10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(10):1190-1203. doi:10.7150/ijbs.11737 This issue Cite
Research Paper
Molecular Physiology of an Extra-renal Cl- Uptake Mechanism for Body Fluid Cl- Homeostasis
1. Institute of Fishery Science, National Taiwan University, Taipei city, Taiwan
2. Institute of Cellular and Organismic Biology, Academia Sinica, Taipei city, Taiwan
3. Department of Life Science, National Taiwan Normal University, Taipei city, Taiwan
Received 2015-1-31; Accepted 2015-7-25; Published 2015-8-15
Abstract
The development of an ion regulatory mechanism for body fluid homeostasis was an important trait for vertebrates during the evolution from aquatic to terrestrial life. The homeostatic mechanism of Cl- in aquatic fish appears to be similar to that of terrestrial vertebrates; however, the mechanism in non-mammalian vertebrates is poorly understood. Unlike in mammals, in which the kidney plays a central role, in most fish species, the gill is responsible for the maintenance of Cl- homeostasis via Cl- transport uptake mechanisms. Previous studies in zebrafish identified Na+-Cl- cotransporter (NCC) 2b-expressing cells in the gills and skin as the major ionocytes responsible for Cl- uptake, similar to distal convoluted tubular cells in mammalian kidney. However, the mechanism by which basolateral ions exit from NCC cells is still unclear.
Of the in situ hybridization signals of twelve members of the clc Cl- channel family, only that of clc-2c exhibited an ionocyte pattern in the gill and embryonic skin. Double in situ hybridization/immunocytochemistry confirmed colocalization of apical NCC2b with basolateral CLC-2c. Acclimation to a low Cl- environment increased mRNA expression of both clc-2c and ncc2b, and also the protein expression of CLC-2c in embryos and adult gills. Loss-of-function of clc-2c resulted in a significant decrease in whole body Cl- content in zebrafish embryos, a phenotype similar to that of ncc2b mutants; this finding suggests a role for CLC-2c in Cl- uptake. Translational knockdown of clc-2c stimulated ncc2b mRNA expression and vice versa, revealing cooperation between these two transporters in the context of zebrafish Cl- homeostasis. Further comparative genomic and phylogenetic analyses revealed that zebrafish CLC-2c is a fish-specific isoform that diverged from a kidney-predominant homologue, in the same manner as NCC2b and its counterparts (NCCs).
Several lines of molecular and cellular physiological evidences demonstrated the cofunctional role of apical NCC2b and basolateral CLC-2c in the gill/skin Cl- uptake pathway. Taking the phylogenetic evidence into consideration, fish-specific NCC2b and CLC-2c may have coevolved to perform extra-renal Cl- uptake during the evolution of vertebrates in an aquatic environment.
Keywords: ionocyte, CLC chloride channel, NCC, coevolution, zebrafish
Introduction
Chloride (Cl-) is one of the most abundant biological anions, and Cl- transporters have been demonstrated to make a major contribution to many cellular processes, including regulating excitability in nerve and muscle, facilitating transepithelial solute transport, and affecting the volume and pH of organelles [1]. Most studies of the mechanisms of body fluid Cl- homeostasis have been conducted using mammals, in which distal convoluted tubules (DCTs) in the kidney reabsorb 5%-10% of the total filtered Na+ and Cl- under normal situations [2]. Na+ and Cl- are raised above electrochemical equilibrium within the DCT cell by the apical Na+-Cl- cotransporter (NCC, SLC12A3) [3]. Na+ appears to exit cells across the basolateral membrane via the ubiquitous Na+-K+-ATPase (NKA). Previous studies demonstrated that one major pathway by which Cl- crosses the basolateral cell membrane is via a member of the CLC family of Cl- channels. For example, Vandewalle et al. (1997) used an antibody recognizing both CLC-K1 and CLC-K2 to demonstrate their abundance in the DCT basolateral membrane [4]. Yoshikawa et al. (1999) adopted in situ hybridization and immunocytochemical techniques to identify the CLC-K expression sites. Whereas CLC-K1 was expressed in the medulla, CLC-K2 was expressed throughout the thick ascending limb, DCT, and connecting tubules [5]. Mutations in human genes encoding CLC-Ka and CLC-Kb caused renal salt wasting and deafness [6]. Moreover, CLC-K1 knockout mice exhibited impaired urine concentration and nephrogenic diabetes insipidus; it is unknown whether such symptoms would also be observed in CLC-K2 knockout mice, as these mice die prematurely [7]. The above results demonstrate the involvement of CLC-Ks in renal DCT Cl- reabsorption. Knowledge regarding this apical NCC-basolateral CLC-K-mediated Cl- uptake pathway in non-mammalian species is very limited [1].
Vertebrates originated from marine organisms; however, some primitive vertebrates (e.g. lampreys) invaded freshwater (FW) habitats before the emergence of their terrestrial relatives [8, 9]. The mechanisms by which body fluid ionic (Cl- and other ions) homeostasis is maintained through ion absorption may have developed in fish following their invasion of FW [9, 10], and these mechanisms of fish (e.g. the FW teleosts) appear to be similar to those of terrestrial vertebrates. In fish, the gills (or the skin during embryonic stages), rather than the kidney, perform the majority of the ion transport associated with body fluid ionic homeostasis [9, 10]. Recently, an extra-renal Cl- uptake mechanism was first identified in certain teleosts that have developed a specific group of ionocytes expressing apical NCC2 (SLC12A10) transporters [11-13]. According to phylogenetic tree analyses, NCC2 orthologues are included in a fish-specific cluster, which diverges from the kidney-predominant NCC clusters; furthermore, the fish NCCs (SLC12A3), like their mammalian orthologues, are mainly expressed in the kidney (Fig. S1 and Table S2) [11-14]. However, the identity and function of the basolateral CLC Cl- channels which cooperate with apical NCC2s during transepithelial Cl- uptake in fish gills/skin remain unclear. Recent studies investigating evolutionary rate covariation demonstrated coevolution of interacting or cofunctional proteins, based on similar (or correlations in) phylogenetic trees and expression levels [15]. Such discoveries raise the possibility that fish-specific CLC members may have also coevolved to participate in fish-specific NCC-mediated Cl- uptake.
Few studies have investigated CLCs in fish species. Tilapia CLC-3 and -5 were cloned and found to be expressed in various organs, including the gills; in vitro functional analysis suggested these CLCs function as intracellular Cl- channels [16]. CLC-3 was also found to be expressed in the gills and kidney of sea bass [17]. However, the data obtained by using sea bass is somewhat conflicting; the expression of CLC-3 mRNA was stimulated by SW (sea water), while Western blot data (with an anti-rat CLC-3 antibody) indicated an increase of CLC-3 protein after transfer from SW to FW [17]. In addition, analysis with the same heterologous antibody revealed higher protein expression in the gills of pufferfish and tilapia acclimated to FW or an ion-deficient environment [18, 19]. Notably, mammalian CLC-3 has been determined to be an intracellular endosomal/lysosomal Cl-/H+-exchanger [20]. Taking all of the above into consideration, we hypothesize that there are other candidates responsible for basolateral Cl- uptake by the NCC2b-mediated Cl- uptake mechanism in fish gill/skin.
The purpose of the present study was to further explore the extra-renal pathway of Cl- uptake in fish. The study was designed to answer the following specific questions: (1) is/are there (a) CLC Cl- channel(s) specifically expressed in the NCC2b- expressing ionocytes of fish gill/skin? (2) Is/are the identified CLC candidate(s) involved in transepithelial Cl- uptake in fish gill/skin? (3) Is/are the identified CLC candidate(s) cofunctional with NCC2b during Cl- uptake in fish gill/skin? (4) Is/ are the CLC candidate(s) fish-specific isoform(s) divergent from the kidney-predominant homologues (similar to NCC2b), thereby reflecting possible coevolution? Zebrafish was previously used to identify NCC2b (SLC12A10.2)-expressing ionocytes [12], and zebrafish NCC2b has been determined to be a major player in Cl- uptake, with a redundant role in Na+ uptake, in the gills and skin [12, 21]. Such earlier findings, combined with the availability of genetic databases for phylogenetic analyses and the applicability of genetic manipulation for functional analyses, led us to use zebrafish as the model for the present study.
Materials and Methods
Experimental animals
The AB strain of zebrafish was obtained from stocks of the Institute of Cellular and Organismic Biology, Academia Sinica. Fish were kept in local tap water (normal FW) with a circulating system at 28.5°C under a 14:10-h light-dark photoperiod. Fish were fed artificially-bred brine shrimp. The experimental protocols were approved by the Academia Sinica Institutional Animal Care and Utilization Committee (approval no. RFIZOOHP220782).
Acclimation experiment
High-Cl- (10 mM) and low-Cl- (0.04 mM) artificial FW were prepared with double-deionized water (Milli-RO60; Millipore, Temecula, CA) supplemented with adequate CaSO4∙2H2O, MgSO4∙7H2O, NaCl, Na2SO4, K2HPO4, and KH2PO4. Other ion concentrations ([Ca2+] = 0.18-0.20 mM; [Mg2+] = 0.18-0.21 mM; and [K+] = 0.16- 0.19 mM) and the pH (6.7- 6.9) of the media were similar to those in the local tap water, except the Na+ was adjusted to 9.98-10.03 mM to minimize disturbance to zebrafish Cl- uptake. Adult zebrafish were acclimated to high- or low- Cl- artificial FW for 1 wk, and the embryos were acclimated for 3-5 days. The artificial waters were replaced every day to keep the ion concentrations and pH stable.
Data mining and bioinformatic analyses
Peptide sequences of CLC homologues in zebrafish, other fish species, and mammalian species were collected from the relevant genome databases (of the Ensembl Genome Browser system; Release 79, March 2015). In silico predictions of full-length zebrafish CLC homologues obtained from the genome were carefully confirmed using the NCBI database. To verify the membership of identified candidates in the core CLC-2 family, the deduced amino acid sequences of predicted and cloned (see below) zebrafish CLC homologues were aligned with other CLC orthologue amino acid sequences available in the ENSEMBL database using ClustalW, and then subjected to phylogenetic inferences using the neighbor-joining (NJ) method. One thousand bootstrap replicate analyses were carried out with Mega 6. Physical gene maps of the verified CLC-2 loci were scaled based on assemblies of the Ensembl Genome Browser. Genes located up- and downstream in the conserved synteny of zebrafish CLC-2 orthologues were subjected to BLAST searches against mammalian genomes to determine the highest score.
Preparation of total RNA
Appropriate amounts of zebrafish tissues were collected and soaked in Trizol reagent (Invitrogen, Carlsbad, CA) for homogenization, and total RNA was then purified following the manufacturer's protocol. The amount of total RNA was determined using a NanoDrop ND-1000 (Thermo Scientific, San Francisco, CA), and the RNA quality was checked by electrophoresis through an RNA-denaturing gel. Total RNA pellets were stored at -20°C.
Cloning of zebrafish clc isoforms
The mRNA was purified from total RNA of zebrafish tissues with a commercial kit (Oligitex; Qiagen, Hilden, Germany). The cDNA templates for cloning and rapid amplification of cDNA ends (RACE) were generated from mRNA using a SuperScript III reverse transcriptase kit (Invitrogen) and a Smart RACE cDNA amplification kit (Clontech, Mountain View, CA), following the respective manufacturers' protocols. For PCR amplification, 3 μl of cDNA were used as a template in a 50 μl final reaction volume containing 0.25 mM dNTP, 2.5 units Ex-Taq polymerase (Takara, Shiga, Japan), and 0.2 μM of each primer. Primer sets were obtained using a bioinformatics method, and sequences are shown in Table S1. The PCR products obtained were subcloned into a pGEM-T Easy vector (Promega, Madison, WI), and the amplicons were sequenced to confirm their identity. The specific primers used for 5' and 3' RACE were designed based on the partial sequences obtained from PCR using the primer sets listed in Table 1. The thermal program used for RACE PCR was that recommended by the manufacturer, and RACE PCR products were also subcloned into the pGEM-T Easy vector and sequenced.
Specific primer sets used for RACE PCR
| Gene | Primer Sequence |
|---|---|
| clc-2c forward 1 | 5' TCGTCATCGGTGCTGGGTTTGGC 3' |
| clc-2c reverse 1 | 5' CAGCACCGATGACGAAAATGGGAAC 3' |
| clc-2c forward 2 | 5' CAGATGGGGACAGAGGGAGAACAGTAAGA 3' |
| clc-2c reverse 2 | 5' CCGAAGCCCGGTGGAAAAGTGAG 3' |
RT-PCR analyses
For PCR amplification, 1 μl of cDNA was used as template in a 25 μl final reaction volume containing 0.25 μM dNTP, 1.25 units Phusion DNA polymerase (New England Biolabs, Ipswich, MA), and 0.2 μM of each primer. Gene expression of zebrafish clc-1a, clc-1b, clc-2a, clc-2b, clc-2c, clc-3, clc-4, clc-5a, clc-5b, clc-6, clc-7, and clc-k in various organs and embryos of zebrafish were examined by RT-PCR. Total RNA samples were extracted from the blood, brain, gills, eyes, heart, intestines, kidneys, liver, muscles, spleen, ovaries, testes, and embryos of zebrafish. Samples were subjected to RT-PCR analysis with the primer sets shown in Table 2.
Quantitative real-time PCR
Real-time PCR was performed with a LightCycler real-time PCR system (Roche, Penzberg, Germany); the reaction mixture possessed a final volume of 10 μl, containing 5 μl 2X SYBR Green I Master (Roche Applied System), 300 nM of the primer pairs, and 20-30 ng cDNA. The standard curve for each gene was checked in a linear range with RPL13a as an internal control. The primer sets for real-time PCR are shown in Table 3.
Specific primer sequences used for RT-PCR analysis
| Gene | Primer Sequence |
|---|---|
| clc-1a forward | 5' AAACACCGTCTGATTTACCCT 3' |
| clc-1a reverse | 5' GCTCCTTTAGTGCGACCAC 3' |
| clc-1b forward | 5' GGATTATGCTAGTGCCAAGAGT 3' |
| clc-1b reverse | 5' AGAGGTCGAGTCCAGCAAAT 3' |
| clc-2a forward | 5' TTCGTCGCCAAAGTGATTG 3' |
| clc-2a reverse | 5' GGGAAGAAGGCTGCCATAC 3' |
| clc-2b forward | 5' CAGTCACGGAGCGCGTCATCACG 3' |
| clc-2b reverse | 5' TCATCACGCAAGACGGTTTAGACGC 3' |
| clc-2c forward | 5' AACTACTGGCGTGCGTTTC 3' |
| clc-2c reverse | 5' GGTATTGCGTTCGCTCTATC 3' |
| clc-3 forward | 5' TCTGGGCTTTGTCTTTCGC 3' |
| clc-3 reverse | 5' CCGTGGACGCATGACTTCT 3' |
| clc-4 forward | 5' GTGTCTCAGTGGCATTTGGA 3' |
| clc-4 reverse | 5' GGGAAGCCGTTGTAGTCG 3' |
| clc-5a forward | 5' TGGGACCACTTTCAAGGAA 3' |
| clc-5a reverse | 5' GGATAAATAGACCAGACGGCAC 3' |
| clc-5b forward | 5' CCCAATGGCATTCCTAAAC 3' |
| clc-5b reverse | 5' TGTCACAACCAGCACCTCC 3' |
| clc-6 forward | 5' CCAAATACCGCATGAGAAAC 3' |
| clc-6 reverse | 5' CAGCATAGCAGACGCCAC 3' |
| clc-7 forward | 5' GTGATGGAGGCGATGCTG 3' |
| clc-7 reverse | 5' GATGTCGGAGTCCTAGAGCC 3' |
| clc-k forward | 5' TCCAAGCCCTGTTTAAGACC 3' |
| clc-k reverse | 5' CGCAAATTGATGCACCCT 3' |
Specific primer sets used for real-time quantitative PCR analysis
| Gene | Primer Sequence |
|---|---|
| clc-2c forward | 5' ATTGAGAAATGGGAGGAGCA 3' |
| clc-2c reverse | 5' GGCATGGAGCCTGTGATG 3' |
| slc12a10.2 forward | 5' GCCCCCAAAGTTTTCCAGTT 3' |
| slc12a10.2 reverse | 5' TAAGCACGAAGAGGCTCCTTG 3' |
In situ hybridization
The cDNA fragment of clc-2c (1105 bp, nt598 -1705) was obtained by PCR and inserted into a pGEM-T easy vector (Promega). After PCR with the T7 and SP6 primers, the products were subjected to in vitro transcription with T7 and SP6 RNA polymerase, respectively (Roche). The quality and concentration of DIG-labeled RNA probes were examined by separation on RNA gels. Embryos (2, 3, 4, and 5 days post fertilization (dpf)) were fixed in 4% paraformaldehyde in a PBS solution at 4°C for over 16 h. Samples were hybridized with the prepared probe in hybridization buffer overnight at 65°C. On the second day, samples were serially washed with 75% hybridization buffer from 25% to 100%, 2X SSC, and finally with 100% DEPC-PBST, for 10 min each at 65°C. Samples were washed with 0.2X SSC buffer at 70°C overnight. On the third day, samples were washed eight times with DEPC-PBST blocking buffer at room temperature for 15 min each. Samples were subsequently incubated in staining buffer [0.1 M Tris (pH 9.5), 50 mM MgCl2, 0.1 M NaCl, and 0.1% Tween-20] three times at room temperature for 5 min each. Samples were stained in a mixture of NBT (100 mg/ml) and BCIP (50 mg/ml) in 10 ml of staining buffer at room temperature for 10 min to several hours (depending on the gene) in the dark. The reaction was stopped with methanol, and samples were washed several times with DEPC-PBST. Finally, samples were fixed with 4% paraformaldehyde for 20 min, washed twice with PBST for 5 min each, and then stored in PBS at 4°C in a dark box for further examination and analysis. Images were obtained with a stereomicroscope (model SZX-ILLD100; Olympus, Tokyo, Japan) or an upright microscope (Axioplan 2 Imaging; Carl Zeiss, Oberkochen, Germany). For double in situ hybridization/immunocytochemistry, the hybridized samples were further subjected to immunocytochemistry as described below.
Immunocytochemistry (ICC)
ICC analysis of CLC-2c was performed as described previously [22]. The anti- CLC-2c antibody was generated by injecting rabbits with a 17-residue synthetic peptide (ALKRPTACEEQLNNNVY; Genomics, Taipei, Taiwan) of CLC-2c; the resulting antibody was used at a dilution of 1:50. Embryos were washed twice with 1X PBST and incubated with an Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG) antibody (1:300 dilution with PBS; Molecular Probes, Invitrogen, Carlsbad, CA) for 2 h at room temperature. After being washed with 1X PBST twice, embryos were incubated with a polyclonal antibody against the N-terminal domain (IKKSRPSLDVLRNPPDD; Genomics) of zebrafish NCC2b (diluted 1:100) at 4°C overnight [21]. Embryos were then washed twice with 1X PBST and incubated with an Alexa Fluor 568 goat anti-rabbit immunoglobulin G (IgG) antibody (1:300 dilution with PBS; Molecular Probes, Invitrogen, Carlsbad, CA) for 2 h at room temperature. Samples were subsequently incubated with DAPI (1:100 with PBST) for 15 min at room temperature, before being washed three times with 1X PBST for 15 min/wash. After rinsing in 1X PBST, embryos were stained with 5 mg/ml DAPI (1:300 dilution with PBST; Life Technologies, Grand Island, NY) for 15 min. Finally, embryos were washed three times with 1X PBST (15 min/wash). Images were acquired with a Leica TCS-SP5 confocal laser scanning microscope (Leica Lasertechnik, Heidelberg, Germany) or an upright microscope (Axioplan 2 Imaging; Carl Zeiss).
Morpholino knockdown
A morpholino-modified antisense oligonucleotide (MO; Open Biosystems, Huntsville, AL) with the sequence 5'-GCCCAAAATGAAGCCCAGGATCATG-3' was used to target +186 to +210 of the coding region of zebrafish clc-2c; a second MO (Open Biosystems, Huntsville, AL), 5'-TTGCCAAAATCAGCCTCTCCCATAT-3', was used to target +74 to +98 of the coding region of zebrafish slc12a10.2. Standard control oligonucleotides (5'-CCTCTTACCTCAGTTACAATTTATA-3'; Gene Tools, Philomath, OR) have no target and no significant biological activity, and were thus used as the control MO. The MOs were re-suspended in 1X Danieau solution, stored at -20°C as a stock solution, and diluted before use to the desired concentrations (1.0, 2.0, or 4.0 ng/embryo). The MO (2.0 ng/embryo) was injected into embryos at the one to four-cell stage using a gas-driven microinjection apparatus (ASI, Eugene, OR). After injection with the clc-2c MO, embryos at 4 and 5 dpf were sampled to measure mRNA and protein.
Whole body Cl- contents
Fifteen zebrafish larvae were briefly rinsed in deionized water and then pooled as one sample. For Cl- content measurements, samples were homogenized with 1 ml deionized water and centrifuged at 13,000 rpm for 30 min. Supernatant was collected, and thereafter mixed with Hg(SCN4) (0.3 g in 95% ethanol) and NH4Fe(SO4)∙12 H2O (30 g in 135 ml 6 N HNO3) solutions for analysis. Cl- concentration was measured by the ferricyanide method with a double-beam spectrophotometer (model U-2000; Hitachi). Standard solutions of Cl- (Merck, Darmstadt, Germany) were used to generate the standard curves.
Western blot analysis. Forty larvae were pooled and homogenized as one sample. Protein samples were separated via 10% SDS-PAGE electrophoresis (100 μg/well). After transfer of the proteins to a polyvinylidene difluoride membrane (Millipore), the blots were incubated with antibodies against CLC-2c, at concentrations of 1:450. The membranes were subsequently incubated with an alkaline-phosphatase-conjugated goat anti-rabbit IgG (dilute 1:2500, Jackson Laboratories), and then developed with 5-bromo-4-chloro-3-indolyphosphate/nitroblue tetrazolium. β-actin was used as an internal control. Western blot images were captured by using the UVP BioSpectrum 600 Image System (UVP Inc., CA, USA). The intensities of CLC-2c protein and β-actin bands were quantified using the software Image-Pro Plus (Media Cybernetics, Maryland, USA).
Statistical analyses
Values are presented as means ± SD and were compared using Student's t-test.
Results
Molecular and phylogenetic characterization of zebrafish CLC family genes
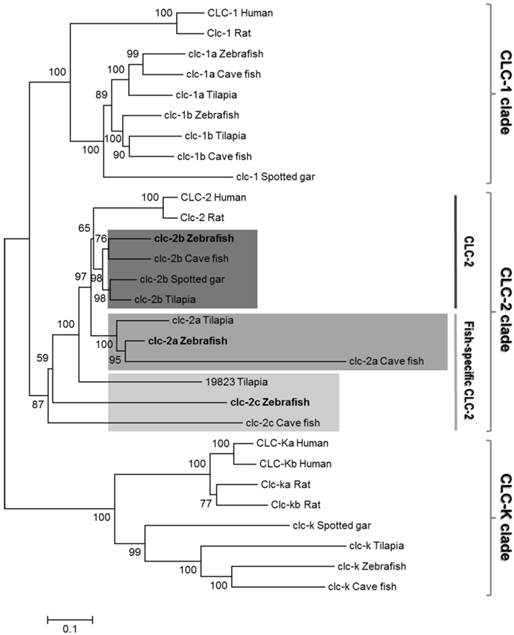
Six zebrafish genes (clc-1a, clc-1b, clc-2a, clc-2b, clc-2c, and clc-k) of the clc family were predicted based on analysis using the NCBI and Ensembl genome database (release 79). To confirm the nature of these zebrafish clc genes, phylogenetic analysis was performed by using the neighbor-joining method (Fig. 1), and their genomic loci were compared between species by using genome sequence databases (Fig. S2 and Fig. S3). Phylogenetic analysis revealed that the six members of the zebrafish CLC family can be classified into three main branches. The two subtypes of CLC-2 (zebrafish CLC-2a and CLC-2b) formed a monophyletic group; zebrafish CLC-2a and CLC-2b were placed in the conventional CLC-2 (CLC-2b) and fish-specific CLC-2 (CLC-2a) group (according to [23]), respectively, and such placement was derived based on multiple bioinformatics data, as shown below. The novel zebrafish clc-2c gene was found to nest in an outgroup with other fish species (Fig. 1). The clc-2c gene (the major target of the following experiments) was cloned and sequenced from full-length cDNAs containing the 5' and 3' untranslated regions. The zebrafish clc-2c gene (accession no. ENSDARG00000060439) is located on chromosome 15 (genome database ZV9; Fig. S2), and is predicted to encode a protein of 810 amino acids. Zebrafish CLC-2a and CLC-2b have 44% and 53% identity with CLC-2c, respectively, while CLC-2a and CLC-2b exhibit 67% identity (Table S3). The zebrafish clc-2a gene (accession no. ENSDARG00000062427) is located on chromosome 2 (genome database ZV9; Fig. S2), and the zebrafish clc-2b gene (accession no. ENSDARG00000074681) is located on Scaffold (genome database ZV9; Fig. S2). As compared with their human counterparts, zebrafish CLC-2a, CLC-2b, and CLC-2c have 63%, 69%, and 44% identity, respectively (Table S3).
The gene loci of clc-2a, -2b, and -2c are located on different chromosomes, indicating these three CLC-2 isoforms have their own respective syntenies in zebrafish. To establish whether the three zebrafish CLC-2 proteins were properly annotated (in accordance with the instructions provided by ZFIN), we closely examined the syntenic regions of the three isoforms. The zebrafish orthologues of the neighboring genes located in the conserved syntenies of human (Homo sapiens), Xenopus (Xenopus tropicalis), and spotted gar (Lepisosteus oculatus) CLC-2 were all examined using the available genome databases (Figs. S2 and S3). The genomic locus of heat shock protein b12 (HSPB12), which neighbors CLC-2b, was conserved among all teleosts examined. Moreover, three genes neighboring CLC-2 in spotted gar (genes encoding (i) family with sequence similarity 131 member A (FAM131A), (ii) thrombopoietin (THPO), and (iii) eukaryotic translation initiation factor 4 gamma 1 (EIF4G1)) were also present on zebrafish chromosome 2, together with clc-2a. Another CLC-2 neighboring gene, which encodes the chordin (bone morphogenetic protein antagonist) (CHD), was also found to be adjacent to the zebrafish orthologue of clc-2c and teleost clc-2b genes. However, orthologues of zebrafish clc-2c were not identified by screening neighboring genomic proteins (such as CHD and integrin-linked kinase-associated serine/threonine phosphatase (ILKAP)) in other teleosts. In addition, the loci containing the genes encoding the proteins neighboring zebrafish CLC-2 were found to form a paralogon in spotted gar, a primitive FW fish.
The mRNA expression patterns of the zebrafish clc Cl- channel family
The mRNA expressions of the zebrafish clc Cl- channel family in various tissues were examined by RT-PCR, with β-actin and RPL13a as internal controls. As shown in Fig. 2a, CLC Cl- channel family members exhibited different expression patterns in the adult zebrafish tissues. While clc-3, -4, -5a, -5b, -6, and -7 were expressed in most tissues, clc-1a, -1b, -2a, -2c, and clc-k were expressed in only one or two tissues. Of the twelve isoforms, ten (clc-1a,-2a, -2b, -2c, -3, -4, -5a, -5b, -6, and -7) were expressed in the gills. The clc-2c gene was mainly expressed in the gill, with considerably lower expression in the kidney; clc-2b, meanwhile, was obviously expressed in the spleen, with slight expression in the gills and muscle.
A phylogenetic tree of CLC chloride channel family proteins. The relationships among conventional CLC-2 and fish-specific CLC-2 are shown. The zebrafish genome contains not only one clc-2 gene but also the fish-specific clc-2 gene, which is specifically expressed in the yolk sac membrane and gill. Zebrafish clc chloride channels are shown in bold. The phylogenic tree was constructed using the neighbor-joining method with ClustalW [51] and MEGA6. Numbers indicate bootstrap values and the scale bar represents a genetic distance of 0.05 amino-acid substitutions per site. The NCBI or Ensembl accession numbers are as listed in Table S1.
The mRNA expression of the clc chloride channel family in zebrafish. (a) RT-PCR analysis of the mRNA expression levels of the clc chloride family in various tissues of adult zebrafish. β-actin and RPL13a were used as internal controls. (b) Whole mount in situ hybridization with a specific clc-2c RNA probe. Embryos at 2 (b), 3 (c), 4 (d), and 5 days postfertilization (dpf) (e), and gills (f) were examined. clc-2c mRNA signals were observed in the skin of yolk sac from 2 dpf (b-e) and were also clearly present in the gill epithelial cells (f). Arrows indicate clc-2c mRNA signals. Scale bar, 100 μm.
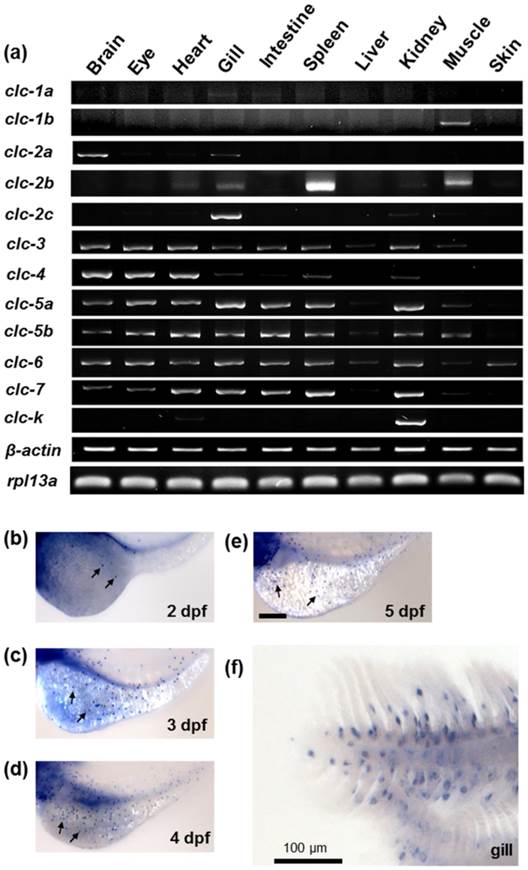
Whole mount in situ hybridization was used to examine the mRNA expression of clc gene family members in zebrafish embryos at different developmental stages. Of the family members cloned in the present study, only clc-2c exhibited an ionocyte pattern [24] of mRNA expression in the embryonic skin of zebrafish; consequently, all subsequent experiments focused on the clc-2c isoform. As shown in Figs. 2b-f, clc-2c was expressed in a certain group of ionocytes in the embryonic skin and adult gills of zebrafish; clc-2c mRNA began to be expressed in skin ionocytes at 2 days post-fertilization (dpf) (Fig. 2b). Following embryonic development, clc-2c signals increased throughout the entire yolk and yolk tube (Figs. 2b-e), and the expression was also clearly visible in the gill filament and lamella of zebrafish (Fig. 2f).
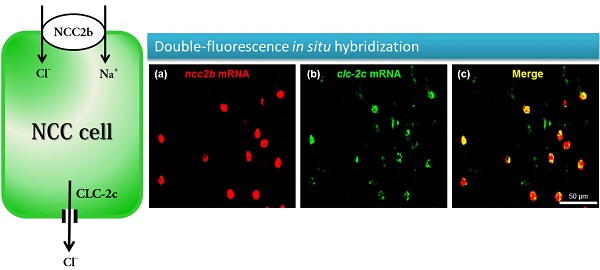
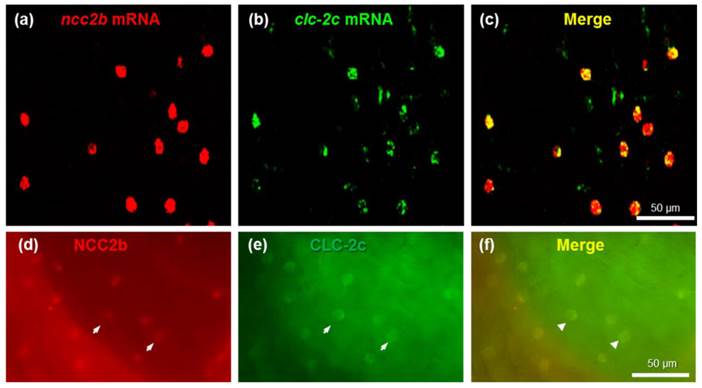
Double-fluorescence in situ hybridization (a-c) and double-fluorescence immunocytochemistry (d-f) in the yolk sac of 5 dpf zebrafish embryos. (a): ncc2b mRNA (red signal). (b): clc-2c mRNA (green signal). (c): Merged image of (a) and (b). Scale bar, 50 μm. (d): NCC2b protein (red signal). Arrows indicate NCC2b signals. (e): CLC-2c protein (green signal). Arrows indicate CLC-2c signals. (f): Merged image of (d) and (e). Arrowheads indicate merged signals of NCC2b and CLC-2c. Scale bar, 50 μm.
Double labeling of CLC-2c protein and nuclei staining using 4',6-diamidino-2-phenylindole (DAPI). (a): CLC-2c protein (green signal). (b): DAPI (blue signal). (c): Colocalization of (a) and (b). (d): z-plane image of (c), obtained by confocal microscopic image analysis. Scale bar, 10 μm.
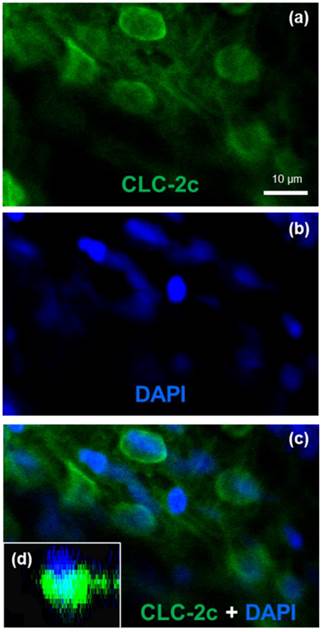
Localization of clc-2c mRNA and protein in zebrafish ionocytes
To identify the types of ionocytes expressing clc-2c, we performed double-fluorescence in situ hybridization (Figs. 3a-c) and double-fluorescence ICC (Figs. 3d-f) on the yolk sac of 5 dpf zebrafish embryos. As shown in Figs. 3a-c, clc-2c mRNA signals were colocalized with ncc2b (slc12a10.2), a marker for the type of Na+-Cl--cotransporter (NCC) ionocyte in zebrafish [12]. Consistent with the mRNA results, CLC-2c protein signals were also colocalized with those of NCC2b (Figs. 3d-f).
Cellular localization of CLC-2c in zebrafish ionocytes
To identify the cellular localization of the transporter in zebrafish NCC cells, we conducted double-fluorescence ICC by staining cells with 4,6-diamidino-2-phenylindole (DAPI, a marker for nucleus staining) and an antibody against CLC-2c, and examining the samples under a confocal microscope (Fig. 4). Z-plane images revealed a DAPI-labeled nucleus with an apical membrane at the upper side, and CLC-2c protein signals were located at the lateral and lower sides of the nucleus (Fig. 4d), indicating basolateral localization of CLC-2c in the cell.
The effect of translational knockdown of clc-2c on its protein expression in 5 dpf zebrafish embryos (a) and whole body Cl- contents of 3 dpf embryos (b). (a): Western blot signal of CLC-2c (~100 kDa molecular mass) was significantly decreased in CLC-2c morphants (clc-2c MO) as compared with that in the control group (ctrl MO). The histogram shows band densitometric values; CLC-2c protein expression was decreased in embryos injected with 2.0 ng MO (compared to embryos injected with ctrl MO). Data are shown as means ± SE (n = 6). *Student's t-test (p < 0.05), significantly different from the respective control. (b): Whole body Cl- contents were significantly decreased in CLC-2c morphants as compared with those in the control group. Data are shown as means ± SE (n = 6-8). *Student's t-test (p < 0.05), significantly different from the respective control.
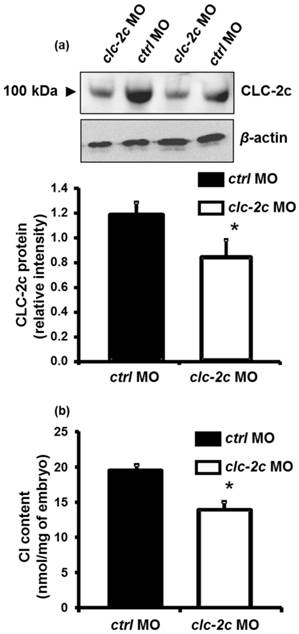
Effects of clc-2c knockdown on whole body Cl- contents in zebrafish embryos
To determine the role of CLC-2c in the Cl- uptake function of zebrafish NCC cells, we performed loss-of-function experiments with specific clc-2c MOs. The phenotypes and behaviors of the clc-2c morphants appeared normal compared with embryos which were injected with control MOs. Western blot revealed that the protein signal (~100 kDa) was considerably decreased in embryos injected with clc-2c MO (2.0 ng/embryo) at 5 dpf as compared with levels in the control groups, confirming the effectiveness and specificity of the MO (Fig. 5a). Ultimately, whole body ion contents in zebrafish embryos reflect the ion net fluxes. Therefore, we measured whole body Cl- contents of zebrafish embryos injected with the clc-2c MO (2.0 ng/embryo) at 3 dpf. As shown in Fig. 5b, the Cl- content in morphants was significantly lower than the control morphants, indicating a disruption in Cl- uptake.
Effects of altering environmental Cl- concentrations on mRNA expression of clc-2c or related transporter genes in zebrafish gills and embryos
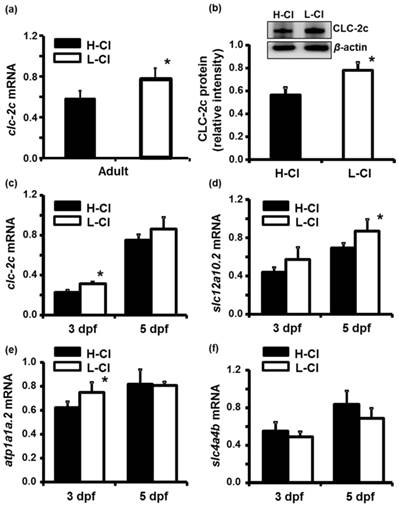
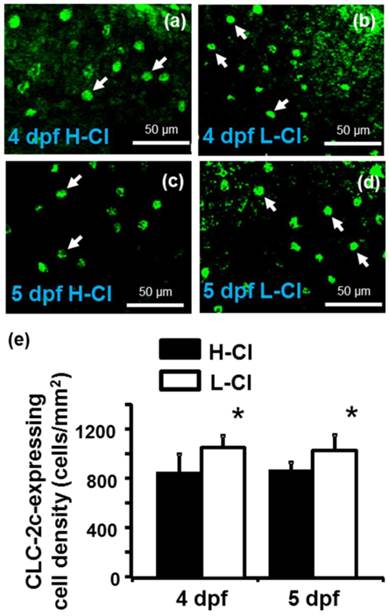
Real-time PCR was conducted to examine clc-2c mRNA expression in the gills of zebrafish acclimated to different Cl- concentrations (0.04 and 10 mM) for 1 week; environmental ion levels were found to have a clear effect on clc-2c expression (Fig. 6a). A low-Cl- environment was found to result in up-regulation of clc-2c mRNA expression in zebrafish gills (~1.32-times higher than the high-Cl- environment; Fig. 6a). Western blot against CLC-2c in the gills of adult zebrafish provided supporting data (Fig. 6b). We proceeded to examine the effects of a low-Cl- environment on the mRNA levels of slc12a10.2 (NCC2b), clc-2c, atp1a1a.2 (NKA alpha-1 subunit subtype), and slc4a4b (sodium bicarbonate cotransporter 1b) in zebrafish embryos at 3 or 5 dpf. Real-time PCR was used to show that the expression levels of both clc-2c (Fig. 6c) and atp1a1a.2 (Fig. 6e) were significantly increased in 3 dpf zebrafish embryos under low-Cl- as compared with those under high-Cl-. On the other hand, low-Cl- did not result in increased expression of slc12a10.2 until 5 dpf (Fig. 6d), and did not affect slc4a4b mRNA expression at either time point (Fig. 6f). The mRNA results were supported by observations of CLC-2c protein expression. As shown in Figs. 7a-e, the cell densities of CLC-2c-expressing ionocytes were increased in 4 and 5 dpf embryos of low-Cl- environments while compared to those kept under a high-Cl- environment, as determined using ICC (Figs. 7a-e).
Interactions of Cl- transporters in NCC-expressing cells
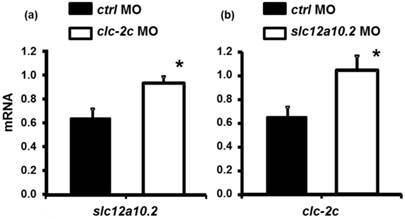
As shown in Fig. 8a, loss-of-function of clc-2c increases the expression of slc12a10.2 mRNA as compared to those in control morphants. A similar result was observed in slc12a10.2 morphants - clc-2c mRNA expression was increased as compared to the control morphants (Fig. 8b).
Effects of high-Cl- (H-Cl) and low-Cl- (L-Cl) artificial waters on mRNA and/or protein expression of CLC-2c and other transporters in adult gills and embryos. (a): clc-2c mRNA in the gills with 7d acclimation. (b): Western blot and densitometric analysis of CLC-2c protein in the gills with 7d acclimation. (c): clc-2c mRNA in embryos. (d): slc12a10.2 in embryos. (e): atp1a1a.2 (NKA alpha-1 subunit subtype) in embryos. (f): slc4a4b (sodium bicarbonate cotransporter 1b) mRNA in embryos. The mRNA expression levels of clc-2c, slc12a10.2, and atp1a1a.2 were higher in L-Cl than in H-Cl, while that of slc4a4b showed no significant difference. Data are shown as means ± SE (n = 3-5). *Student's t-test (p < 0.05), significantly different from the respective control.
Effects of high-Cl- (H-Cl) and low-Cl- (L-Cl) artificial waters on the cell densities of CLC-2c-expressing ionocytes in embryos. (a, b): Immunocytochemical images of CLC-2c (arrow) in embryos acclimated for 4 d. Scale bar, 50 μm. (c, d): Immunocytochemical images of CLC-2c (arrow) in embryos acclimated for 5 d. Scale bar, 50 μm. (e): Comparison of CLC-2c-expressing cell numbers in 4 and 5 dpf zebrafish embryos raised in artificial waters with different Cl- concentrations (n = 6-8). Data are shown as means ± SE. * Student's t-test (p < 0.05), significantly different from the respective control.
The effect of the clc-2c MO (2.0 ng/embryo) on slc12a10.2 mRNA expression (a) and effects of the slc12a10.2 MO (2.0 ng/ embryo) on clc-2c mRNA expression (b) in 5 dpf zebrafish embryos. (a): Real-time PCR analysis of slc12a10.2 mRNA expression in clc-2c morphants. (b): Real-time PCR analysis of clc-2c mRNA expression in slc12a10.2 morphants. Data are shown as means ± SE (n = 4-5). *Student's t-test (p < 0.05), significantly different from the respective control.
Discussion
In the present study, twelve CLC family members, clc-1a, clc-1b, clc-2a, clc-2b, clc-2c, clc-3, clc-4, clc-5a, clc-5b, clc-6, clc-7, and clc-k, were identified in zebrafish; of these, only clc-2c was expressed in ncc2b (slc12a10.2)-expressing ionocytes in the gills and skin. Loss-of-function of clc-2c via translational knockdown resulted in decreased Cl- content in zebrafish embryos. Furthermore, a low-Cl- environment enhanced both the mRNA and protein expression of clc-2c. Knockdown of clc-2c expression resulted in up-regulation of slc12a10.2 mRNA and vice versa, indicating that these two transporters cooperate in zebrafish Cl- homeostasis. Furthermore, zebrafish CLC-2c, similar to NCC2b, has a distinct role from its kidney-predominant homologue (CLC-K and NCC, respectively), suggesting neofunctionalization of these genes enabled the development of an extra-renal Cl- uptake mechanism. The twelve CLC chloride channel members possess different tissue expression patterns in zebrafish adults, based on our RT-PCR data (Fig. 2a). The expression of clc-1a is too low for its expression pattern to be effectively described by using RT-PCR. Other members of this family, including clc-3, -4, -5a, -5b, -6, and -7, were broadly expressed in most tissue studies, similar to their counterparts in mammals [20]. On the other hand, the expression of clc-1b, -2a, -2c, and -k were confined to distinct tissues. The clc-1b gene was mainly expressed in the muscle, as in mammals [20], clc-2a mainly in the brain and at a low level in the gills, and clc-2b mostly in the spleen and muscle. Notably, clc-2c and clc-k were specifically expressed in the gills and kidney, respectively, in a similar manner to ncc2b (slc12a10.2) and the kidney-predominant form of ncc (slc12a3), which exhibit predominant expression in these two organs, respectively [12]. This result suggested that CLC-2c, like mammalian CLC-K, was likely to be involved in branchial Cl- uptake [25]. Therefore, our subsequent experiments focused on CLC-2c.
Protein and mRNA localization experiments supported our hypothesis that CLC-2c protein has a role in Cl- uptake. In situ hybridization with specific probes was used to screen the CLC gene family, demonstrating that only clc-2c shows a reminiscent pattern of ionocytes in adult gill and embryonic skin (Figs. 2b-f). Double in situ hybridization and ICC experiments further indicate both mRNA and protein signals of clc-2c were specifically localized in NCC-expressing ionocytes in the skin of zebrafish embryos (Fig. 3). Subsequent double-labeling experiments detected basolateral localization of CLC-2c in ionocytes (Fig. 4). These colocalization findings suggest that CLC-2c is involved in the Cl- uptake mechanism of zebrafish NCC ionocytes. The NCC ionocytes in zebrafish gill and skin have been demonstrated to perform Cl- (and Na+) uptake, similar to mammalian DCT cells, which express apical NCC and basolateral CLC-Kb, and are responsible for NaCl reabsorption [4, 26-28]. Further experiments provided several lines of physiological and molecular evidence supporting a role for CLC-2c in the Cl- uptake mechanism in NCC ionocytes. Acclimation to low-Cl- artificial water was reported to enhance Cl- uptake [12, 29-31] and stimulate mRNA and protein expression of clc-2c in the adult gills and embryos of zebrafish (Figs. 6-7). A further line of evidence was obtained from the finding that clc-2c knockdown reduced body Cl- content (Fig. 6a). In mammals, CLC-2 loss-of-function was found to impair extracellular ion homeostasis [32]; a mutation of clc-k also resulted in severe renal salt loss [20]. Taken together, these findings support the role of the zebrafish CLC-2c Cl- channel in transepithelial Cl- uptake via regulation of basolateral Cl- efflux. Indeed, a recent electrophysiological study characterized the functional properties of the three zebrafish CLC members (CLC-2a, -2b, and -2c) by detecting inward rectified Cl- current of the transporters overexpressed in Xenopus oocytes (however, CLC-2c was not appropriately expressed due to technical problems) [23].
CLC-K is the major isoform participates in the transepithelial Cl- transport that is associated with body fluid Cl- homeostasis in mammals [25]. Nevertheless, several mammalian studies have suggested that CLC-2 is also involved in body fluid Cl- regulation. Gyomorey (2000) demonstrated that CLC-2 protein is predominantly localized to the tight junction complex between adjacent intestinal epithelial cells [33]. Subsequent electrophysiological evidence, coupled with knowledge of its basolateral localization in the distal colon, indicated that CLC-2 may mediate basolateral Cl- exit during the NaCl absorption process [34, 35]. CLC-2 was also identified to contribute to transepithelial Cl- secretion in both rat and human airways [36]. A recent study by Kato and colleagues (2011) reported that CLC-2, NKA, and a K+ channel (Kir 7.1) are major players in the basolateral step, once Na+ and Cl- are apically absorbed in the mammalian intestine [37]. These findings further support the hypothesis that, like its mammalian orthologues, zebrafish CLC-2c plays an important role in the transepithelial Cl- uptake mechanism of NCC ionocytes.
The roles of CLC Cl- channels in the mechanisms underlying transepithelial Cl- secretion/reabsorption and related diseases in mammalian kidneys have been well studied [1]. However, the function of CLC-2c in vertebrates other than mammals is remained unknown. The fish clc-3 gene was first cloned from tilapia, and reported to be expressed in various osmoregulatory organs, including gills [38]. Certain later studies suggested that CLC-3 is responsible for absorbing Cl- in the gills for body fluid Cl- homeostasis in FW-acclimated fishes. Use of a heterologous antibody identified the CLC-3 protein is localized to the basolateral membrane of a certain group of gill ionocytes; furthermore, its expression was reported to be stimulated by a hypoosmotic or low-Cl- environment in pufferfish, milkfish, and tilapia [18, 39, 40]. In sea bass, higher level of CLC-3 protein was observed in FW than in SW gills; however, higher mRNA expression was found in the SW gills, conflicting with the protein data [17]. The apparent contradiction between the present CLC-2c and earlier CLC-3 results appears to be due to the differences in the antibodies used and the mRNA data provided. Our novel antibody was designed against a zebrafish CLC-2c-specific region. The heterologous antibody used in earlier studies was raised against a fragment of the rat CLC-3 sequence that is located at a conserved region of CLC Cl- channel proteins, raising the possibility that it may have been cross-reactive with other CLC isoforms. The possibility that this antibody may recognize other CLC Cl- channel members, and not just CLC-3, should be taken into consideration. Moreover, the mRNA and protein data described here for CLC-2c are consistent, supporting our conclusions. We also note that CLC-3, like other intracellular CLCs, is considered to be a Cl-/H+-exchanger [41]. The CLC-3 is located in the intracellular compartment and functions in the acidification of synaptic vesicles and endosomes [20]. Nevertheless, the possibility that certain fish species (sea bass, tilapia, etc.) employ different CLC paralogues used in their gill transepithelial Cl- uptake mechanisms cannot be excluded.
The present study also provides evidence for the cooperation of CLC-2c, NCC2b, and other transporters in the context of transepithelial Cl- transport in NCC ionocytes. Acclimation to low-Cl- artificial FW stimulates not only clc-2c expression, but also slc12a10.2 (NCC2b) and atp1a1a.2 (NKA alpha-1 subunit subtype) (Figs. 6-7). Furthermore, loss-of-function of clc-2c resulted in up-regulated mRNA expression of ncc2b, while knockdown ncc2b caused an enhancement of clc-2c (Fig. 8), suggesting mutual compensation between these transporters. By integrating the previous findings with our present results, we are able to more comprehensively interpret the mechanisms of Cl- uptake in zebrafish NCC-expressing cells [9, 12, 42-44]. In the apical membrane, NCC2b transports ambient Na+ and Cl- into cells [12]. To achieve transepithelial Cl- uptake, basolateral CLC-2c extrudes cytosolic Cl- out of the cells, probably down the electric gradient created by basolateral NKA (atp1a1a.2, the specific isoform expressed in NCC ionocytes [45]). This Cl- uptake mechanism within zebrafish skin/gill ionocytes appears to be similar to that in mammalian DCT cells, which involves apical NCC and basolateral CLC-Kb [2].
It remains formally possible that in addition to NCC-mediated CLC-2c Cl- uptake, other pathways involving SLC26 anion transporters may also participate in Cl- uptake in fish acclimated to FW. Several SLC26 anion transporters (SLC26A3, -4, and/or -6) were found to be localized to certain groups of ionocytes in zebrafish and the euryhaline Atlantic stingray, and were proposed to contribute to Cl- uptake through apical Cl-/HCO3- exchange [46-48]. One may ask whether CLC member(s) or other transporters are responsible for the basolateral transport of the SLC26-mediated Cl- uptake pathways. In zebrafish, SLC26s are mostly expressed in Na+-K+-ATPase-rich ionocytes [46]. According to the present colocalization experiments in zebrafish, of the 12 members, only CLC-2c shows an ionocyte pattern and is specifically expressed in NCC-expressing ionocytes (Figs. 2-4); these cells are different from the Na+-K+-ATPase-rich type ionocytes [12]. It seems possible that the hypothetical association of SLC26s with candidates other than the CLC member(s) may be involved in (a) separate extra-renal Cl- uptake pathway(s) in FW fish, and this issue demands further investigation.
The present study utilized a comparative genomics approach to show that the three zebrafish CLC-2 genes are located in genomic regions that may share a common evolutionary history (Fig. S2). Our in silico analyses indicated that the zebrafish CLC-2 genes and their neighboring protein families apparently form a paragon among teleosts. The paragon is especially apparent in spotted gar (L. oculatus), and may have undergone tandem duplication during vertebrate evolution. As a possible consequence, clc-2b and clc-2c (accession no. ENSAMXG00000011800) in cave fish (Astyanax mexicanus) are derived from the same chromosomal region, with neighboring genomic loci. On one hand, the syntenies of CLC-2b were conserved between teleosts that have this isoform, e.g. cave fish (A. mexicanus), tilapia (Oreochromis niloticus), spotted green pufferfish (Tetraodon nigroviridis), zebrafish (D. rerio), and platyfish (CLC-2c, accession no. ENSXMAG00000008109) (Xiphophorus maculatus). Based on the phylogenetic analysis, as well as the comprehensive genomic examination of the conserved syntenies of CLC-2b in other vertebrate species, it appears that teleost CLC-2b may be functionally conserved and clustered with CLC-2, the latter having been well-studied in other higher vertebrates (Fig. 1) [34]. On the other hand, it is now evident that CLC-2c and its genomic neighbors have undergone numerous gene re-arrangement events, resulting in various permutations of the ancestral genomic arrangement. However, this novel isoform has only been identified in zebrafish and cave fish, and not in the other teleosts examined. Moreover, the genomic synteny analysis and phylogenetic examination collectively indicate that the CLC-2b genes in zebrafish and cave fish may also be derived from the ancestral CLC-2. Consistent with this hypothesis, our phylogenetic tree analysis (Fig. 1) identified two CLC-2 clusters: conventional CLC-2 and fish-specific CLC-2. The data on zebrafish CLC-2c and mammalian CLC-2 [34, 35, 37] (see above) suggest that the divergent CLC-2 paralogues may mediate Cl- absorption in gnathostoma, in tissues or organs other than the kidney, in which Cl- absorption is mainly carried out by CLC-Ks (in a cluster divergent from the CLC-2 paralogues).
Recent studies of evolutionary rate covariation have demonstrated coevolution of many interacting proteins among different plant and animal species; furthermore, such coevolution or correlated evolution was observed not only in paired proteins with physical contact, but also in those with cofunctions or correlated expression levels [49]. The fish-specific Cl- transporters, CLC-2c and NCC2b, may also have undergone coevolution or correlated evolution. As apparent from our phylogenetic tree, zebrafish CLC-2c and its fish orthologues have diverged from the kidney cluster, which includes CLC-K orthologues from fishes and mammals (Fig. 1). The CLC-K orthologues are specifically expressed and function in the kidneys of mammals and zebrafish (expression data only; Fig. 2a.), while zebrafish CLC-2c is predominantly expressed in the gills (and the skin in embryonic stages), and serves as an extra-renal transporter for Cl- uptake (see above). In a similar manner to CLC-2c, zebrafish NCC2b is also included in a fish-specific cluster which diverged from the kidney-predominant NCC cluster [12] (Fig. S1), and specifically performs gill/skin Cl- uptake [12]. Indeed, interacting protein families have been shown to coevolve, with the interacting members of different families displaying similar phylogenetic trees [50]. Although evolutionary rate covariation data were not examined in the present study, the similar phylogenetic trees and the correlated expression levels suggest that zebrafish CLC-2c and NCC2b, the two cofunctional transporters, may have coevolved to perform extra-renal Cl- uptake, which was one of the pre-requisites to allow vertebrates to maintain their body fluid homeostasis on the land. It is important for our understanding of vertebrate evolution to determine whether the two cofunctional transporters also underwent coevolution or a correlated evolution event in other fish species; answering this question will necessitate further studies in additional species.
Supplementary Material
Tables S1-S3, Figures S1-S3.
Acknowledgements
We thank the Institute of Cellular and Organismic Biology Core Facility and the Taiwan Zebrafish Core Facility for technical support during the experiments. This work was supported by grants (to P.-P.H.) from Academia Sinica and the Ministry of Science and Technology of Taiwan.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jentsch TJ, Stein V, Weinreich F, Zdebik AA. Molecular structure and physiological function of chloride channels. Physiological reviews. 2002;82:503-68 doi:10.1152/physrev.00029.2001
2. Reilly RF, Ellison DH. Mammalian distal tubule: physiology, pathophysiology, and molecular anatomy. Physiological reviews. 2000;80:277-313
3. Velazquez H, Good DW, Wright FS. Mutual dependence of sodium and chloride absorption by renal distal tubule. The American journal of physiology. 1984;247:F904-11
4. Vandewalle A, Cluzeaud F, Bens M, Kieferle S, Steinmeyer K, Jentsch TJ. Localization and induction by dehydration of ClC-K chloride channels in the rat kidney. The American journal of physiology. 1997;272:F678-88
5. Yoshikawa M, Uchida S, Yamauchi A, Miyai A, Tanaka Y, Sasaki S. et al. Localization of rat CLC-K2 chloride channel mRNA in the kidney. The American journal of physiology. 1999;276:F552-8
6. Schlingmann KP, Konrad M, Jeck N, Waldegger P, Reinalter SC, Holder M. et al. Salt wasting and deafness resulting from mutations in two chloride channels. The New England journal of medicine. 2004;350:1314-9 doi:10.1056/NEJMoa032843
7. Matsumura Y, Uchida S, Kondo Y, Miyazaki H, Ko SB, Hayama A. et al. Overt nephrogenic diabetes insipidus in mice lacking the CLC-K1 chloride channel. Nature genetics. 1999;21:95-8 doi:10.1038/5036
8. Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I. et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311-6 doi:10.1038/nature12027
9. Hwang PP, Lin LY. Gill ion transport, acid-base regulation and nitrogen excretion. In: (ed.) Evans D, Claiborne JB, Currie S. Fourth edition. Boca Raton: CRC Press. 2014
10. Evans DH, Piermarini PM, Choe KP. The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiological reviews. 2005;85:97-177 doi:10.1152/physrev.00050.2003
11. Inokuchi M, Hiroi J, Watanabe S, Lee KM, Kaneko T. Gene expression and morphological localization of NHE3, NCC and NKCC1a in branchial mitochondria-rich cells of Mozambique tilapia (Oreochromis mossambicus) acclimated to a wide range of salinities. Comparative biochemistry and physiology Part A, Molecular & integrative physiology. 2008;151:151-8 doi:10.1016/j.cbpa.2008.06.012
12. Wang YF, Tseng YC, Yan JJ, Hiroi J, Hwang PP. Role of SLC12A10.2, a Na-Cl cotransporter-like protein, in a Cl uptake mechanism in zebrafish (Danio rerio). American journal of physiology Regulatory, integrative and comparative physiology. 2009;296:R1650-60 doi:10.1152/ajpregu.00119.2009
13. Hsu HH, Lin LY, Tseng YC, Horng JL, Hwang PP. A new model for fish ion regulation: identification of ionocytes in freshwater- and seawater-acclimated medaka (Oryzias latipes). Cell and tissue research. 2014;357:225-43 doi:10.1007/s00441-014-1883-z
14. Campean V, Kricke J, Ellison D, Luft FC, Bachmann S. Localization of thiazide-sensitive Na(+)-Cl(-) cotransport and associated gene products in mouse DCT. American journal of physiology Renal physiology. 2001;281:F1028-35
15. Clark NL, Alani E, Aquadro CF. Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome research. 2012;22:714-20 doi:10.1101/gr.132647.111
16. Miyazaki H, Kaneko T, Uchida S, Sasaki S, Takei Y. Kidney-specific chloride channel, OmClC-K, predominantly expressed in the diluting segment of freshwater-adapted tilapia kidney. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15782-7 doi:10.1073/pnas.242611099
17. Bossus M, Charmantier G, Blondeau-Bidet E, Valletta B, Boulo V, Lorin-Nebel C. The ClC-3 chloride channel and osmoregulation in the European sea bass, Dicentrarchus labrax. Journal of comparative physiology B, Biochemical, systemic, and environmental physiology. 2013;183:641-62 doi:10.1007/s00360-012-0737-9
18. Tang CH, Lee TH. The effect of environmental salinity on the protein expression of Na+/K+-ATPase, Na+/K+/2Cl- cotransporter, cystic fibrosis transmembrane conductance regulator, anion exchanger 1, and chloride channel 3 in gills of a euryhaline teleost, Tetraodon nigroviridis. Comparative biochemistry and physiology Part A, Molecular & integrative physiology. 2007;147:521-8 doi:10.1016/j.cbpa.2007.01.679
19. Tang CH, Lee TH. Ion-deficient environment induces the expression of basolateral chloride channel, ClC-3-like protein, in gill mitochondrion-rich cells for chloride uptake of the tilapia Oreochromis mossambicus. Physiological and biochemical zoology: PBZ. 2011;84:54-67 doi:10.1086/657161
20. Stauber T, Weinert S, Jentsch TJ. Cell biology and physiology of CLC chloride channels and transporters. Comprehensive Physiology. 2012;2:1701-44 doi:10.1002/cphy.c110038
21. Chang WJ, Wang YF, Hu HJ, Wang JH, Lee TH, Hwang PP. Compensatory regulation of Na+ absorption by Na+/H+ exchanger and Na+-Cl- cotransporter in zebrafish (Danio rerio). Frontiers in zoology. 2013;10:46. doi:10.1186/1742-9994-10-46
22. Inoue D, Wittbrodt J. One for all--a highly efficient and versatile method for fluorescent immunostaining in fish embryos. PloS one. 2011;6:e19713. doi:10.1371/journal.pone.0019713
23. Perez-Rius C, Gaitan-Penas H, Estevez R, Barrallo-Gimeno A. Identification and characterization of the zebrafish ClC-2 chloride channel orthologs. Pflugers Archiv: European journal of physiology. 2014 doi:10.1007/s00424-014-1614-z
24. Hsiao CD, You MS, Guh YJ, Ma M, Jiang YJ, Hwang PP. A positive regulatory loop between foxi3a and foxi3b is essential for specification and differentiation of zebrafish epidermal ionocytes. PloS one. 2007;2:e302. doi:10.1371/journal.pone.0000302
25. Uchida S, Sasaki S, Nitta K, Uchida K, Horita S, Nihei H. et al. Localization and functional characterization of rat kidney-specific chloride channel, ClC-K1. The Journal of clinical investigation. 1995;95:104-13 doi:10.1172/JCI117626
26. Estevez R, Boettger T, Stein V, Birkenhager R, Otto E, Hildebrandt F. et al. Barttin is a Cl- channel beta-subunit crucial for renal Cl- reabsorption and inner ear K+ secretion. Nature. 2001;414:558-61 doi:10.1038/35107099
27. Kobayashi K, Uchida S, Mizutani S, Sasaki S, Marumo F. Intrarenal and cellular localization of CLC-K2 protein in the mouse kidney. Journal of the American Society of Nephrology: JASN. 2001;12:1327-34
28. Kobayashi K, Uchida S, Okamura HO, Marumo F, Sasaki S. Human CLC-KB gene promoter drives the EGFP expression in the specific distal nephron segments and inner ear. Journal of the American Society of Nephrology: JASN. 2002;13:1992-8
29. Lin LY, Hwang PP. Modification of morphology and function of integument mitochondria-rich cells in tilapia larvae (Oreochromis mossambicus) acclimated to ambient chloride levels. Physiological and biochemical zoology: PBZ. 2001;74:469-76 doi:10.1086/322159
30. Chang IC, Wei YY, Chou FI, Hwang PP. Stimulation of Cl- uptake and morphological changes in gill mitochondria-rich cells in freshwater tilapia (Oreochromis mossambicus). Physiological and biochemical zoology: PBZ. 2003;76:544-52 doi:10.1086/375438
31. Hwang PP, Lee TH. New insights into fish ion regulation and mitochondrion-rich cells. Comparative biochemistry and physiology Part A, Molecular & integrative physiology. 2007;148:479-97 doi:10.1016/j.cbpa.2007.06.416
32. Bosl MR, Stein V, Hubner C, Zdebik AA, Jordt SE, Mukhopadhyay AK. et al. Male germ cells and photoreceptors, both dependent on close cell-cell interactions, degenerate upon ClC-2 Cl(-) channel disruption. The EMBO journal. 2001;20:1289-99 doi:10.1093/emboj/20.6.1289
33. Gyomorey K, Yeger H, Ackerley C, Garami E, Bear CE. Expression of the chloride channel ClC-2 in the murine small intestine epithelium. American journal of physiology Cell physiology. 2000;279:C1787-94
34. Catalan M, Cornejo I, Figueroa CD, Niemeyer MI, Sepulveda FV, Cid LP. ClC-2 in guinea pig colon: mRNA, immunolabeling, and functional evidence for surface epithelium localization. American journal of physiology Gastrointestinal and liver physiology. 2002;283:G1004-13 doi:10.1152/ajpgi.00158.2002
35. Catalan M, Niemeyer MI, Cid LP, Sepulveda FV. Basolateral ClC-2 chloride channels in surface colon epithelium: regulation by a direct effect of intracellular chloride. Gastroenterology. 2004;126:1104-14
36. Lipecka J, Bali M, Thomas A, Fanen P, Edelman A, Fritsch J. Distribution of ClC-2 chloride channel in rat and human epithelial tissues. American journal of physiology Cell physiology. 2002;282:C805-16 doi:10.1152/ajpcell.00291.2001
37. Kato A, Romero MF. Regulation of electroneutral NaCl absorption by the small intestine. Annual review of physiology. 2011;73:261-81 doi:10.1146/annurev-physiol-012110-142244
38. Miyazaki H, Uchida S, Takei Y, Hirano T, Marumo F, Sasaki S. Molecular cloning of CLC chloride channels in Oreochromis mossambicus and their functional complementation of yeast CLC gene mutant. Biochemical and biophysical research communications. 1999;255:175-81 doi:10.1006/bbrc.1999.0166
39. Tang CH, Hwang LY, Lee TH. Chloride channel ClC-3 in gills of the euryhaline teleost, Tetraodon nigroviridis: expression, localization and the possible role of chloride absorption. The Journal of experimental biology. 2010;213:683-93 doi:10.1242/jeb.040212
40. Tang CH, Hwang LY, Shen ID, Chiu YH, Lee TH. Immunolocalization of chloride transporters to gill epithelia of euryhaline teleosts with opposite salinity-induced Na+/K+-ATPase responses. Fish physiology and biochemistry. 2011;37:709-24 doi:10.1007/s10695-011-9471-6
41. Matsuda JJ, Filali MS, Volk KA, Collins MM, Moreland JG, Lamb FS. Overexpression of CLC-3 in HEK293T cells yields novel currents that are pH dependent. American journal of physiology Cell physiology. 2008;294:C251-62 doi:10.1152/ajpcell.00338.2007
42. Dymowska AK, Hwang PP, Goss GG. Structure and function of ionocytes in the freshwater fish gill. Respiratory physiology & neurobiology. 2012;184:282-92 doi:10.1016/j.resp.2012.08.025
43. Hwang PP, Chou MY. Zebrafish as an animal model to study ion homeostasis. Pflugers Archiv: European journal of physiology. 2013;465:1233-47 doi:10.1007/s00424-013-1269-1
44. Hwang PP, Lee TH, Lin LY. Ion regulation in fish gills: recent progress in the cellular and molecular mechanisms. American journal of physiology Regulatory, integrative and comparative physiology. 2011;301:R28-47 doi:10.1152/ajpregu.00047.2011
45. Liao BK, Chen RD, Hwang PP. Expression regulation of Na+-K+-ATPase alpha1-subunit subtypes in zebrafish gill ionocytes. American journal of physiology Regulatory, integrative and comparative physiology. 2009;296:R1897-906 doi:10.1152/ajpregu.00029.2009
46. Bayaa M, Vulesevic B, Esbaugh A, Braun M, Ekker ME, Grosell M. et al. The involvement of SLC26 anion transporters in chloride uptake in zebrafish (Danio rerio) larvae. The Journal of experimental biology. 2009;212:3283-95 doi:10.1242/jeb.033910
47. Perry SF, Vulesevic B, Grosell M, Bayaa M. Evidence that SLC26 anion transporters mediate branchial chloride uptake in adult zebrafish (Danio rerio). American journal of physiology Regulatory, integrative and comparative physiology. 2009;297:R988-97 doi:10.1152/ajpregu.00327.2009
48. Piermarini PM, Verlander JW, Royaux IE, Evans DH. Pendrin immunoreactivity in the gill epithelium of a euryhaline elasmobranch. American journal of physiology Regulatory, integrative and comparative physiology. 2002;283:R983-92 doi:10.1152/ajpregu.00178.2002
49. Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS genetics. 2014;10:e1004108. doi:10.1371/journal.pgen.1004108
50. Goh CS, Bogan AA, Joachimiak M, Walther D, Cohen FE. Co-evolution of proteins with their interaction partners. Journal of molecular biology. 2000;299:283-93 doi:10.1006/jmbi.2000.3732
51. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 1994;22:4673-80
52. Hartmann AM, Tesch D, Nothwang HG, Bininda-Emonds OR. Evolution of the cation chloride cotransporter family: ancient origins, gene losses, and subfunctionalization through duplication. Molecular biology and evolution. 2014;31(Suppl 1):434-47 doi:10.1093/molbev/mst225
Author contact
![]() Corresponding author: pphwangsinica.edu.tw
Corresponding author: pphwangsinica.edu.tw

 Global reach, higher impact
Global reach, higher impact