10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(10):1236-1247. doi:10.7150/ijbs.12118 This issue Cite
Research Paper
Defective Initiation of Liver Regeneration in Osteopontin-Deficient Mice after Partial Hepatectomy due to Insufficient Activation of IL-6/Stat3 Pathway
1. State Key Laboratory of Oncogenes and Related Genes, Renji-Med X Clinical Stem Cell Research Center, Department of Liver Surgery, Ren Ji Hospital, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China
2. Laboratory of liver diseases, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Bethesda, MD, 20892, USA
Received 2015-3-13; Accepted 2015-7-29; Published 2015-8-21
Abstract
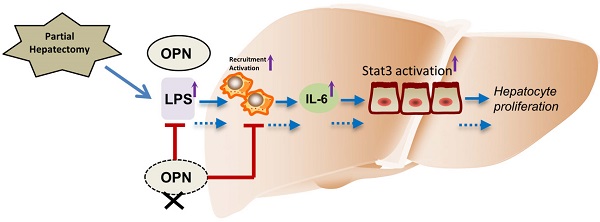
The initial process in liver regeneration after partial hepatectomy involves the recruitment of immune cells and the release of cytokines. Osteopontin (OPN), a pro-inflammatory protein, plays critical roles in immune cell activation and migration. Although OPN has been implicated in the pathogenesis of many liver diseases, the role of OPN in liver regeneration remains obscure. In the present study, we found that serum and hepatic OPN protein levels were significantly elevated in wild-type (WT) mice after partial hepatectomy (PHx) and that bile ductal epithelia were the major cell source of hepatic OPN. Compared to WT mice, OPN knockout (KO) mice exhibited delayed liver regeneration after PHx. This delay in OPN-/- mice was attributed to impaired hepatic infiltration of macrophages and neutrophils, decreased serum and hepatic IL-6 levels, and blunted activation of macrophages after PHx. Furthermore, we demonstrate that the attenuated activation of macrophages is at least partially due to decreased hepatic and portal vein LPS levels in OPN-/- mice. In response to decreased IL-6 levels, the activation of signal transducer and transcription (Stat) 3 was reduced in hepatocytes of OPN-/- mice compared to WT mice after PHx. Consequently, hepatic activation of the downstream direct targets of IL6/Stat3, such as c-fos, c-jun, and c-myc, was also suppressed post-PHx in OPN-/- mice compared to WT mice. Collectively, these results support a unique role for OPN during the priming phase of liver regeneration, in which OPN enhances the recruitment of macrophages and neutrophils, and triggers hepatocyte proliferation through Kupffer cell-derived IL-6 release and the downstream activation of Stat3.
Keywords: Osteopontin, partial hepatectomy, liver regeneration, Kupffer cell, IL-6, Stat3
Introduction
The liver is an important multifunctional organ that has a remarkable capacity to regenerate. Any acute or chronic liver injury can result in liver regeneration. Following liver injury, most hepatocytes exit G0 phase and re-enter the cell cycle to generate new hepatocytes. Liver regeneration is an orchestrated process that is regulated by multiple inflammatory cytokines, growth factors and hormones. Inflammatory cytokines, such as IL-6 and TNFα, play critical roles in the initiation of liver regeneration. During liver regeneration, IL-6 binds to its receptor, IL-6R, and further activates signal transducer and activator of transcription (Stat) 3 signaling pathways, resulting in increased hepatocyte survival and proliferation [1, 2]. By activating NF-κB, TNFα also facilitates the priming of hepatocyte proliferation [3, 4]. However, the mechanism by which the production of these inflammatory cytokines is regulated during liver regeneration remains elusive.
Despite of the potent regenerative capacity of the liver, nearly one million deaths per year are associated with chronic liver failure, indicating that, in these cases, liver regeneration was not sufficient to compensate for the loss of hepatocytes or to restore liver function. Therefore, novel therapeutic targets for enhancing liver regeneration are definitely desired by those patients suffering from fatal liver injuries.
Osteopontin (OPN), which is also known as secreted phosphoprotein 1 (Spp1) and early T-lymphocyte activation 1 (Eta-1), is a multifunctional protein that is involved in hepatic inflammation and diverse liver diseases [5, 6]. OPN is significantly induced in macrophages after stimulation by cytokines such as IL-6, TNFα, IL-1β and IFNγ [7, 8]. Interestingly, OPN can function as a ligand to induce the secretion of IL-12 and TNFα from peritoneal macrophages [9]. In addition, OPN has been suggested as a mediator for macrophage activation, migration and infiltration [9, 10]. It has been reported that OPN promotes the migration of Kupffer cells isolated from normal rats, suggesting that OPN may act as a chemoattractant to recruit macrophages in the liver [11]. Earlier studies indicated that the absence of OPN decreased macrophage infiltration in the liver after liver injury [12, 13]. However, whether OPN modulates liver regeneration by regulating liver inflammation or macrophage infiltration has not been determined.
The aim of this study was to investigate the role of OPN in liver regeneration and to understand the underlying mechanisms. In the present study, we demonstrate that OPN is an important regulator that facilitates the priming phase of liver regeneration following PHx. The advantages conferred OPN during liver regeneration are attributed to its functions in enhancing hepatic recruitment of macrophages and neutrophils, inducing IL-6 production, and subsequently promoting Stat3 activation in hepatocytes.
Materials and Methods
Animals
All animal studies were approved by the Institutional Animal Care and Use Committees of Renji Hospital Clinical Stem Cell Research Center and Shanghai Jiao Tong University. OPN knockout (OPN-/-) mice (B6.Cg-Spp1tm1Blh/J) and their wild-type (WT) control C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME).
Partial Hepatectomy Model
Eight- to ten-week-old male OPN-/- and age-matched WT mice were subjected to two-thirds partial hepatectomy (PHx) as previously described [14]. Briefly, after anesthetization, the left lateral lobe and the median lobe plus gall bladder were removed. The mice were sacrificed at various time points after surgery. Mice were injected intraperitoneally with BrdU (Sigma-Aldrich, St. Louis, MO) at 50 mg/kg body weight 2 hours before sacrificed. The regenerated liver was weighed, and the liver-body weight ratio was calculated. After anesthetization, both liver tissues and blood plasma from the portal vein 3 hours after sham surgery or PHx were collected and analyzed for endotoxin using an endotoxin test kit (Houshiji, Xiamen, China) according to the manufacturer's protocol. Serum was collected for tests, including alanine transaminase (ALT) and aspartate aminotransferase (AST) analyses using microplate test kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer's protocols. The regenerated liver tissues were collected for molecular and biochemical analyses.
Histological Analysis and Immunohistochemistry
The livers were harvested and fixed in 4% paraformaldehyde for at least 24 hours. Liver samples, embedded in paraffin, were cut into 5-μm-thick sections. Liver tissue sections were stained with hematoxylin-eosin (H&E) using standard procedures. For immunohistochemistry, sections were rehydrated and processed for an antigen-unmasking procedure, then incubated with primary antibodies overnight at 4 °C, followed by horseradish peroxidase-conjudated secondary antibodies. For all tissues, sections were counterstained with hematoxylin. Six to ten images from random fields in each section were taken with a light microscope (Axio Imager A1; Zeiss), and at least three mice per group were subjected to each experiment. Image-Pro Plus 6.0 was used for image analysis of sections. The primary antibodies used were goat anti-OPN (R&D Systems, Minneapolis, MN), mouse anti-BrdU, mouse anti-PCNA (Cell Signaling, Boston, MA), rabbit anti-MPO (BioCare Medical, Concord, CA), and rat anti-F4/80 (AbD Serotec, Oxford, UK).
Western Blot Analysis
Liver tissues and cell samples were homogenized in RIPA buffer (Thermo Scientific, Rockford, IL) containing a protease inhibitor cocktail (Calbiochem, Raleigh, NC). Protein concentrations were quantified with a BCA protein assay kit (Thermo Scientific, Rockford, IL) according to the manufacturer's manual. Fifty micrograms of total protein from each tissue or cell lysate sample were loaded and separated by gel electrophoresis and then transferred to nitrocellulose membranes. After blocking, membranes were incubated with primary antibodies at 4 °C overnight under shaking conditions. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies. Detection and quantification of protein bands were performed using the ChemiDocTM XRS+ System with Image LabTM Software (Bio-Rad). The primary antibodies used were rabbit anti-phospho-Stat3 (Tyr705), rabbit anti-Stat3, rabbit anti-phospho-IκBα (Ser32), mouse anti-IκBα (Cell Signaling), anti-β-actin (Sigma).
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was purified from approximately 30 mg liver samples or cell lysates according to the manufacturer's protocol (Qiagen, Valencia, CA), and then 500 ng mRNA was reverse-transcribed to cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Rockford, IL). Relative quantitative gene expression was measured via real-time PCR using a ViiATM 7 Real-Time PCR System (Life Technologies) and a SYBR Green PCR Kit (TaKaRa Biotechnology, Dalian, China). GAPDH expression was used as an internal standard. The primer sequences are as follows (5'-3'): GAPDH sense: AGGCCGGTGCTGAGTATGTC, anti-sense: TGCCTGCTTCACCACCTTCT; PCNA sense: GTGGAGCAACTTGGAATCCC, anti-sense: GGTTACCGCCTCCTCTTCTT; IL-6 sense: TGTTCTCTGGGAAATCGTGGA, anti-sense: TTTCTGCAAGTGCATCATCGT; TNFα sense: TTCTATGGCCCAGACCCTCA, anti-sense: TTTGCTACGACGTGGGCTAC; MCP-1 sense: CCCTAAGGTCTTCAGCACCT, anti-sense: ACTGTCACACTGGTCACTCC; F4/80 sense: TCTGCAGTGTCAGCTCAGAA, anti-sense: GTATGCCATGATGCTTGCCA; MPO sense: CTCGATGGAATGGGGAGAAGC, anti-sense: CCAACACCAAGGGCAGGTAGTC; c-fos sense: TACTACCATTCCCCAGCCGA, anti-sense: GCTGTCACCGTGGGGATAAA; c-jun sense: CGATGCCCTCAACGCC, anti-sense: CTTAGGGTTACTGTAGCCGTAGGC; c-myc sense: ATGTTGCGGTCGCTACGTC, anti-sense: AGAAGTTGCCACCGCCG; HGF sense: CAAGCAATCCAGAGGTACGC, anti-sense: CCATGGGACCTCTGTAGCTT; IL-22 sense: CAGGTTCCAGCCCTACAT, anti-sense: CAGCCTTCTGACATTCTTCT; and IL-22Rα1 sense: TTCCTCGGCTCCATCACA, anti-sense: AGCGTCTTCACTCGGCAC.
Flow Cytometry Analysis
Liver leukocytes were isolated as previously described [15] from livers of mice with minor modifications. In brief, liver tissues were passed through a 70-μm cell strainer (BD Falcon) in Hanks' balanced salt solution. The cell suspension was centrifuged at 500 ×g for 10 minutes. The cell pellet, including hepatocyte debris, was re-suspended in 5 ml of 35% Percoll (GE Healthcare Life Science), and the cell suspension was centrifuged at 500 ×g for 15 minutes at room temperature. The cell pellet containing the leukocytes was harvested and re-suspended in 5 ml of ACK Lysing Buffer (Gibco) to remove red blood cells. After incubated for 5 minutes, cells were washed twice and counted. The resulting leukocytes were incubated with APC-conjugated anti-CD45, PE-conjugated anti-CD11b, PerCP Cyanine5.5-conjugated anti-F4/80 (eBioscience, San Diego, CA), FITC-conjugated anti-Gr-1 (Miltenyi Biotec, Bergisch Gladbach, Germany) and analyzed using a BD FACSCalibur flow cytometer. The purity of the isolated leukocytes was over 90%. The number of neutrophils and macrophages per liver was calculated by multiplying the percentage of each population from the flow cytometric data by the total number of leukocytes per liver.
Cell Isolation and Culture
Primary cells were isolated by perfusion of the liver with Gey's balanced salt solution containing collagenase (Sigma). In brief, the liver was perfused via the portal vein at 5 ml per minute with EGTA buffer for 8 minutes. Subsequently, the liver was perfused with perfusion buffer containing collagenase at 5 ml per minute for 10 minutes. The liver was then dissociated in digestion buffer containing collagenase and filtered with a 70-μm cell strainer. Hepatocytes were collected by centrifugation at 400 rpm for 5 minutes. The supernatant containing the non-parenchymal cells was further cleared by another centrifugation at 400 rpm for 5 minutes. Non-parenchymal cells were pelleted by centrifugation at 650 ×g for 10 minutes. HSC were purified by a 12.5% OptiPrep gradient (Sigma) and collected for analysis. Kupffer cells were purified by gradient centrifugation using 17% OptiPrep. The cell fraction including Kupffer cells was gently aspirated and plated in non-collagen coated culture plates for 20 minutes. Kupffer cells attached to the plate were collected for analysis. The purity of isolated Kupffer cells or HSCs was over 90%.
The isolated hepatocytes were counted and seeded in culture plates. After cell spreading, the culture medium was deprived of fetal bovine serum for 24 hours and cells were subjected to proliferation analysis in the presence of 50 nM of recombinant mouse OPN (rmOPN) (R&D Systems) stimulation according to dose-response (5-500 nM) experiments. Cells were then subjected to immunofluorescence for PCNA after 48 hours of treatment, and cell lysates were collected for PCNA mRNA analysis following 24 hours of treatment. Isolated Kupffer cells were cultured in RPMI 1640 medium supplemented with 10% FBS (Gibco). After spreading overnight, cells were starved and treated with 100 nM of rmOPN for the indicated time according to dose-response (5-500 nM) experiments. Cell lysates were collected for mRNA analysis and cell supernatants were collected for ELISA analysis.
Immunofluorescence
Frozen liver sections (5-μm thick) and cells were fixed. Nonspecific antibody binding was blocked by 5% goat serum and 0.5% Triton X-100 in PBS. After aspirating away the blocking buffer, cells were incubated with mouse anti-PCNA, rabbit anti-desmin (Cell Signaling, Boston, MA), goat anti-OPN (R&D Systems), rabbit anti-CK19 (Proteintech, Chicago, IL), or rat anti-F4/80 (AbD Serotec) antibody diluted in PBS containing 1% bovine serum albumin and 0.5% Triton X-100 overnight at 4 °C. After washing in PBS, cells were incubated with fluorescence-conjugated secondary antibodies at room temperature for 1 hour. DAPI was used to stain cell nuclei. Six to ten images were acquired for each sample and the percentage of positive nuclei was calculated.
Enzyme-Linked Immunosorbent Assay
ELISA kits were used to determine liver and serum OPN levels (USCN Life Science, Wuhan, China) and IL-6 and TNFα levels in the serum and the cell supernatants (NeoBioscience Technology, Shenzhen, China) according to the manufacturer's protocols.
Statistical Analysis
Results are expressed as the mean ± SEM. The results were assessed using two-tailed, unpaired or paired Student's t-tests. P<0.05 was considered statistically significant (*=P<0.05/**=P<0.01/***=P<0.001).
Results
OPN levels increase in the serum and regenerating liver after PHx
To investigate the role of OPN in liver regeneration, we first measured OPN protein levels in the serum and liver after PHx. OPN levels significantly increased in both the serum and liver after 3 hours and reached the peak at 12 hours post-PHx (Fig. 1A and B). We then sought to determine which cell type contributed to OPN production during liver regeneration. In WT adult mouse livers, consistent with previous studies [12, 16], OPN was selectively expressed in epithelial cells around bile ducts (Fig. 1C). A significant increased staining intensity was observed for OPN in these epithelial cells early after PHx (Fig. 1C). To further define these cells, double immunofluorescence staining with anti-OPN and anti-F4/80 (a specific marker for macrophages), anti-desmin (a specific marker for HSCs) or anti-CK19 (a specific marker for cholangiocytes) antibodies was performed. There was no co-localization between OPN and markers of Kupffer cells or HSCs (Fig. S1A and B), suggesting that OPN is not expressed by these cell types after PHx. Interestingly, CK19 and OPN were highly co-localized 3 hours after PHx (Fig. 1D). Although CK19 also serves as a marker of oval cells, given that oval cells were usually absent from the normal liver controls and are normally activated 5-10 days after PHx [17, 18], we concluded that the CK19-positive cells that were labeled at 3 hours after PHx were mainly BECs (Fig. 1D).
OPN deletion delays hepatocyte proliferation after PHx
OPN knockout (OPN-/-) mice were used to determine the in vivo function of OPN during liver regeneration. There was no difference in liver weight, body weight or the liver-body weight ratio between 8-week-old WT and OPN-/- mice, suggesting that depletion of OPN did not impact liver growth in adult mice (Fig. S2A). Histological analyses also revealed that the liver from OPN-/- mice displayed normal histology (Fig. S2B).
We then performed two-thirds partial hepatectomy in both WT and OPN-/- mice and collected sera and liver tissues at the indicated time points after PHx. Serum ALT and AST levels were greatly increased at 12 hours post-PHx in both WT and OPN-/- mice, but no significant difference in serum ALT or AST levels was observed between WT and OPN-/- mice (Fig. 2A and B). BrdU incorporation assays indicated that WT mice displayed peak hepatocyte proliferation at 40 hours after PHx, but the proliferative peak in OPN-/- mice was delayed to 48 hours after PHx (Fig. 2C). Similar results were obtained by staining with PCNA, which is a marker for proliferating cells (Fig. 2D). In addition, the difference in the liver-to-body weight ratio between WT and OPN-/- mice was significant at 40 hours after PHx, but the ratio became comparable at 48 hours after PHx (Fig. 2E). These findings indicate that compared to WT, OPN-/- mice exhibit delayed liver regeneration after PHx.
OPN does not induce hepatocyte proliferation directly
To uncover the mechanism by which OPN depletion causes delayed hepatic proliferation, we examined whether OPN has a direct effect on hepatocyte proliferation. Freshly isolated hepatocytes were treated with recombinant mouse OPN (rmOPN) and the proliferative activity of hepatocytes was determined by measuring PCNA levels. As shown in Fig. 3A, comparable PCNA mRNA levels were detected in hepatocytes that were treated with rmOPN at the indicated concentrations. In addition, there was no difference in PCNA-positive hepatocytes between the control and rmOPN-treated groups (Fig. 3B and C). These results suggest that OPN does not induce hepatocyte proliferation directly.
OPN levels increase in the serum and regenerating liver after PHx. (A) Serum and (B) liver OPN levels were determined by ELISA at the indicated time points after PHx in WT mice (n = 4-10). The data are shown as the mean ± SEM. *P<0.05, **P<0.01, ***P<0.001 compared with 0 hour. (C) Immunohistochemistry of OPN in livers from WT mice and livers at 3 and 12 hours after PHx (200× magnification). At least three experiments were performed with similar results. (D) Immunofluorescence analysis showed co-staining of OPN and CK19 at 3 hours after PHx (200× magnification). Three independent experiments were performed with similar results.
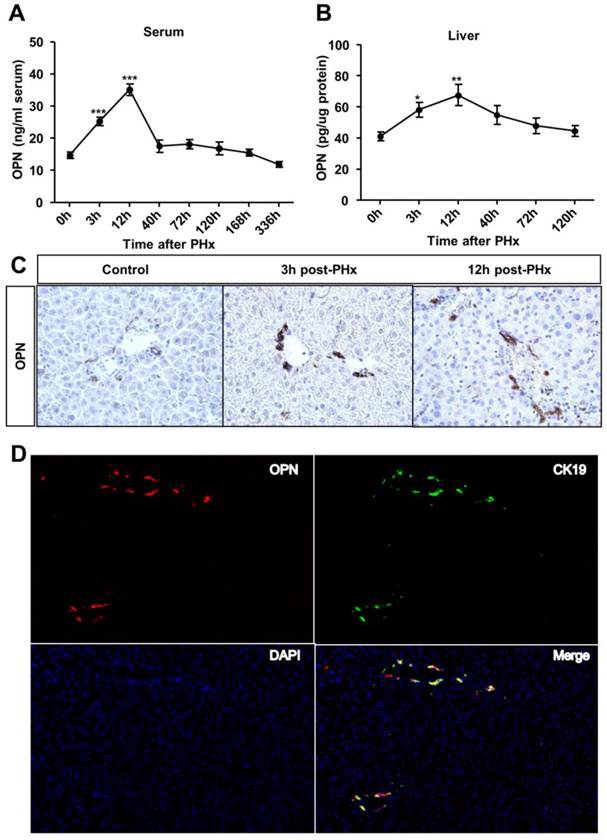
OPN deletion delays the peak of hepatocyte proliferation during liver regeneration after PHx. (A) Serum ALT and (B) AST activities were determined at different time points after PHx (n = 3-8). The data are shown as the mean ± SEM. (C) BrdU incorporation were measured in liver sections at the indicated time points after PHx (200× magnification). Quantitative analysis of BrdU labeling was performed (n = 3-7). The data are shown as the mean ± SEM. *P<0.05. (D) PCNA immunolabeling in liver sections after PHx (200× magnification). The number of PCNA positive cells in remnant liver lobes was counted in at least 5 fields per liver section (n = 3-7). The data are shown as the mean ± SEM. *P<0.05, **P<0.01. (E) The liver-to-body weight ratio was determined at the indicated time points after PHx (n = 3-8). The data are shown as the mean ± SEM. *P<0.05.
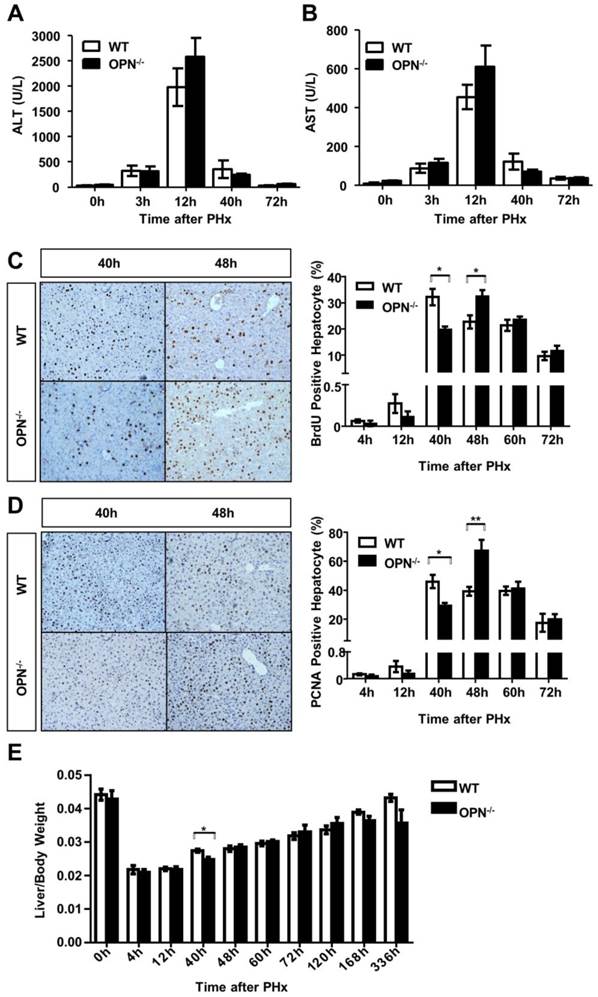
OPN does not stimulate hepatocyte proliferation in vitro. Primary hepatocytes isolated from WT mice were treated with rmOPN. (A) Hepatocyte lysates were collected 24 hours after OPN treatment and analyzed for PCNA mRNA expression (n = 3). The data are shown as the mean ± SEM. (B) Immunofluorescence for PCNA was performed 48 hours after treatment with 50 nM rmOPN (200× magnification). Three independent experiments were performed with similar results. (C) Quantitative analysis of PCNA immunolabeling was performed (n = 3). The data are shown as the mean ± SEM. RFC: relative fold change.
Loss of OPN impairs hepatic recruitment of macrophages and neutrophils and IL-6 expression during the priming phase of liver regeneration
Previous studies have reported that OPN is a key regulator of inflammatory processes, which indicates that OPN is not only considered to be a pro-inflammatory cytokine but also a potent chemoattractant for macrophages and neutrophils [19, 20]. Thus, to further investigate the association between OPN deletion and delayed liver regeneration, we studied the correlation between OPN and the inflammatory response in regenerating livers. FACS analysis of liver immune cells was performed at 3 hours post-PHx in WT and OPN-/- mice. Compared to WT mice, OPN-/- mice showed significantly impaired hepatic recruitment of macrophages and neutrophils 3 hours after PHx (Fig. 4A, Fig. S3A and B). Reduced hepatic recruitment of macrophages and neutrophils in OPN-/- mice was also confirmed by immunohistochemical staining and qPCR at the indicated time points post PHx (Fig. 4B-E). Consistent with a reduction in hepatic recruitment of macrophages and neutrophils, impaired induction of MCP-1, also known as monocyte chemotactic and activating factor, was also observed in the livers of OPN-/- mice 3 hours after PHx (Fig. S3C). Interestingly, compared to WT mice, although the hepatic recruitment of macrophages was restored, the liver infiltration of neutrophils was maintained at a very low level in OPN-/- mice 12 hours after PHx (Fig. 4B and C), suggesting that the hepatic recruitment of these two cell types was modulated by different chemokines. Given the critical roles of IL-6 and TNFα in the initiation of liver regeneration after PHx, we then measured the protein and mRNA levels of those two cytokines in the serum and liver. Compared to WT mice, OPN-depleted mice showed a significant reduction in IL-6 at 3 hours after PHx (Fig 5A and B). Although significantly reduced induction of TNFα mRNA was observed in the liver in OPN-/- mice compared to WT mice at 3 hours after PHx, we failed to confirm this difference at the protein level in serum (Fig. S4A and B).
OPN depletion impairs Kupffer cell activation after PHx
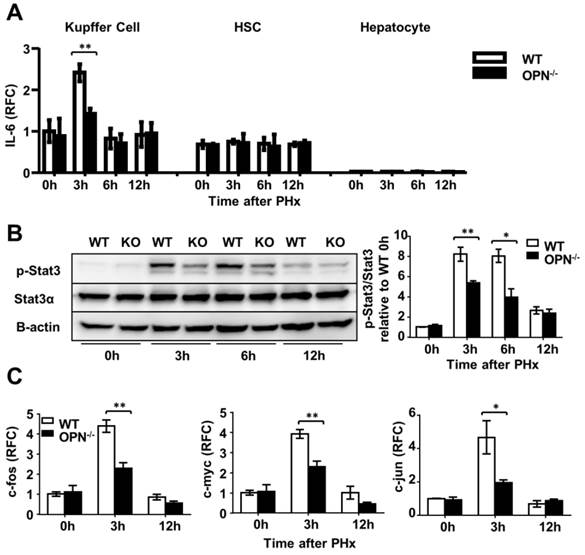
Kupffer cells are the major source of IL-6 during liver regeneration [21]. Although impaired hepatic recruitment of macrophages could contribute to reduced IL-6 levels in OPN-/- mice after PHx, other possibilities should not be ruled out. One possibility is that OPN might directly drive Kupffer cells to produce IL-6, and the other is that Kupffer cells in WT mice are more activated than those in OPN-/- mice after PHx. We isolated Kupffer cells from WT livers and treated them with 100 nM rmOPN. Quantitative PCR and ELISA assays were performed to examine changes in IL-6 at the mRNA and protein levels respectively. rmOPN treatment slightly increased IL-6 production but significantly stimulated TNFα expression in Kupffer cells (Fig. 5C and D, S4C and D), suggesting that the direct function of OPN on Kupffer cells may not attribute to reduced IL-6 levels in OPN-/- mice post-PHx. To examine the in vivo status of Kupffer cell activation, we isolated Kupffer cells, HSCs and hepatocytes from the livers of WT and OPN-/- mice after PHx at the indicated time points. IL-6 levels were measured by qPCR. Compared to WT counterparts, Kupffer cells from OPN-/- mice produced significantly less IL-6 at 3 hours after PHx (Fig. 6A). No induction of IL-6 was observed in either the HSCs or the hepatocytes after PHx (Fig. 6A). Similarly, Kupffer cells from WT mice produced a greater amount of TNFα than those from OPN-/- mice 3 hours after PHx (Fig. S4E). These results indicate that Kupffer cells are more activated in WT mice compared to OPN-/- mice 3 hours after PHx and suggest that the decrease in serum IL-6 levels in OPN-/- mice after PHx is due to both impaired liver infiltration of macrophages and the reduced activation of Kupffer cells.
OPN depletion reduces gut-derived endotoxin release into portal circulation
Endotoxin has been reported as an essential factor that contributes to the initiation of liver regeneration by binding to TLR4 to activate intracellular signaling pathways, which finally induces Kupffer cells to produce IL-6 and TNFα [4, 22, 23]. To answer why WT Kupffer cells were more activated than OPN-/- cells after PHx, we measured the levels of gut-derived endotoxin in portal circulation and the liver. We found that the concentrations of hepatic and plasma LPS were significantly higher in WT mice compared to OPN-/- mice 3 hours after PHx (Fig. S5A and B), suggesting that different LPS concentrations may contribute to the differential activation of Kupffer cells in WT and OPN-/- mice after PHx. Because LPS can activate the NF-κB pathway in Kupffer cells, resulting in the production of IL-6 and increased phosphorylation of IκBα, indicating the activation of the NF-κB pathway [24], we then examined phosphorylated IκBα levels in both WT and OPN-/- Kupffer cells. As expected, phosphorylation of IκBα was significantly impaired in OPN-/- Kupffer cells compared to WT counterparts 3 hours post-PHx (Fig. S5C). These results clearly indicate that OPN indirectly affects Kupffer cell activation by modulating LPS release into portal circulation after PHx.
OPN depletion inhibits Stat3 activation in hepatocytes in vivo
The Stat3 pathway plays a critical role in liver regeneration by promoting hepatocyte survival and liver repair. Given the key role of IL-6 in activating the Stat3 pathway, we then measured indicators of the Stat3 signaling pathway in hepatocytes after PHx. In response to reduction in IL-6 induction in OPN-/- mice, significant decreased Stat3 phosphorylation was observed in OPN-/- liver tissues compared to WT tissues (Fig. 6B). IL-22 has also been shown to play an important role in promoting liver regeneration by activating Stat3 [25]. We quantified the mRNA expression of IL-22 and IL-22Rα1 by qPCR. Three hours after PHx, the IL-22 mRNA level was increased 1.5-fold in WT livers and no IL-22 induction was observed in OPN-/- livers (Fig. S6A). No induction of IL-22Rα1 was detected in either WT or OPN-/- mice 3 hours after PHx (Fig. S6B). Compared to OPN-/- mice, WT mice presented 1.5-fold and 10-fold higher IL-22 and IL-6 levels respectively 3 hours after PHx (Fig. 5B, Fig. S6A). Therefore, it seems that IL-6, but not IL-22, plays the primary role in activating Stat3 in our model.
To further confirm that OPN depletion impairs the activation of Stat3, the expression of the downstream genes c-fos, c-myc and c-jun was measured. As shown in Fig. 6C, all three of these immediate early genes were markedly higher in WT livers compared to OPN-/- livers 3 hours after hepatectomy. Given that HGF, a mitogenic and anti-apoptotic factor, is synthesized in response to IL-6 in the liver [26, 27], we then measured HGF levels in the liver 3 hours after PHx. We found that the level of HGF in WT mice was 1.5-fold higher than that in OPN-/- mice (Fig. S7). Collectively, these results indicate that by impairing hepatic recruitment of macrophages and blunting Kupffer cell activation, the loss of OPN consequently reduces IL-6 production and in turn attenuates the activation of the Stat3 pathway to impair the priming phase of liver regeneration.
Discussion
Although a number of studies have suggested that OPN plays important roles in liver fibrosis, alcoholic hepatitis, non-alcoholic fatty liver disease and hepatocellular carcinoma [28-31], thus far, its role in liver regeneration has not been determined. The close relationship between OPN and hepatology implies that OPN, a multifunctional cytokine, may act as a regulator during liver regeneration. In this study, we identified OPN act as a novel positive regulator in liver regeneration. In the case of PHx, depletion of OPN significantly delayed the proliferative peak of hepatocytes. We demonstrated that OPN depletion leads to impaired liver recruitment of macrophages and neutrophils, blunts the activation and inhibits IL-6 production in Kupffer cells, and consequently attenuates the activation of the Stat3 pathway in hepatocytes. Therefore, OPN is an additional molecule that may promote the initiation of liver regeneration.
Loss of OPN impairs the recruitment of macrophages and neutrophils into the regenerating liver. (A) Liver leukocytes were isolated and analyzed at 0 and 3 hours post-PHx, and the number of liver leukocytes (Neutrophil: Gr-1high CD11b+; Macrophage: CD11bint F4/80+) was calculated by multiplying the percentage of each population determined by the FACS data by the total number of leukocytes per liver (n = 4). The data are shown as the mean ± SEM. *P<0.05. Immunohistochemistry for (B) MPO and (C) F4/80 were performed in mouse liver sections at 3, 6, and 12 hours post-PHx (400× magnification). Quantitative analysis was performed (n = 3-5). The data are shown as the mean ± SEM. *P<0.05, **P<0.01. (D) MPO and (E) F4/80 relative expression levels were analyzed in livers after PHx (n = 3-4). The data are shown as the mean ± SEM. *P<0.05. HPF: high power field; RFC: relative fold change.
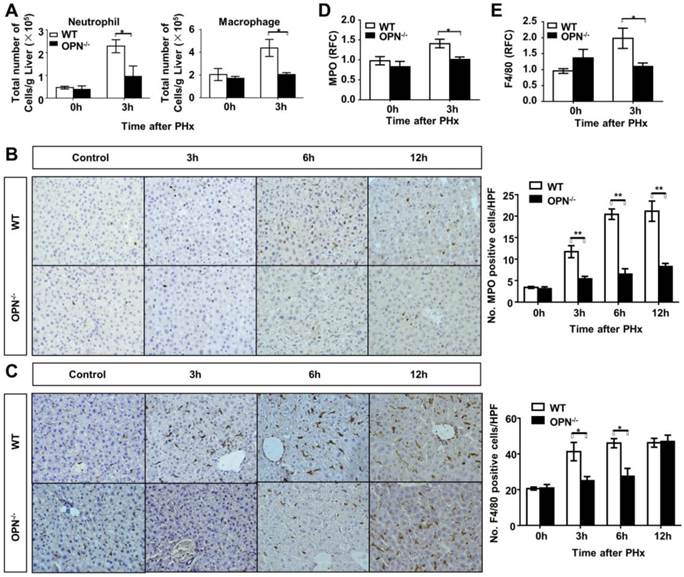
OPN facilitates IL-6 release in the regenerating liver and isolated Kupffer cells. (A) Serum IL-6 levels were determined by ELISA after PHx (n = 4-6). The data are shown as the mean ± SEM. *P<0.05. (B) IL-6 mRNA expression in the liver was analyzed by qPCR (n = 3-5). The data are shown as the mean ± SEM. *P<0.05. (C) Kupffer cells were isolated from WT mouse livers and treated with 100 nM rmOPN. Cell culture supernatants were subjected to ELISA for IL-6 (n = 3). The data are shown as the mean ± SEM. (D) Cell lysates were collected for quantitative analysis of IL-6 mRNA (n = 3). The data are shown as the mean ± SEM. *P<0.05. RFC: relative fold change.
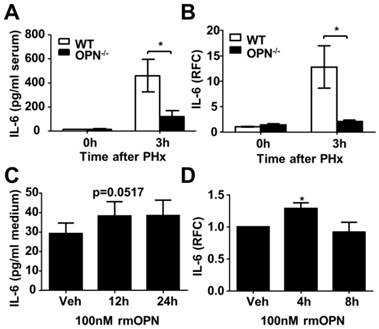
OPN facilitates IL-6 production in Kupffer cells and promotes liver regeneration via activation of the Stat3 pathway. (A) Kupffer cells, hepatic stellate cells and hepatocytes were isolated from WT and OPN-/- mice after PHx and collected for quantitative analysis of IL-6 mRNA (n = 3-6). The data are shown as the mean ± SEM. **P<0.01. (B) Phosphorylation of Stat3 in regenerating livers was analyzed by western blot. Densitometric analysis of immunoblots is shown (n = 3). The data are shown as the mean ± SEM. *P<0.05, **P<0.01. (C) mRNA expression of c-fos, c-myc, and c-jun was analyzed by qPCR (n = 3-6). The data are shown as the mean ± SEM. *P<0.05, **P<0.01. RFC: relative fold change.
Consistent with a previous finding [16], we observed that OPN is exclusively expressed in BECs in normal livers. Furthermore, after PHx, the majority of induced OPN was located in the areas of the bile ductal epithelium. Here, we used CK19 as the specific marker for BECs, as shown in Fig. 1D. It has been reported that oval cells also stain positive for CK19, but they are usually absent in a normal liver and are generally activated 5-10 days post-PHx [17, 18]. Hence, we believe that the CK19-positive cells observed at 3 hours post-PHx are mainly BECs. Therefore, we concluded that BECs are primarily responsible for OPN production after PHx.
Compared to the control group, we observed a delay of hepatocyte proliferative peak after PHx in mice deficient in OPN; however, we did not observe a proliferative advantage in hepatocytes treated with recombinant mouse OPN (rmOPN) in vitro, suggesting that OPN does not directly stimulate hepatocyte proliferation. Interestingly, a recent study reported that recombinant human OPN (rhOPN) decreased primary hepatocyte proliferation but did not alter cell viability [32]. Given that the recombinant mouse OPN shares only 64% amino acid sequence identity with human OPN, we suspect that this discrepancy might be due to different types of rOPN. Further studies examining whether OPN can directly affect hepatocyte proliferation are warranted.
In the present study, we demonstrate that ablation of OPN impaires recruitment of macrophages and neutrophils into the liver during the priming stage following PHx, which may contribute to the reduced IL-6 levels observed in OPN-/- mice. We also detected that WT Kupffer cells produce more IL-6 in vivo at 3 hours after PHx compared to OPN-/- Kupffer cells, suggesting that, in addition to the OPN-mediated hepatic infiltration of macrophages, OPN may also enhance IL-6 production in Kupffer cells, either directly or indirectly. Notably, in vitro direct incubation of Kupffer cells with rmOPN slightly induced IL-6 production, which raises an interesting question: how does OPN modulate the remarkable IL-6 induction in Kupffer cells after PHx? We then examined LPS levels in the liver and in portal circulation. Significantly higher LPS levels were detected in both in WT compared to OPN-/- mice, suggesting that after PHx, OPN may affect LPS release into the portal vein and subsequently modulate IL-6 induction in Kupffer cells. Intriguingly, in an alcohol-induced liver injury mouse model, a recent study found that OPN blocks the gut-derived LPS into the liver by binding to LPS [33]. Therefore, we propose that OPN might affect the stimulation of LPS release in different experimental models.
The IL-6/Stat3 pathway may affect the induction of nearly 36 percents of the immediate early genes involved in the regeneration of the liver [23, 34]. Among them, c-fos, c-myc and c-jun play pivotal roles in liver regeneration. c-fos is positively transactivated by the Stat DNA binding element; c-myc transcriptionally regulates cyclin D1, the best marker for the G1-to-S transition in the regenerating liver; and c-jun acts as a positive factor in cell growth [1]. In OPN-/- mice, the expression of all three of these genes was repressed due to the reduced activation of the Stat3 pathway, which is the key process that activates hepatocytes to enter the mitosis [35].
In summary, to the best of our knowledge, we have for the first time demonstrated the positive regulatory role of OPN in liver regeneration. Deletion of OPN delays the peak of hepatocyte proliferation but does not impair overall liver regeneration. This delay in proliferation in OPN-/- mice post-PHx seems to be results of a decrease in IL-6 production, which is attributed to the impaired hepatic recruitment of macrophages and the reduced LPS release to portal circulation observed in OPN-/- mice after PHx. Thus, our study sheds additional light on the molecular mechanisms that regulate the process of liver regeneration.
Supplementary Material
Figures S1-S7.
Abbreviations
OPN: osteopontin; IL: interleukin; Stat3: signal transducer and transcription 3; TNFα: tumor necrosis factor α; IL-6R: IL-6 receptor; IL-22R: IL-22 receptor; NF-κB: nuclear factor κB; Spp1: secreted phosphoprotein 1; Eta-1: early T-lymphocyte activation 1; IFNγ: interferon γ; PHx: partial hepatectomy; BrdU: 5-Bromo-2'-deoxyuridine; ALT: alanine transaminase; AST: alanine transaminase; PCNA: proliferating cell nuclear antigen; MPO: myeloperoxidase; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; HSC: hepatic stellate cell; BEC: biliary epithelial cell; LPS: lipopolysaccharide; HGF: hepatocyte growth factor; CK19: cytokeratin 19; RFC: relative fold change; rmOPN: recombinant mouse OPN; rhOPN: recombinant human OPN; SEM: standard error of mean; HPF: high power field.
Acknowledgements
We thank Dr. Bin Gao from National Institutes of Health, Bethesda, MD for editing the manuscript. This work was supported by the National Natural Science Foundation of China (31300742 to X Kong, 81372233 to H. Wu, 8147224 to Q Xia, 81130038 and 81372189 to WQ Gao), and the Shanghai Education Committee (Eastern Scholar Program) to X Kong, the Chinese Ministry of Science and Technology (2012CB966800 to WQ Gao), the Shanghai Health Bureau Key Joint Efforts Foundation (2013ZYJB001) to Q Xia, the Shanghai Health Bureau Key Discipline and Specialty Foundation and the KC Wong Foundation to WQ Gao.
Conflict of Interests
All authors have read the manuscript and declared that there is no conflict of interests.
References
1. Cressman DE, Greenbaum LE, DeAngelis RA, Ciliberto G, Furth EE, Poli V. et al. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379-83
2. Streetz KL, Luedde T, Manns MP, Trautwein C. Interleukin 6 and liver regeneration. Gut. 2000;47:309-12
3. Webber EM, Bruix J, Pierce RH, Fausto N. Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology. 1998;28:1226-34
4. Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45-53
5. Ramaiah SK, Rittling S. Pathophysiological role of osteopontin in hepatic inflammation, toxicity, and cancer. Toxicological sciences: an official journal of the Society of Toxicology. 2008;103:4-13
6. Nagoshi S. Osteopontin: Versatile modulator of liver diseases. Hepatology research: the official journal of the Japan Society of Hepatology. 2014;44:22-30
7. Ogawa D, Stone JF, Takata Y, Blaschke F, Chu VH, Towler DA. et al. Liver x receptor agonists inhibit cytokine-induced osteopontin expression in macrophages through interference with activator protein-1 signaling pathways. Circulation research. 2005;96:e59-67
8. Nakamachi T, Nomiyama T, Gizard F, Heywood EB, Jones KL, Zhao Y. et al. PPARalpha agonists suppress osteopontin expression in macrophages and decrease plasma levels in patients with type 2 diabetes. Diabetes. 2007;56:1662-70
9. Weber GF, Zawaideh S, Hikita S, Kumar VA, Cantor H, Ashkar S. Phosphorylation-dependent interaction of osteopontin with its receptors regulates macrophage migration and activation. Journal of leukocyte biology. 2002;72:752-61
10. Giachelli CM, Lombardi D, Johnson RJ, Murry CE, Almeida M. Evidence for a role of osteopontin in macrophage infiltration in response to pathological stimuli in vivo. The American journal of pathology. 1998;152:353-8
11. Kawashima R, Mochida S, Matsui A, YouLuTu ZY, Ishikawa K, Toshima K. et al. Expression of osteopontin in Kupffer cells and hepatic macrophages and Stellate cells in rat liver after carbon tetrachloride intoxication: a possible factor for macrophage migration into hepatic necrotic areas. Biochemical and biophysical research communications. 1999;256:527-31
12. Lorena D, Darby IA, Gadeau AP, Leen LL, Rittling S, Porto LC. et al. Osteopontin expression in normal and fibrotic liver. altered liver healing in osteopontin-deficient mice. Journal of hepatology. 2006;44:383-90
13. He CY, Liang BB, Fan XY, Cao L, Chen R, Guo YJ. et al. The dual role of osteopontin in acetaminophen hepatotoxicity. Acta pharmacologica Sinica. 2012;33:1004-12
14. Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nature protocols. 2008;3:1167-70
15. Liu ZX, Govindarajan S, Kaplowitz N. Innate immune system plays a critical role in determining the progression and severity of acetaminophen hepatotoxicity. Gastroenterology. 2004;127:1760-74
16. Brown LF, Berse B, Van de Water L, Papadopoulos-Sergiou A, Perruzzi CA, Manseau EJ. et al. Expression and distribution of osteopontin in human tissues: widespread association with luminal epithelial surfaces. Molecular biology of the cell. 1992;3:1169-80
17. Lowes KN, Brennan BA, Yeoh GC, Olynyk JK. Oval cell numbers in human chronic liver diseases are directly related to disease severity. The American journal of pathology. 1999;154:537-41
18. Apte U, Thompson MD, Cui S, Liu B, Cieply B, Monga SP. Wnt/beta-catenin signaling mediates oval cell response in rodents. Hepatology. 2008;47:288-95
19. Lund SA, Giachelli CM, Scatena M. The role of osteopontin in inflammatory processes. Journal of cell communication and signaling. 2009;3:311-22
20. Wang KX, Denhardt DT. Osteopontin: role in immune regulation and stress responses. Cytokine & growth factor reviews. 2008;19:333-45
21. Selzner N, Selzner M, Odermatt B, Tian Y, Van Rooijen N, Clavien PA. ICAM-1 triggers liver regeneration through leukocyte recruitment and Kupffer cell-dependent release of TNF-alpha/IL-6 in mice. Gastroenterology. 2003;124:692-700
22. Cornell RP. Gut-derived endotoxin elicits hepatotrophic factor secretion for liver regeneration. The American journal of physiology. 1985;249:R551-62
23. Taub R. Liver regeneration: from myth to mechanism. Nature reviews Molecular cell biology. 2004;5:836-47
24. Cui J, Chen Y, Wang HY, Wang RF. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Human vaccines & immunotherapeutics. 2014;10:3270-85
25. Ren X, Hu B, Colletti LM. IL-22 is involved in liver regeneration after hepatectomy. American journal of physiology Gastrointestinal and liver physiology. 2010;298:G74-80
26. Sun R, Jaruga B, Kulkarni S, Sun H, Gao B. IL-6 modulates hepatocyte proliferation via induction of HGF/p21cip1: regulation by SOCS3. Biochemical and biophysical research communications. 2005;338:1943-9
27. Kang LI, Mars WM, Michalopoulos GK. Signals and cells involved in regulating liver regeneration. Cells. 2012;1:1261-92
28. Urtasun R, Lopategi A, George J, Leung TM, Lu Y, Wang X. et al. Osteopontin, an oxidant stress sensitive cytokine, up-regulates collagen-I via integrin alpha(V)beta(3) engagement and PI3K/pAkt/NFkappaB signaling. Hepatology. 2012;55:594-608
29. Morales-Ibanez O, Dominguez M, Ki SH, Marcos M, Chaves JF, Nguyen-Khac E. et al. Human and experimental evidence supporting a role for osteopontin in alcoholic hepatitis. Hepatology. 2013;58:1742-56
30. Shang S, Plymoth A, Ge S, Feng Z, Rosen HR, Sangrajrang S. et al. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology. 2012;55:483-90
31. Syn WK, Agboola KM, Swiderska M, Michelotti GA, Liaskou E, Pang H. et al. NKT-associated hedgehog and osteopontin drive fibrogenesis in non-alcoholic fatty liver disease. Gut. 2012;61:1323-9
32. Wang X, Lopategi A, Ge X, Lu Y, Kitamura N, Urtasun R. et al. Osteopontin induces ductular reaction contributing to liver fibrosis. Gut. 2014;63:1805-18
33. Ge X, Leung TM, Arriazu E, Lu Y, Urtasun R, Christensen B. et al. Osteopontin binding to lipopolysaccharide lowers tumor necrosis factor-alpha and prevents early alcohol-induced liver injury in mice. Hepatology. 2014;59:1600-16
34. Li W, Liang X, Leu JI, Kovalovich K, Ciliberto G, Taub R. Global changes in interleukin-6-dependent gene expression patterns in mouse livers after partial hepatectomy. Hepatology. 2001;33:1377-86
35. Li W, Liang X, Kellendonk C, Poli V, Taub R. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. The Journal of biological chemistry. 2002;277:28411-7
Author contact
![]() Corresponding authors: Xiaoni Kong, PhD., Department of Liver Surgery, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, 160 Pujian Road, Shanghai, China. 200127. Telephone: +862134206284; E-mail: xiaonikongedu.cn; xiaoni-kongcom or Wei-Qiang Gao, PhD., Stem Cell Research Center, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, 160 Pujian Road, Shanghai, China. 200127. Telephone: +862168383917; Fax: +862168383916; E-mail: gao.weiqiangedu.cn
Corresponding authors: Xiaoni Kong, PhD., Department of Liver Surgery, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, 160 Pujian Road, Shanghai, China. 200127. Telephone: +862134206284; E-mail: xiaonikongedu.cn; xiaoni-kongcom or Wei-Qiang Gao, PhD., Stem Cell Research Center, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, 160 Pujian Road, Shanghai, China. 200127. Telephone: +862168383917; Fax: +862168383916; E-mail: gao.weiqiangedu.cn

 Global reach, higher impact
Global reach, higher impact