10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(9):1036-1048. doi:10.7150/ijbs.12020 This issue Cite
Research Paper
Identification and Characterization of Candidate Chemosensory Gene Families from Spodoptera exigua Developmental Transcriptomes
College of Plant Protection, Nanjing Agricultural University, Nanjing 210095, China /Key Laboratory of Integrated Management of Crop Diseases and Pests (Nanjing Agricultural University), Ministry of Education, China
Abstract
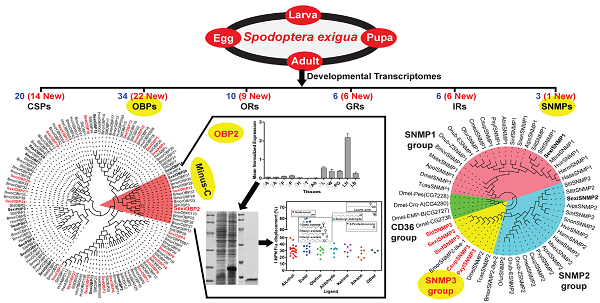
Insect chemosensory genes have been considered as potential molecular targets to develop alternative strategies for pest control. However, in Spodoptera exigua, a seriously polyphagous agricultural pest, only a small part of such genes have been identified and characterized to date. Here, using a bioinformatics screen a total of 79 chemosensory genes were identified from a public transcriptomic data of different developmental stages (eggs, 1st to 5th instar larvae, pupae, female and male adults), including 34 odorant binding proteins (OBPs), 20 chemosensory proteins (CSPs), 22 chemosensory receptors (10 odorant receptors (ORs), six gustatory receptors (GRs) and six ionotropic receptors (IRs)) and three sensory neuron membrane proteins (SNMPs). Notably, a new group of lepidopteran SNMPs (SNMP3 group) was found for the first time in S. exigua, and confirmed in four other moth species. Further, reverse transcription PCR (RT-PCR) and quantitative real time PCR (qPCR) were employed respectively to validate the sequences and determine the expression patterns of 69 identified chemosensory genes regarding to sexes, tissues and stages. Results showed that 67 of these genes could be detected and reconstructed in at least one tissue tested. Further, 60 chemosensory genes were expressed in adult antennae and 52 in larval heads with the antennae, whereas over half of the genes were also detected in non-olfactory tissues like egg and thorax. Particularly, S. exigua OBP2 showed a predominantly larval head-biased expression, and functional studies further indicated its potentially olfactory roles in guiding food searching of larvae. This work suggests functional diversities of S. exigua chemosensory genes and could greatly facilitate the understanding of olfactory system in S. exigua and other lepidopteran species.
Keywords: Spodoptera exigua, developmental transcriptome, chemosensory gene, expression pattern, fluorescence competitive binding assay

 Global reach, higher impact
Global reach, higher impact