331 target gene at 3' UTR of the reporter were constructed for the dual-luciferase assay. The luciferase activity of plasmid containing the 3' UTR of CDCA7 was markedly lower than that of the control (Fig. 3A). To generate 3' UTR mutant of CDCA7 mRNA containing mutations of conserved miR-4331 binding site, the site-directed mutagenesis was performed using the DNA sequence of wild-type 3' UTR as the template. Two binding sites of miR-4331 seed sequence at CDCA7 3' UTR were respectively mutated with the 3-bp substitution (Fig. 3B). To determine the direct interaction between miR-4331 and 3' UTR of CDCA7 mRNA, the porcine CDCA7 3' UTR or its mutant was respectively subcloned into the 3' UTR of a Renilla luciferase gene in a luciferase reporter psiCHECK-2 (Fig. 3C). The constructs were co-transfected into PK-15 cells with miR-4331 mimics, miR-4331 inhibitors, or negative control. Compared with the control, introduction of exogenous miR-4331 decreased the CDCA7-WT reporter activity by 64%, while overexpression and knockdown of miR-4331 using miR-4331 mimics and inhibitors did not affect CDCA7-mut reporter activity (Fig. 3D). To assess the specificity of miR-4331 binding, miR-4331 mimics, 4331 inhibitor, and CDCA7-WT reporter are co-transfected. The results showed that the miR-4331 mimics decreased the CDCA7-WT reporter activity and the miR-4331 mimics and inhibitors had specific binding activity (Fig. 3E). To determine whether miR-4331 inhibits CDCA7 expression, miR-4331 mimics or mimics control was transfected into PK-15 cells, and CDCA7 expression was assessed by western blot. Introduction of exogenous miR-4331 decreased expression of CDCA7 in PK-15 cells compared with the control (Fig. 3F). Knockdown of endogenous miR-4331 using miR-4331 inhibitors failed to increase the CDCA7 expression (Fig. 3G). This is likely due to the lower level of endogenous miR-4331 in PK-15 cells. Moreover, we observed that TGEV infection decreased CDCA7 protein levels in PK-15 cells (Fig. 3H), while downregulation of endogenous miR-4331 using miR-4331 inhibitors reversed the effect of TGEV infection on CDCA7 expression (Fig. 3I). Together, our data conclusively demonstrate that CDCA7 is a direct target of miR-4331 in PK-15 cells.
Identification of miR-4331 target. (A) The effect of miR-4331 on 3' UTRs of 16 potential target genes measured by dual-luciferase reporter assay (normalized to firefly). (B) Bioinformatic prediction of interaction between miR-4331 and the 3' UTRs of swine CDCA7. For each schematic, the upper sequence is the binding site of miR-4331 in 3ʹUTRs of swine CDCA7, the middle is the sequence of mature miR-4331, and the lower sequence is the mutated sequence of CDCA7 3ʹ UTR. The seed sequence is underlined. (C) Schematic overview of the putative binding sites or mutations of miR-4331 binding-sites in 3' UTR of CDCA7 mRNA. The locations of the potential binding sites or their mutations are presented by blank boxes. (D) The CDCA7 luciferase reporter construct was co-transfected with miR-4331 mimics (or negative control) or miR-4331 inhibitors (or negative control) into PK-15 cells (normalized to the firefly luciferase activity). Data are expressed as relative luciferase activities to control. (E) The specific binding activity of miR-4331 mimics and inhibitors (normalized to the firefly luciferase activity). (F) Western blot analysis of the effect of miR-4331CDCA7 in cells transfected with miR-4331 mimics or mimics control. (G) Western blot analysis of CDCA7 in cells transfected with miR-4331 inhibitors or inhibitors control. (H) Western blot analysis of endogenous CDCA7 protein level in TGEV-infected cells at 24 hpi. (I) Western blot analysis of endogenous CDCA7 protein level in TGEV-infected cells at 24 hpi by knockdown of endogenous miR-4331 using miR-4331 inhibitors. Data represent means ±S.D. of three independent experiments. * P < 0.05 in comparison with the control. ** P < 0.01 in comparison with the control.
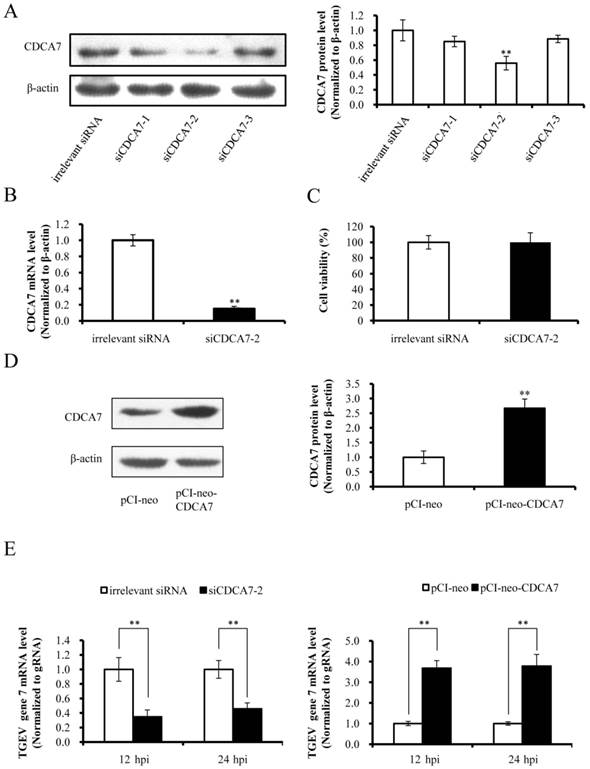
Effect of CDCA7 on TGEV transcription of gene 7
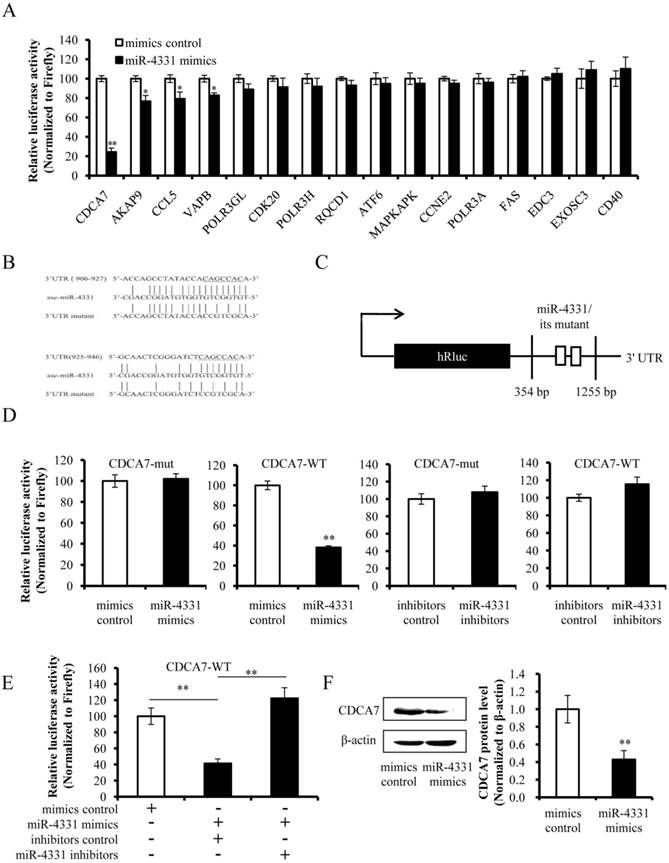
The miRNAs regulate target gene expression via binding to the 3' UTR of target mRNA, and one miRNA may target multiple genes. We confirmed that miR-4331 suppressed transcription of gene 7 and targeted CDCA7 directly, but whether miR-4331 inhibits transcription of gene 7 by targeting CDCA7 is unclear. To test the effect of artificial siRNA of CDCA7, three siRNAs with different sequences were respectively transfected into PK-15 cells and the effects of silencing were assessed by western blot analysis. The data indicated that siCDCA7-2 was the most effective siRNA (Fig. 4A). The mRNA level of CDCA7 was significantly silenced by siCDCA7-2 (Fig. 4B), however the viability of cells was not affected by siCDCA7-2 in comparison with irrelevant siRNA (Fig. 4C). To detect the overexpression effect of pCI-neo-CDCA7, we measured the protein level of CDCA7 at 48 hpt using western blotting. As expected CDCA7 expression level in transfected group increased compared with the control (Fig. 4D). To investigate the effect of CDCA7 on the transcription of TGEV gene 7, we detected transcription level of gene 7 using real-time PCR. The results showed that silence of CDCA7 using siCDCA7-2 decreased transcription of gene 7 at 12 and 24 hpi and that overexpression of CDCA7 facilitated transcription of gene 7 at 12 and 24 hpi (Fig. 4E). To understand the relevance of CDCA7 expression in viral infection, we determined the effect of CDCA7 silencing and overexpression on TGEV gRNA, other sgmRNAs, and viral titers. The results showed that CDCA7 had no effect on TGEV gRNA level, other sgmRNAs level, and viral titers (Data are not shown).
Taken together, the data presented here demonstrated that TGEV infection induced miRNAs profiling changes and, among these altered miRNAs the up-regulated miR-4331 induced by TGEV infection inhibited transcription of gene 7 via directly targeting CDCA7.
Discussion
The major findings in this study are that TGEV infection leads to the change of cellular miRNAs expression profile, and altered miRNAs regulate transcription of TGEV gene 7 through targeting cellular CDCA7. Increasing evidence shows that miRNAs play important roles in virus-host interactions including viral transcription, cell apoptosis, and antiviral effects of host [26]. In this study, we examined the connection between miRNAs and TGEV infection and assessed global expression patterns of cellular miRNAs. Based on the findings of this study, we proposed that miR-4331 expression altered by TGEV infection is involved in regulating transcription of TGEV gene 7.
The infection of coronavirus, such as SARS-CoV and infectious bronchitis virus (IBV), affects many cellular processes such as viral replication, host innate immunity or signaling pathways [27]. In some cases, the altered processes are directed by the virus for its own advantage and other altered processes are cellular defense responses to viral infection. In this study, we observed that the expression levels of cellular miRNAs are regulated positively or negatively by TGEV infection. The miRNAs regulate viral infection through interacting with targets. Therefore, the miRNAs and targets form a complicated regulatory network during viral infection which suggests that this greatly expands the range of possible virus-host interactions. This is in good agreement with the notion that miRNAs have evolved as a necessary and critical component to regulate host-virus interactions [28].
Effect of CDCA7 on transcription of TGEV gene 7. (A) Silencing effect of CDCA7 siRNA on CDCA7 expression. PK-15 cells were transfected with CDCA7-specific siRNA or irrelevant siRNA and analyzed by western blot. (B) The effect of siCDCA7-2 on CDCA7 at mRNA level. The CDCA7 mRNA level was reduced at 24 h hpt measured by real-PCR (Normalized to β-actin). (C) The effect of siCDCA7-2 on PK-15-cell viability. The cells were incubated after transfecting with 50 nM siCDCA7-2 for 48 h. Cell viability was evaluated by CCK-8 assay. (D) The overexpression effect of CDCA7 using pCI-neo-CDCA7. PK-15 cells were transfected with pCI-neo-CDCA7 or pCI-neo vector, and the expression level of CDCA7 was assessed by western blot at 48 hpt. (E) The effect of CDCA7 on transcription of TGEV gene 7. Knockdown of CDCA7 using siRNA decreased transcription of gene 7 at 12 and 24 hpi. Overexpression of CDCA7 increased transcription of gene 7 at 12 and 24 hpi. Data represent mean ±S.D. of three independent experiments. * P < 0.05 in comparison with the control. ** P < 0.01 in comparison with the control.
In this study, we observed that TGEV altered the miRNAs profile and miR-4331 was involved in regulating the interplay between TGEV and PK-15 cells by modulating transcription of TGEV gene 7. It was reported that SARS-CoV infection resulted in differentially expressed miRNAs profile and some of altered miRNAs regulated the interaction between SARS-CoV and host [9]. These indicate coronaviruses may affect cellular miRNAs expression and the altered miRNAs are able to regulate viral infection.
A key observation in this study was that miR-4331 suppresses transcription of TGEV gene 7. TGEV gene 7, a non-structure gene, contributes to viral invasion and inhibits apoptosis in host cells [13]. Previously we reported that TGEV induces apoptosis by multiple pathways [29-31]. These observations suggest that miR-4331 might be involved in regulating apoptosis through gene 7. Therefore it will be important to detect effects of miR-4331 on apoptosis.
miR-4331 was detected in porcine intestinal and milk by deep sequencing [32, 33], but the current studies related to miR-4331 are very few. To identify what gene miR-4331 will target to regulate the transcription of TGEV gene7, the potential targets of miR-4331 were predicted using TargetScan and miRanda. More than 600 potential targets were obtained (Supplementary Table S1). Since TGEV genes were not the potential targets of miR-4331, miR-4331 must target the cellular genes. We identified the potential targets and obtained a new finding that exogenous miR-4331 directly interacts with the 3' UTR of CDCA7 mRNA and suppresses CDCA7 protein level. CDCA7 is a transcriptional regulator that is directly targeted and activated by transcription factor E2F1 [34]. The E2F1 plays a crucial role in controlling cell cycle and action of tumor suppressor proteins [35]. In addition, CDCA7 is a target gene of c-MYC and the phosphorylated CDCA7 regulates c-MYC-dependent apoptosis and transformation [36]. Therefore it will be very important to discover the regulatory mechanism of CDCA7 during TGEV infection.
In conclusion, the results of the present study provide evidence that TGEV infection resulted in altered profiles of miRNAs in PK-15 cells and the differentially expressed miR-4331 was involved in regulation of TGEV transcription by targeting cellular CDCA7. These findings will give insight into the regulatory mechanism of miRNA to transcription of TGEV gene 7. In addition, this opens a new avenue to target host cellular factors using siRNA technologies to combat TGEV transcription.
There are limitations in this study that need to be addressed and resolved in future studies. The miRNAs play regulatory role at post-transcriptional level [37]. An miRNA may target multiple genes, and one gene may be targeted by many miRNAs [38, 39]. For miR-4331, it is supposed to take part in regulating the TGEV infection process via targeting other genes. For the whole study, numerous differentially expressed miRNAs and a quantity of targets of these miRNAs form a complex regulatory network to modulate viral-host interactions at any stage of TGEV infection. Therefore, more functions of altered miRNAs on TGEV-host interactions need to uncover and more targets need to identify in the future studies.
Supplementary Material
Supplementary Table S1.
Abbreviations
TGEV: Transmissible gastroenteritis virus; miRNAs: microRNAs; SARS-CoV: severe acute respiratory syndrome coronavirus; CDCA7: cell division cycle-associated protein 7; BASC: bronchoalveolar stem cells; PP1c: phosphatase 1 catalytic subunit; HCV: hepatitis C virus; HIV: human immunodeficiency virus; RNF: ring finger; HRP: Horseradish peroxidase; DMEM: Dulbecco's Minimal Essential Medium; gRNA: genomic RNA; sgmRNA: subgenomic mRNAs; CCK-8: Cell Counting Kit-8; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; PVDF: polyvinylidene difluoride; ECL: enhanced chemiluminescence; MOI: multiplicity of infection; MFI: mean fluorescence intensities.
Acknowledgements
This work was supported by grants from Natural Science Foundation of China (Grant No. 31372401, 31472167), Shaanxi Key and Innovate Group for Science and Technology Program (Grant No. 2013KCT-28), Scientific Research Program of Northwest A&F University (Grant No. Z111021103, 2014YB012, 2013BSJJ015).
We thank Dr. Xiaolong Wang (Northwest A&F University College of Veterinary Medicine, Yangling, P.R. China), Dr. Xiuzhu Sun (Northwest A&F University College of Animal science and Technology, Yangling, P.R. China), and our colleagues for critical reading of the manuscript.
Contributors and authorship
Xiangjun Song and Xiaomin Zhao designed the experiments, interpreted the data and wrote the article. Xiangjun Song and Xiaomin Zhao performed the experiments with assistance and advice from Yong Huang, Hailing Xiang, Wenlong Zhang. Dewen Tong revised the manuscript. All authors have read the manuscript and approved to submit it to this journal.
Competing interests
The authors have declared that no competing interest exists.
References
1. Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annual review of biophysics. 2013;42:217-39 doi:10.1146/annurev-biophys-083012-130404
2. Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S. microRNA: a master regulator of cellular processes for bioengineering systems. Annual review of biomedical engineering. 2010;12:1-27 doi:10.1146/annurev-bioeng-070909-105314
3. Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. International journal of genomics. 2014;2014:970607. doi:10.1155/2014/970607
4. Gu S, Kay MA. How do miRNAs mediate translational repression? Silence. 2010;1:11. doi:10.1186/1758-907X-1-11
5. Guo YE, Steitz JA. Virus Meets Host MicroRNA: the Destroyer, the Booster, the Hijacker. Molecular and Cellular Biology. 2014;34:3780-7 doi:10.1128/mcb.00871-14
6. Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes & development. 2011;25:1881-94 doi:10.1101/gad.17352611
7. Fu M, Gao Y, Zhou Q, Zhang Q, Peng Y, Tian K. et al. Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA. Gene. 2014;536:272-8 doi:10.1016/j.gene.2013.12.012
8. Libri V, Miesen P, van Rij RP, Buck AH. Regulation of microRNA biogenesis and turnover by animals and their viruses. Cell Mol Life Sci. 2013;70:3525-44 doi:10.1007/s00018-012-1257-1
9. Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PloS one. 2009;4:e7837. doi:10.1371/journal.pone.0007837
10. Kim B, Chae C. In situ hybridization for the detection of transmissible gastroenteritis virus in pigs and comparison with other methods. Canadian journal of veterinary research = Revue canadienne de recherche veterinaire. 2001;65:33-7
11. Chae C, Kim O, Choi C, Min K, Cho WS, Kim J. et al. Prevalence of porcine epidemic diarrhoea virus and transmissible gastroenteritis virus infection in Korean pigs. Vet Rec. 2000;147:606-8
12. Penzes Z, Gonzalez JM, Calvo E, Izeta A, Smerdou C, Mendez A. et al. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the purdue virus cluster. Virus genes. 2001;23:105-18
13. Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L. et al. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 2011;7:e1002090. doi:10.1371/journal.ppat.1002090
14. Cruz JL, Becares M, Sola I, Oliveros JC, Enjuanes L, Zuniga S. Alphacoronavirus protein 7 modulates host innate immune response. Journal of virology. 2013;87:9754-67 doi:10.1128/JVI.01032-13
15. Cox EM, Sagan SM, Mortimer SAW, Doudna JA, Sarnow P. Enhancement of hepatitis C viral RNA abundance by precursor miR-122 molecules. RNA. 2013;19:1825-32 doi:10.1261/rna.040865.113
16. Zhang C, Huys A, Thibault PA, Wilson JA. Requirements for human Dicer and TRBP in microRNA-122 regulation of HCV translation and RNA abundance. Virology. 2012;433:479-88 doi:10.1016/j.virol.2012.08.039
17. Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H. et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241-7 doi:10.1038/nm1639
18. Sun G, Li H, Wu X, Covarrubias M, Scherer L, Meinking K. et al. Interplay between HIV-1 infection and host microRNAs. Nucleic acids research. 2012;40:2181-96 doi:10.1093/nar/gkr961
19. Stewart CR, Marsh GA, Jenkins KA, Gantier MP, Tizard ML, Middleton D. et al. Promotion of Hendra virus replication by microRNA 146a. J Virol. 2013;87:3782-91 doi:10.1128/JVI.01342-12
20. Ding L, Chen G.D, Xu X.G, Tong D.W. Isolation and identification of porcine transmissible gastroenteritis virus Shaanxi strain and sequence analysis of its N gene. Chin J Vet Med. 2011;47:09-11 doi:10.1016/j.jviromet.2010.02.010
21. Lin Y, Zeng Y, Zhang F, Xue L, Huang Z, Li W. et al. Characterization of microRNA expression profiles and the discovery of novel microRNAs involved in cancer during human embryonic development. PLoS ONE. 2013;8:e69230
22. Chen C. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic acids research. 2005;33:e179-e doi:10.1093/nar/gni178
23. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402-8 doi:10.1006/meth.2001.1262
24. Mateos-Gomez PA, Zuniga S, Palacio L, Enjuanes L, Sola I. Gene N proximal and distal RNA motifs regulate coronavirus nucleocapsid mRNA transcription. J Virol. 2011;85:8968-80 doi:10.1128/JVI.00869-11
25. Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nature Protocols. 2007;2:924-32 doi:10.1038/nprot.2007.132
26. tenOever BR. RNA viruses and the host microRNA machinery. Nature reviews Microbiology. 2013;11:169-80
27. Zhong Y, Tan YW, Liu DX. Recent progress in studies of arterivirus- and coronavirus-host interactions. Viruses. 2012;4:980-1010 doi:10.3390/v4060980
28. Zhou R, Rana TM. RNA-based mechanisms regulating host-virus interactions. Immunological reviews. 2013;253:97-111 doi:10.1111/imr.12053
29. Ding L, Xu XG, Huang Y, Li ZC, Zhang K, Chen GD. et al. Transmissible gastroenteritis virus infection induces apoptosis through FasL- and mitochondria-mediated pathways. Veterinary microbiology. 2012;158:12-22 doi:DOI 10.1016/j.vetmic.2012.01.017
30. Ding L, Zhao XM, Huang Y, Du Q, Dong F, Zhang HL. et al. Regulation of ROS in transmissible gastroenteritis virus-activated apoptotic signaling. Biochemical and biophysical research communications. 2013;442:33-7 doi:DOI 10.1016/j.bbrc.2013.10.164
31. Ding L, Huang Y, Du Q, Dong F, Zhao XM, Zhang WL. et al. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochemical and biophysical research communications. 2014;445:497-503 doi:DOI 10.1016/j.bbrc.2014.02.039
32. Sharbati S, Friedlander MR, Sharbati J, Hoeke L, Chen W, Keller A. et al. Deciphering the porcine intestinal microRNA transcriptome. BMC genomics. 2010;11:275. doi:10.1186/1471-2164-11-275
33. Chen T, Xi Q-Y, Ye R-S, Cheng X, Qi Q-E, Wang S-B. et al. Exploration of microRNAs in porcine milk exosomes. BMC genomics. 2014;15:100. doi:10.1186/1471-2164-15-100
34. Goto Y, Hayashi R, MuramatSU T, Ogawa H, Eguchi I, Oshida Y. et al. JPO1/CDCA7, a novel transcription factor E2F1-induced protein, possesses intrinsic transcriptional regulator activity. Bba-Gene Struct Expr. 2006;1759:60-8 doi:DOI 10.1016/j.bbaexp.2006.02.004
35. Korotayev K, Ginsberg D. Many pathways to apoptosis: E2F1 regulates splicing of apoptotic genes. Cell Death Differ. 2008;15:1813-4 doi:Doi 10.1038/Cdd.2008.155
36. Gill RM, Gabor TV, Couzens AL, Scheid MP. The MYC-Associated Protein CDCA7 Is Phosphorylated by AKT To Regulate MYC-Dependent Apoptosis and Transformation. Mol Cell Biol. 2013;33:498-513 doi:Doi 10.1128/Mcb.00276-12
37. Stroynowska-Czerwinska A, Fiszer A, Krzyzosiak WJ. The panorama of miRNA-mediated mechanisms in mammalian cells. Cellular and Molecular Life Sciences. 2014;71:2253-70 doi:10.1007/s00018-013-1551-6
38. Seddiki N, Brezar V, Ruffin N, Lévy Y, Swaminathan S. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology. 2014;142:32-8 doi:10.1111/imm.12227
39. Rossetti S, Sacchi N. RUNX1: A MicroRNA Hub in Normal and Malignant Hematopoiesis. International journal of molecular sciences. 2013;14:1566-88 doi:10.3390/ijms14011566
Author contact
![]() Corresponding author: Prof. Dewen Tong. Telephone: +86-29-87091622; Fax: +86-29-87091032; E-mail: dwtongedu.cn
Corresponding author: Prof. Dewen Tong. Telephone: +86-29-87091622; Fax: +86-29-87091032; E-mail: dwtongedu.cn

 Global reach, higher impact
Global reach, higher impact