ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2015; 11(11):1296-1305. doi:10.7150/ijbs.12463 This issue Cite
Research Paper
Transgenic Cotton Plants Expressing Double-stranded RNAs Target HMG-CoA Reductase (HMGR) Gene Inhibits the Growth, Development and Survival of Cotton Bollworms
College of Plant Science and Technology, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, Hubei 430070, P.R. China
Abstract
RNA interference (RNAi) has been developed as a powerful technique in the research of functional genomics as well as plant pest control. In this report, double-stranded RNAs (dsRNA) targeting 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) gene, which catalyze a rate-limiting enzymatic reaction in the mevalonate pathway of juvenile hormone (JH) synthesis in cotton bollworm, was expressed in cotton plants via Agrobacterium tumefaciens-mediated transformation. PCR and Sothern analysis revealed the integration of HMGR gene into cotton genome. RT-PCR and qRT-PCR confirmed the high transcription level of dsHMGR in transgenic cotton lines. The HMGR expression both in transcription and translation level was significantly downregulated in cotton bollworms (helicoverpa armigera) larvae after feeding on the leaves of HMGR transgenic plants. The transcription level of HMGR gene in larvae reared on transgenic cotton leaves was as much as 80.68% lower than that of wild type. In addition, the relative expression level of vitellogenin (Vg, crucial source of nourishment for offspring embryo development) gene was also reduced by 76.86% when the insect larvae were fed with transgenic leaves. The result of insect bioassays showed that the transgenic plant harboring dsHMGR not only inhibited net weight gain but also delayed the growth of cotton bollworm larvae. Taken together, transgenic cotton plant expressing dsRNAs successfully downregulated HMGR gene and impaired the development and survival of target insect, which provided more option for plant pest control.
Keywords: 3-hydroxy-3-methylglutaryl coenzyme A reductase(HMGR), cotton bollworm, RNA interference, transgenic cotton, double-stranded RNAs, pest control.
Introduction
Cotton (Gossypium hirsutum) is an important fiber and economic crop around the world, which shows conspicuous significance in crop production. Pests and pathogenic funguses pose main stress for the productivity and quality of cotton. Currently, the major pest in cotton production is cotton bollworm (Helicoverpa armigera). In spite of the wide cultivation of transgenic insect-resistant BT cotton showing tremendous economic and social superiorities [1], the mutations of bollworm gene between generations and the selections resulting from BT insect-resistant proteins endow bollworm with resistance to transgenic BT crops [2-6]. Hence transgenic insect-resistance crops with alternative strategies is desirable.
RNA interference (RNAi), an effective gene-silencing mechanism in eukaryotes, has been discovered in Caenorhabditis elegans [7] for the first time and then developed as an effective insect resistant system in a wide variety of plant species [8, 9]. Double-strand RNA (dsRNA) can be produced by interior transcription, transposon, artificial transgenesis and RNA virus infection, which are recognized and decomposed into small interfering RNAs (siRNA) by endoribonuclease Dicer. RNA-induced silencing complex (RISC), including siRNA and several enzymes such as endonucleases, exonucleases and helicases, exhibits the function of nucleases to recognize and cleave specific target RNA [10]. Therefore, RNAi can be artificially used to inhibit the expression of endogenous gene. Sharing common molecular mechanism of sequence-specific gene silencing in a wide variety of species, RNAi triggered by exogenous dsRNA has been developed as one of the most efficient tools for the research of gene function [11]as well as pest control [12-16]. The dsRNA produced by transgenic plants against key gene of pests has been regarded as safeguard that endows transgenic resistant-pest plants with new innovation [17, 18].
Cotton bollworm undergoes complex molting process in its life cycle, whose growth and development are coordinately regulated by hydroxyecdysone [19-22] and juvenile hormone (JH) [22]. Analogues of JH have been synthesized artificially and brought new originalities to the basic research and development of pesticide [23]. Successful applications of hormone analogues related to molting process showed that impairment or overdoing of crucial hormones could be a new strategy for the integrated pest management (IPM) of insects belonging to the Phylum Arthropoda [24]. So, the crucial genes or transcription factors involved in the hormone biosynthesis pathways are inclined to be used as ideal targets when RNAi technology is applied to pest control [25]. JH is synthesized via the mevalonate pathway in insects, in which mevalonate is one of the most important intermediates. HMG-CoA, the precursor of mevalonate pathway, is obliged to undergo three enzymatic reactions to be converted into mevalonate, and HMGR catalyzes and regulates the last reaction [26]. Therefore, HMGR correlates a rate-limiting step in the biosynthesis of mevalonate, emerging as a promising target for the RNAi technology for insect control [27, 28]. In addition, the biosynthesis of vitellogenin, the crucial nourishment for offspring embryo development, also can be inhibited by the downregulation of HMGR gene in B.germanica [27].
In our previous report, a 3329-bp full-length cDNA (GenBank accession no. GU584103) of HaHMGR gene has been cloned by RACE technology. A 1176-bp fragment in the coding sequence of HaHMGR was amplified from the cDNA of cotton bollworm and used to generate dsRNA. By injecting dsHMGR into the abdomen of 2-day-old pupa, we observed that the number of eggs laid was decreased and the relative expression of both HMGR and Vg gene was downregulated in the tested larvae [27]. Therefore, we expect to create transgenic cotton plants that produces double-strand RNAs of HMGR gene for pest control. In this report, two HMGR fragments were selected as RNAi target sites and expressed in transgenic cotton plants. The results of real-time PCR showed the expression of both HMGR and Vg in cotton bollworm were downregulated when the larvae fed on the transgenic cotton leaves producing dsRNA-HMGR and the development of these tested larvae was dramatically impaired.
Materials and Methods
Plant materials
In this study, an elite cotton cultivar YZ1 [29, 30] was used for Agrobacterium mediated genetic transformation. Insect eggs and larvae of cotton bollworm (Heliothis armigera) were provided by Prof. Changying Niu from College of Plant Science and Technology of Huazhong Agriculture University.
Gene cloning, vector construction and cotton transformation
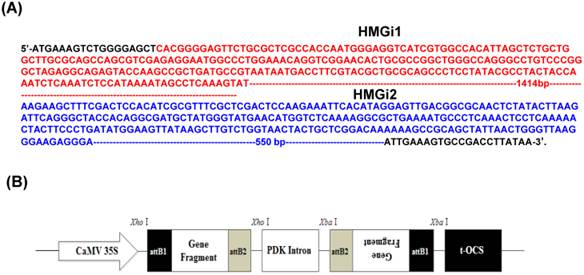
Two target fragments (namely HMGi1 and HMGi2) in the coding sequence of HMGR gene (GenBank accession no. GU584103) were chosen for RNAi targets including a 455bp and a 874bp fragments, as shown in Figure 1A respectively. Two pairs of primers with attB1 and attB2 adaptors were designed according to target fragments. All the sequences of primers for HMGi1 and HMGi2 were displayed in Table 1. The purified PCR products were incorporated into vector pHellsgate4 according to the manufacturer's recommendations [31], which harboring a NPT II gene as the select marker. The T-DNA region of the plastid was shown in Figure 1B.Then the constructed vectors were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation, whose positivity was verified by PCR. The Agrobacterium tumefaciens-mediated transformation of cotton was applied according to our previous reports [29, 32-34].
Molecular analysis for the putative transgenic cotton plants
Putative transgenic cotton plant lines were identified by PCR and Southern blot. Genomic DNA was extracted from tender leaves of T0 generation transgenic and wild-type plants by using Plant Genomic DNA Kit (Tiangen Biotech, China). Two pair of primers for PCR test, HMGi1 and HMGi2, were shown in Table 1 and designed according to target fragments, producing a 455bp product for HMGi1 and a 874bp product for HMGi2. For Southern blot, 20 μg genomic DNA was digested with Hind III-HF for 72 hours and then was separated on 0.8 % agarose gel by electrophoresis. Separated DNA was blotted onto Hybond-N+ nylon membranes (Amersham-Pharmacia, USA) and incubated at 80°C for 2 h in an oven. 100 ng PCR product of NPTII was labeled with Digoxin by using DIG high prime DNA labeling and detection starter kit (Roche, Germany).
Target sequences of HMG gene for RNA interference and plasmid vector for cotton genetic transformation. (A) Two sequences of HMG gene were used for RNAi target sequences in this experiment. The HMGi 1 sequence was indicated in red font and the HMGi 2 sequence was indicated in blue font. (B) Schematic representation of T-DNA region of the expression cassette of HMGR RNA interference vector.
Primer used in this study
| Primer set | Primer sequence |
|---|---|
| Primers used for vector construction | (vHMGi1-F) 5'-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAGGAGCTCACGGGGAGTTCTG-3' |
| (vHMGi1-R) 5'-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTTCCAACCCTCTTACCCTGGT-3' | |
| (vHMGi2-F) 5'- GGGGACAAGTTTGTACAAAAAAGCAGGCTGCTCACGAATGCAGACAATGGATA-3' | |
| (vHMGi2-R) 5'- GGGGACCACTTTGTACAAGAAAGCTGGGTCTAAGGTCGGCACTTTCAAGGTG-3' | |
| Primers used for PCR test and RT-PCR for transgenic plants | (HMGi1-F) 5'-GGAGCTCACGGGGAGTTCTG-3' |
| (HMGi1-R) 5'-CTTCCAACCCTCTTACCCTGGT-3' | |
| (HMGi2-F) 5'-TCACGAATGCAGACAATGGATA-3' | |
| (HMGi2-R) 5'-TAAGGTCGGCACTTTCAAGGTG-3' | |
| Primers used for real-time PCR test for transgenic plants | (qHMGi1-F) 5'-GCTTTATTTTCACTTCAGCTGTCG-3' |
| (qHMGi1-R) 5'-AACCCTCTTACCCTGGTCTTCC-3' | |
| (qHMGi2-F) 5'-CCCTCAAACTCCTCAAAAACTACTT-3' | |
| (qHMGi2-R) 5'-TTTCCCTCTTCCCTTAACCCA-3' | |
| (qub7-F) 5'-GACTCTACTCAATCCCCACCAGCCT-3' | |
| (qub7-R) 5'-CCGCACCTTAGCCGACTACAACATC-3' |
Detection the transcription level of dsRNA in transgenic cotton leaves by RT-PCR and qRT-PCR
RT-PCR and qRT-PCR were performed to detect the production of dsRNA in different transgenic cotton lines. Total RNA was extracted from tender leaves of T1 positive transgenic and negative control plants. Three μg RNA was reverse-transcribed to cDNA with the M-MLV reverse transcriptase (Promega, Madison, WI, USA) with Oligo (dT) 15 primer (Rromega, Madison, WI, USA). All qRT-PCR tests were accomplished with ABI Prism 7500 (Applied Biosystems, Foster City,CA, USA) under the following condition: 95°C for 30 sec, followed by 40 cycle of 95°C for 5 sec and 60°C for 35 sec.
Feeding bioassay for cotton bollworm larvae with transgenic and negative control cotton plant leaves
For analyzing the expression inhibition of HMGR and Vg in the target cotton bollworm larvae, five first-instar larvae were reared on the second leaves from the top of T1 transgenic and negative control plants. When larvae developed to third instars, they were collected respectively and used for total RNA extraction. For the analysis of larva mortality, 20 newly-hatched cotton bollworm larvae were reared on the tender leaves of negative control and two positive transgenic lines. The lethality of larvae in different lines was detected every 12 hours. This test was repetitiously performed for three times. The student's t-test was used to perform the statistical analyses of the data. For net weight gain analysis, 10 two-instar larvae were reared on second leaves from the top of T1 positive and negative control plants. Every larva was fed separately to avoid the cannibalism. Each larva was weighed every 24 hours individually and fresh leaves were uninterruptedly supplied. To detect the HMGR protein, larvae were reared on negative and positive leaves when they were in the third instars. Three larvae were collected every 24 hours and used to perform protein assay by ELISA.
Quantification of HMGR and Vg expression by qRT-PCR in larvae after feeding on transgenic leaves
In order to detect the change of HMGR and Vg expression in larvae after feeding on transgenic plants expressing dsRNA-HMGR, qRT-PCR were set for the tested larvae according to the previous description[35]. Total RNA was extracted from tested larvae using Trizol (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. RT-PCR and qRT-PCR processes were performed following the same protocol used for dsRNA detection in leaves described previously. Primers used in this test were as our early work [35].
Quantification of HMGR protein by ELISA
The HMGR protein concentration of cotton bollworm larvae was measured by ELISA. The HMGR ELISA kit used here was purchased from Senbeijia Nanjing Biotechnology Co., Ltd. Larvae reared on negative and positive transgenic leaves according to the previous statement were collected respectively every 24 hours and treated according to product instruction manual. The test was repetitiously performed for three times. The student's t-test was used to perform the statistical analyses of the data.
Results
Genetic transformation and molecular analyses of HMGi1 and HMGi2 cotton plants
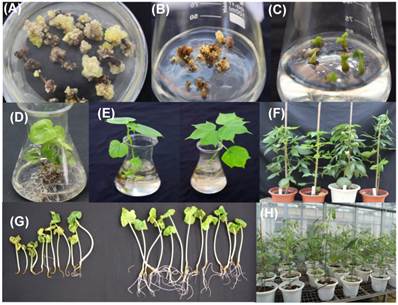
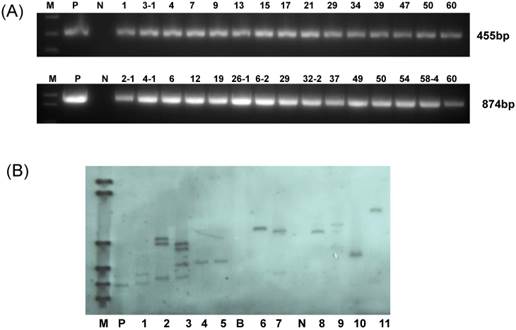
In this experiment, YZ-1, an elite cotton cultivar was used for genetic transformation. Following callus induction (Fig.2A), somatic embryogenesis (Fig.2B) and plant regeneration were realized (Fig.2C-D). All regenerated plantlets were cultured in nutrients-containing water for acclimation (Fig.2E) and then transferred to pots (Fig.2F). Totally, 30 independent positive transgenic T0 plants (15 for each vector of HMGi1 and HMGi2 respectively) were verified by PCR analysis (Fig.3A). Most of these T0 plants were fertile and exhibited normal growth and phenotypes when compared to the wild type control plants containing no constructs. A Kanamycin resistant analysis [30] was performed to screen theT1 population from the positive transgenic T0 plants. As shown in Figure 2G, two types of root phenotypes were observed after the kanamycin test in that some plantlets showed a strong root system with a tap root and lateral roots, and other plantlets had a weak root phenotype without lateral roots (Fig. 2G). A subsequent PCR analysis revealed that only the plants with strong branch roots were PCR positive. The PCR data were in accordance with the root phenotype, suggesting that the antibiotics selection as described here is an effective method for the primary screening of transgenic progeny. Totally, 6 T0 transgenic lines contained a single copy T-DNA insertion and 5 lines had multiple T-DNA copies as verified by the Southern blot data (Fig. 3B). All these PCR and Southern blot positive T0 regeneration plants exhibited normal phenotype and were selected for further analysis.
Creating transgenic cotton plants by Agrobactrium-mediated genetic transformation. (A) Callus induction on selective media containing kanamycin. (B) Embryogenic callus emerged from non-embryogenic callus. (C) Somatic embryos and young plantlets developed from embryogenic callus. (D), (E)and(F) Regenerated putative transgenic plants cultured in rooting media, water and soil. (G)Identification the positive and negative plants of T1 progeny on selective media containing kanamycin. (H)Positive T1 transgenic progeny cultivated in greenhouse.
Higher expression of dsRNA-HMGR in transgenic cotton plants revealed by RT-PCR and qRT-PCR
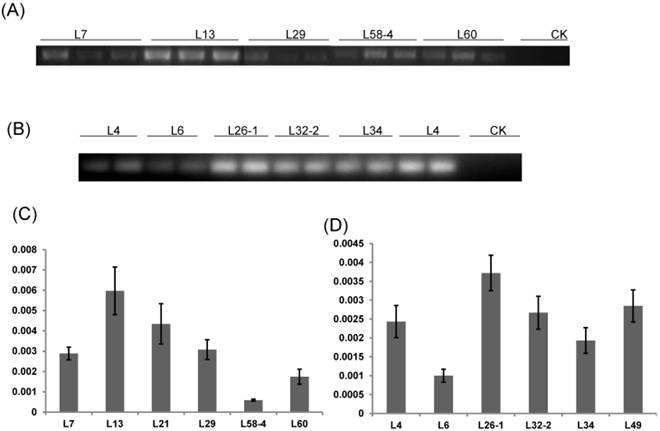
RT-PCR and qRT-PCR were performed to detect the expression level of dsRNA-HMGR in T1 lines from each positive T0 transgenic plant. The result of RT-PCR showed that dsHMGR was expressed in all 11 positive lines (5 lines from HMGi1 and 6 from HMGi2, Fig. 4A and B), while the transcription levels of dsRNAs in these lines appeared diversity, which were quantified by qRT-PCR. All the semi-quantitative results from RT-PCR were confirmed by qRT-PCR suggesting that the dramatic variation of dsRNAs expression existed in different transgenic lines (Fig. 4C and D). For example, the dsHMGR expression level in line L13 of HMGi1 was 9.13 times higher than in the line L58-4.Three lines with relatively higher expression level of dsRNAs were selected for further insect feeding bioassay to analyze the mortality of larvae and the efficiency of RNA interference.
Molecular analysis for the putative transgenic cotton plants. (A) PCR analysis for HMGi1 and HMGi2 putative transgenic plants. M:Marker; N:Negative control; P: Positive control; Numbers marked above the gel indicating the corresponding T0 transgenic plants. (B) Southern blotting analysis of transgenic T0 plants. M: DNA molecular weight marker DIG-labeled (0.12-23.1 kb)(Roche, Germany); P: positive control; B: blank lane (no DNA loading); N: negative control plant DNA; Lane: 1-10 different HMGi transgenic lines.
RT-PCR and qRT-PCR analysis of T1 transgenic cotton plants. (A) RT-PCR analysis of HMGi1 transgenic lines. Different transgenic lines expressed dsHMGR while the expression of dsRNA was absent in negative control; CK: null control; numbers marked above the gel indicating corresponding lines. (B) RT-PCR analysis of HMGi2 transgenic lines. Different transgenic lines expressed dsRNA while the expression of double-strand HMGR was absent in negative control. The relative expression of dsHMGRs in the HMGi1 (C) and HMGi2 (D) transgenic lines were verified by qRT-PCR. The lines, with intense signals in electrophoretogram, were verified to show higher expression levels.
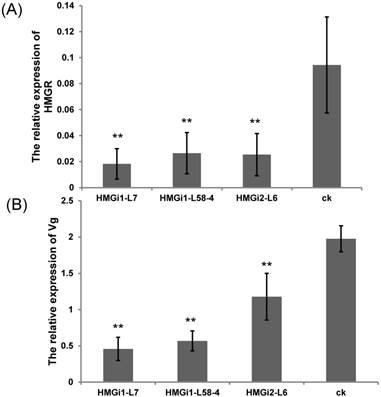
The relative expression of HMGR and vitellogenin gene in cotton bollworm after feeding on T1-generation positive transgenic and control leaves. (A) The expression level of HMGR in cotton bollworm after feeding on the transgenic leaves from three independent lines and control leaves. (B) The expression of vitellogenin gene in cotton bollworm after feeding on the transgenic leaves from three transgenic lines and control leaves. The expressions of HMGR and Vg gene in cotton bollworm were conspicuously downregulated by the transgenic plant derived dsHMGR, in spite of the variation between lines. The test was repetitiously performed for three times. The student's t-test was used to perform the statistical analyses of the data. **statistically significant at 0.01.
Transcription of endogenous HMGR and Vg genes in tested larvae were dramatically suppressed by transgenic cotton plants derived dsHMGR
For the analysis of HMGR and Vg transcription inhibition in cotton bollworm larvae, five first-instar larvae were fed with T1 transgenic and negative control plant leaves till third instars. Then, all the tested larvae were collected respectively and used for qRT-PCR analysis. The expression levels of endogenous HMGR and Vg genes in cotton bollworms were detected by qRT-PCR. Results showed that the transcription levels of endogenous HMGR in larvae reared on transgenic plant leaves expressing dsHMGR were significantly (P<0.01) lower than that on negative leaves (Fig. 5A). Among three tested lines by bioassay, the RNAi efficiency of line HMGi1-7 was found to be higher than other lines. The relative transcription level of HMGR gene in larvae reared on HMGi1-7 transgenic cotton leaves was as much as 80.68% lower than that of wild type. Likewise, the expression level of Vg gene was also dramatically downregulated when the larvae were fed with all three transgenic lines. Still, the line HMGi1-7 exhibited best performance, which reduced the expression level of Vg gene by 76.86% (Fig. 5B). Taken together, the expression levels of HMGR and Vg genes in larvae reared on all the positive lines tested were significantly downregulated by the transgenic plant-derived dsRNAs.
Transgenic cotton plants expressing dsHMGR impaired survival of cotton bollworm larvae
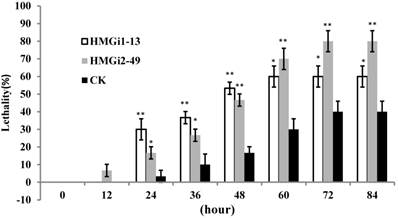
Bioassay of newly-hatched cotton bollworm larvae was performed to testify the lethality by dsHMGR-transgenic plants. The results showed that line HMGi2-49 exhibited higher mortality than line HMGi1-13. However, mortalities resulting from both lines were significantly higher than those of negative control plants at the first 48 hours of bioassay (Fig. 6). After 72 hours, all of the larvae surviving the test kept alive, making the lethality of each group remained unchangeable. Therefore, the results after 96 hours were not illustrated. It was easily to find out that the transgenic cotton leaves expressing dsHMGR caused higher lethality in newly-hatched cotton bollworm larvae, while the survived larvae could keep alive after 72 hours no matter if their food contains dsHMGR or not. A probable explanation is that the tolerance increased with aging may endow larvae with enough ability to bear the lethality brought by dsRNA, while young larvae were sensitive to the dsRNA. In addition, larvae fed on line HMGi2-49 presented higher lethality than HMGi1-13, although the relative expression level of HMGi2-49 was no more than half of HMGi1-13. So we inferred that HMGi2 target sequence in RNA interference vector may be more effective than HMGi1 target sequence on the lethality of young larvae.
Quantification of larvae lethality after feeding on transgenic dsHMGR cotton leaves. For the insect bioassay, 20 newly-hatched cotton bollworm larvae were reared on the detached fresh leaves from null plants as control and two positive transgenic lines. The lethality of different groups was detected every 12 hours. Leaves of positive plants expressing dsRNA-HMGR bring about higher lethality than those of null plants (CK). The test was repetitiously performed for three times. The student's t-test was used to perform the statistical analyses of the data. **statistically significant at 0.01; *statistically significant at 0.05.
Transgenic cotton plants expressing dsHMGR inhibited the growth and development of cotton bollworm
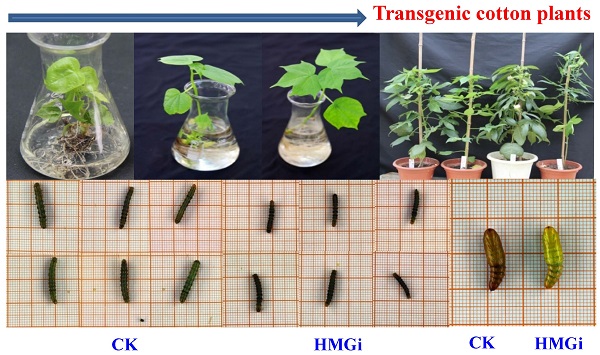
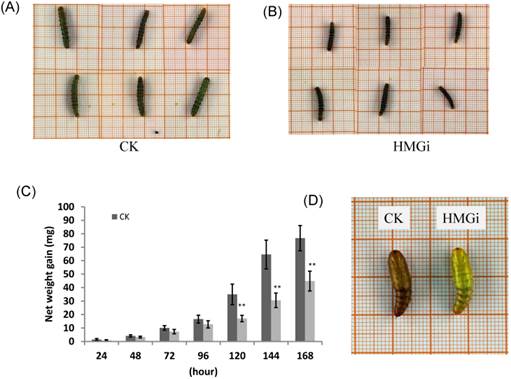
As described previously, newly hatched larvae were very sensitive to plant-derived dsRNA and as much as 80% of tested larvae died 72 h after feeding on the transgenic leaves. So, we also evaluated the effect of transgenic plant derived dsRNAs on the development of relatively older insects (two-instar larvae). Net weight gains and phenotype of two-instar larvae reared on negative control and transgenic leaves were measured to evaluate the suppression efficiency of dsRNAs. The difference on net weight gain between transgenic and control groups was not conspicuous within 96 hours of bioassay, but turned out to be obviously significant (P < 0.01) after 120 hours (Fig. 7C). As is illustrated, the difference was culminated when larvae were fed with negative control and transgenic leaves for 144 hours then decreased a bit 24 hours later, which suggested that the transgenic plant-derived dsRNAs not only inhibited net weight gain but also delayed the growth of cotton bollworm larvae. In the terms of morphological structures, accordingly, the larvae reared on positive transgenic leaves presented smaller body size than those on negative control leaves (Fig. 7A for CK and 7B for HMGi), but any other morphological malformation has not been observed in this experiment. Whether the transgenic plants expressing dsHMGR affect the percentage and time of pupation is not clear based on our available data because most of the tested larvae died before pupation or the larvae number was not even in dsRNAs treated and control group. However, lower frequency of abnormal pupation phenotype was observed and shown in Figure 7D. The larvae reared on transgenic leaves expressing dsHMGRs could pupate on time but their cuticulae were extremely thin and soft (Fig. 7D, on the right), which made the internal structure of pupae can be clearly observed by naked eye. This change may be due to the inhibition effect of plant-delivered dsRNAs on the cuticulae development and the ecdysis of tested larvae.
HMGR accumulation in tested larvae were inhibited by transgenic plants derived dsRNA
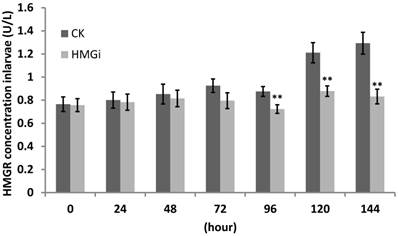
The transcriptional level of HMGR in tested cotton bollworm larvae has been inhibited by transgenic cotton leaves expressing dsHMGR as revealed by RT-and qRT-PCR previously. So, we supposed that HMGR protein accumulation in tested larvae would be suppressed too. For this purpose, ELISA was performed to quantify the HMGR concentration in all tested larvae and the results were summarized in Figure 8. Cotton bollworm larvae fed with negative or positive transgenic cotton leaves did not show obvious variation in HMGR concentration at the first 48 hours of bioassay. However, the difference between transgenic and control groups started to be very significant (P< 0.01) after the larvae have been fed for 96 hours and then the gap was further magnified. The data from 144 hours time point illustrated that the larvae reared on positive transgenic leaves showed 35.71% decrease in the concentration of HMGR, which is significantly lower than in the control group.
The body size, net weight gain and abnormal pupation of cotton bollworm larvae fed on leaves of negative control and positive transgenic cotton leaves. (A) and (B) The larvae feeding on leaves of negative(on the left) and positive(on the right) transgenic plants for 144 h show difference in body size. The growth of cotton bollworms was conspicuously impaired by the ingestion of transgenic leaves expressing dsRNAs. (C) The net weight gain of cotton bollworm larvae fed with transgenic plants expressing ds-HMGRs was impaired. The net weight gain of transgenic and control group did not show significant difference before 96 hours, but became distinct at 120 hours after feeding. The test was repetitiously performed for three times. The student's t-test was used to perform the statistical analyses of the data. **statistically significant at 0.01. (D) The phenotype of pupae pupated from larvae reared on positive and negative transgenic leaves. Larvae fed on leaves expressing dsRNA of HMGR pupated on time but the appearance of their pupae were distinctive from control. Their exocuticle were significantly thinner and softer than control, whose internal structure can be easily observed by naked eye.
Quantification of HMGR enzyme in the larvae feeding on negative and positive transgenic leaves expressing dsRNAs. Third -instar larvae were reared on negative and positive leaves for more than 168 hours. Three larvae were collected every 24 hours and used to perform protein assay by ELISA. Data of ELISA test showed the decrease of HMGR protein concentration by the ingestion of transgenic leaves expressing dsHMGRs. As is illustrated, the inhibiting effect was not distinct within 48 hours, but became conspicuous when larvae had been fed with transgenic leaves for 96 hours. The test was repetitiously performed for three times. The student's t-test was used to perform the statistical analyses of the data. **statistically significant at 0.01.
Discussion
Transgenic pest-resistant plant has been developed for two decades and made enormous contributions to pest control, and the most successful model of which is transgenic Bt cotton. However, the limitations of Bt crops have emerged in several regions or countries. Previous investigators have revealed the resistance to Bt toxins in Busseola fusca [36], Helicoverpa zea [3], Spodoptera frugiperda [37], etc. In addition, Bt crops have specific insecticidal spectrum, which means that only a few insect species from Lepidopterous can be inhibited by Bt toxins. Transgenic pest-resistance crops aiming at sap-sucking insects such as aphid, whitefly, red spider mite need to be designed by novel ideas as Bt crops. In short, the bottleneck of existing Bt crops calls for new strategy in the creation of novel transgenic pest-resistant crops and currently RNAi technique is one of the primary choice of possible strategies.
The application of RNAi in pest control concerns the target genes suppression in insect from specific double-strand RNAs, which has been verified in many insect species. Among the early reports in this area, three methods of dsRNA delivery have been used including injection [38-40], ingestion method [18, 41] and feeding via bacteria expressing dsRNAs [25, 42-44]. Injection method was widely used in this research area for its high efficiency and accuracy in spite of the huge costs. Ingestion method (ingestion of in vitro synthesized dsRNA), another approach to deliver dsRNA into insect, showed potential for application in insect control once dsRNA was verified to down-regulate the target gene expression via ingestion. However, efficiency of these two methods may not be as satisfactory for the instability of dsRNAs when exposed to diverse environments containing Rnase III enzymes. To protect these in vitro synthetic dsRNAs from degradation in the delivering process, bacteria, excellent producer and protector of dsRNAs, were regarded as quality tools. However, all these three methods can hardly be applied for pest control in the field condition. Feeding insect with transgenic plant leaves expressing dsRNA has been developed as an effective method to deliver dsRNA, and first successful application for cotton bollworm control was reported in 2007 [17]. To date, researches using this method have blossomed. Transgenic crops targeting to different insect species such as Helicoverpa armigera [16], whitefly [45], Nilaparvata lugens [46] have been created and genes selected as targets were multifariously involving in detoxification of host allelochemicals, digestion of proteins, larval metamorphosis, etc. Besides, transgenic plants possess relatively stable expression level of dsRNAs between generations, providing insects with continual stress. Most recently, transgenic Arabidopsis plants producing dsHaAK (Helicoverpa armigera arginine kinase) impaired larval growth of Helicoverpa armigera [47]. The ingestion of transgenic plant leaves significantly slowed down the growth of larval, and the transcript levels of target gene HaAK were down-regulated by up to 52%. Surely, dsRNAs delivery based on transgenic plants has greater potential for applications than other delivery methods in pest control.
HMGR, the target gene chosen in this report, plays an important role in larval metamorphosis, growth and development of cotton bollworms. The attendance of HMGR guarantees the biosynthesis of mevalonate, a crucial intermediate metabolite for insects metamorphosis. Our previous work has verified the feasibility of RNAi for HMGR gene in cotton bollworm by injecting in vitro synthesized dsHMGR [35]. So HMGR gene is expected to be as a promising candidate gene for the pest control. In this study, transgenic cotton plants expressing dsHMGR dramatically downregulated the expression of target gene HMGR in both transcription and translation level. Insect bioassay data have verified the inhibiting effects of cotton plant derived dsRNA on survival, growth and development. Notably, the result of qRT-PCR confirmed the downregulation of Vg gene expression, which provided a latent evidence of reproductive barrier for cotton bollworm. The effects brought about by cotton plants expressing dsHMGRs are not intense enough to kill cotton bollworm immediately. But these exogenous dsRNAs endow cotton plants with conspicuous effects on the survival, growth and development of cotton bollworm. The lower lethality of tested larvae may be explained by the limit of interference efficiency such as the barrier in the ingesting of dsRNA and the degradation of dsRNAs in digestive tract of target insects. The second reason for the limited lethal effects of dsRNAs may be due to the lower accumulation of dsRNAs in the transgenic plants. A recent breakthrough in RNAi mediated insect control was achieved by expressing exogenous dsRNAs via chloroplast transformation in potato [48]. In this report, long dsRNAs targeted against the β-actin gene of the Colorado potato beetle can be stably produced and accumulated to as much as 0.4% of the total cellular RNA in potato leaves chloroplasts. Transplastomic potato plants derived dsRNAs were fully protected from herbivores and were lethal to their larvae [48]. Higher accumulation of exogenous toxin or dsRNAs in plant chloroplasts have frequently showed strong lethality to herbivore suggesting that dosage of dsRNA or toxin in transgenic plants was the key factor of its protective function for the host plants [16, 33, 44, 49]. In the current report, the expression level of dsRNAs was not very higher (ranging from 0.001 to 0.006) and varied in different transgenic lines. In the future, increased expression level and stability of plant derived-dsRNAs will substantially improve the efficacy of RNAi for pest control and broaden the application area of this strategy. The third hamper for the application of RNAi concept in plant protection is lack of ideal target genes in herbivore. In major recent reports of plant derived dsRNA mediated RNAi, plant derived dsRNAs only impaired the larvae growth, development and fecundity [16] but did not kill them [50]. This may be due to the target genes selected for RNAi were not fatal for the insect lifecycle or the target genes belong to a big gene family and these homologous genes will replace part of their functions.
RNAi technique is of great significance for pest control, even though the efficiency is not as satisfied as Bt toxins in some cases. Cotton plants expressing dsRNA of HMGR achieved in this research is significantly meaningful as they could hinder the growth and development of cotton bollworms. A significant case is the effect on pupa exocuticle. Larvae fed on transgenic lines expressing dsRNA of HMGR gene could pupate on time but the appearance of pupa was distinctive from control. The pupae pupated from larvae reared on transgenic lines were discovered to possess thinner and softer exocuticle, making the internal structure of pupae can be easily observed by naked eye. Inherent hard exocuticle can keep pupae from mechanical damage to some extent and the weakened exocuticle would lead pupae to be more vulnerable to multifarious injuries in the field. By this means, transgenic lines expressing dsRNA of HMGR may reduce the survival ability of cotton bollworm pupae and inhibit the growth of cotton bollworm population in the long run. Transgenic lines gained in this research may not be applied to breeding as a variety alone, but can be used as a germplasm resource to pyramid with existing Bt cotton by conventional breeding processes to create multiple pest-resistance cotton. In this way, the emergence of pest resistance to Bt cotton may be delayed to some extent. In addition, with progress in insect physiology, genomics and biotechnology, more crucial genes relating to the survival, growth and development of various insect species including sap-sucking pests could be exploited as target genes[51] in RNAi-based pest-resistant breeding in our future work.
Acknowledgements
The work was supported by the National Natural Science Foundation of China (31171592 and 31371673) and Fundamental Research Funds for the Central Universities (2013PY064) to Dr. Shuangxia Jin.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wu KM, Lu YH, Feng HQ, Jiang YY, Zhao JZ. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science. 2008;321:1676-8
2. Bravo A, Soberon M. How to cope with insect resistance to Bt toxins?. Trends in biotechnology. 2008;26:573-9
3. Tabashnik BE, Gassmann AJ, Crowder DW, Carriere Y. Insect resistance to Bt crops: evidence versus theory. Nature biotechnology. 2008;26:199-202
4. Tabashnik BE, Brevault T, Carriere Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol. 2013;31:510-21
5. Gassmann AJ, Petzold-Maxwell JL, Clifton EH, Dunbar MW, Hoffmann AM, Ingber DA. et al. Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proceedings of the National Academy of Sciences. 2014;111:5141-6
6. Brévault T, Heuberger S, Zhang M, Ellers-Kirk C, Ni X, Masson L. et al. Potential shortfall of pyramided transgenic cotton for insect resistance management. Proceedings of the National Academy of Sciences. 2013;110:5806-11
7. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-11
8. Shekhawat UK, Ganapathi TR, Hadapad AB. Transgenic banana plants expressing small interfering RNAs targeted against viral replication initiation gene display high-level resistance to banana bunchy top virus infection. The Journal of general virology. 2012;93:1804-13
9. Wuriyanghan H, Falk BW. RNA Interference towards the Potato Psyllid,, Is Induced in Plants Infected with Recombinant (TMV). PloS one. 2013;8:e66050
10. Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343-9
11. Tang W, He Y, Tu L, Wang M, Li Y, Ruan YL. et al. Down-regulating annexin gene GhAnn2 inhibits cotton fiber elongation and decreases Ca(2+) influx at the cell apex. Plant molecular biology. 2014;85:613-25
12. Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD. et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nature biotechnology. 2006;24:1420-8
13. Huang GZ, Allen R, Davis EL, Baum TJ, Hussey RS. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. P Natl Acad Sci USA. 2006;103:14302-6
14. Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. Journal of insect physiology. 2010;56:227-35
15. Tao XY, Xue XY, Huang YP, Chen XY, Mao YB. Gossypol-enhanced P450 gene pool contributes to cotton bollworm tolerance to a pyrethroid insecticide. Molecular ecology. 2012;21:4371-85
16. Jin S, Singh ND, Li L, Zhang X, Daniell H. Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant biotechnology journal. 2015;13:435-46
17. Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nature biotechnology. 2007;25:1307-13
18. Bautista MA, Miyata T, Miura K, Tanaka T. RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect biochemistry and molecular biology. 2009;39:38-46
19. Zhu JQ, Liu SM, Ma Y, Zhang JQ, Qi HS, Wei ZJ. et al. Improvement of Pest Resistance in Transgenic Tobacco Plants Expressing dsRNA of an Insect-Associated Gene EcR. PloS one. 2012:7
20. Sui YP, Wang JX, Zhao XF. Effects of classical insect hormones on the expression profiles of a lipase gene from the cotton bollworm (Helicoverpa armigera). Insect Mol Biol. 2008;17:523-9
21. Sui YP, Liu XB, Chai LQ, Wang JX, Zhao XF. Characterization and influences of classical insect hormones on the expression profiles of a molting carboxypeptidase A from the cotton bollworm (Helicoverpa armigera). Insect Mol Biol. 2009;18:353-63
22. Sui YP, Wang JX, Zhao XF. The impacts of classical insect hormones on the expression profiles of a new digestive trypsin-like protease (TLP) from the cotton bollworm, Helicoverpa armigera. Insect Mol Biol. 2009;18:443-52
23. Rafaeli A, Bober R. The effect of the juvenile hormone analog, fenoxycarb on the PBAN-receptor and pheromone production in adults of the moth Helicoverpa armigera: an "aging" hormone in adult females?. Journal of insect physiology. 2005;51:401-10
24. Asokan R, Chandra GS, Manamohan M, Kumar NKK, Sita T. Response of various target genes to diet-delivered dsRNA mediated RNA interference in the cotton bollworm, Helicoverpa armigera. J Pest Sci. 2014;87:163-72
25. Xiong Y, Zeng H, Zhang Y, Xu D, Qiu D. Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. International journal of biological sciences. 2013;9:370-81
26. Liu Y, Sheng Z, Jiang RJ, Huang Y, Li S. Progress in juvenile hormone synthesis. Acta Entomologica Sinica. 2007;60:1285-92
27. Feyereisen R, Farnsworth DE. Characterization and regulation of HMG-CoA reductase during a cycle of juvenile hormone synthesis. Molecular and cellular endocrinology. 1987;53:227-38
28. Feyereisen R, Farnsworth DE. Precursor supply for insect juvenile hormone III biosynthesis in a cockroach. Journal of Biological Chemistry. 1987;262:2676-81
29. Jin S, Zhang X, Nie Y, Guo X, Liang S, Zhu H. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plantarum. 2006;50:519-24
30. Li LB, Zhu Y, Jin SX, Zhang XL. Pyramiding Bt genes for increasing resistance of cotton to two major lepidopteran pests: Spodoptera litura and Heliothis armigera. Acta Physiol Plant. 2014;36:2717-27
31. Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM. High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol. 2002;29:1217-25
32. Jin SX, Zhang XL, Liang SG, Nie YC, Guo XP, Huang C. Factors affecting transformation efficiency of embryogenic callus of Upland cotton (Gossypium hirsutum) with Agrobacterium tumefaciens. Plant Cell Tiss Org. 2005;81:229-37
33. Jin SX, Liu GZ, Zhu HG, Yang XY, Zhang XL. Transformation of Upland Cotton (Gossypium hirsutum L.) with gfp Gene as a Visual Marker. J Integr Agr. 2012;11:910-9
34. Liu GZ, Jin SX, Liu XY, Tan JF, Yang XY, Zhang XL. Overexpression of Arabidopsis cyclin D2;1 in cotton results in leaf curling and other plant architectural modifications. Plant Cell Tiss Org. 2012;110:261-73
35. Wang Z, Dong Y, Desneux N, Niu C. RNAi silencing of the HaHMG-CoA reductase gene inhibits oviposition in the Helicoverpa armigera cotton bollworm. PloS one. 2013;8:e67732
36. van Rensburg JBJ. First report of field resistance by the stem borer, Busseola fusca (Fuller) to Bt-transgenic maize. South African Journal of Plant and Soil. 2007;24:147-51
37. Storer NP, Babcock JM, Schlenz M, Meade T, Thompson GD, Bing JW. et al. Discovery and Characterization of Field Resistance to Bt Maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. Journal of economic entomology. 2010;103:1031-8
38. Mutti NS, Park Y, Reese JC, Reeck GR. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. Journal of insect science. 2006:6
39. Brown S, Holtzman S, Kaufman T, Denell R. Characterization of the Tribolium Deformed ortholog and its ability to directly regulate Deformed target genes in the rescue of a Drosophila Deformed null mutant. Development genes and evolution. 1999;209:389-98
40. Hughes CL, Kaufman TC. RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head. Development. 2000;127:3683-94
41. Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O. et al. Control of coleopteran insect pests through RNA interference. Nature biotechnology. 2007;25:1322-6
42. Tian H, Peng H, Yao Q, Chen H, Xie Q, Tang B. et al. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PloS one. 2009;4:e6225
43. Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854
44. Jin S, Zhang X, Daniell H. Pinellia ternata agglutinin expression in chloroplasts confers broad spectrum resistance against aphid, whitefly, lepidopteran insects, bacterial and viral pathogens. Plant biotechnology journal. 2012;10:313-27
45. Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK. Enhanced Whitefly Resistance in Transgenic Tobacco Plants Expressing Double Stranded RNA of v-ATPase A Gene. PloS one. 2014:9
46. Yu R, Xu XP, Liang YK, Tian HG, Pan ZQ, Jin SH. et al. The Insect Ecdysone Receptor is a Good Potential Target for RNAi-based Pest Control. International journal of biological sciences. 2014;10:1171-80
47. Liu F, Wang X-D, Zhao Y-Y, Li Y-J, Liu Y-C, Sun J. Silencing the HaAK Gene by Transgenic Plant-Mediated RNAi Impairs Larval Growth of Helicoverpa armigera. International journal of biological sciences. 2015;11:67
48. Zhang J, Khan SA, Hasse C, Ruf S, Heckel DG, Bock R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science. 2015;347:991-4
49. Jin SX, Kanagaraj A, Verma D, Lange T, Daniell H. Release of Hormones from Conjugates: Chloroplast Expression of beta-Glucosidase Results in Elevated Phytohormone Levels Associated with Significant Increase in Biomass and Protection from Aphids or Whiteflies Conferred by Sucrose Esters. Plant physiology. 2011;155:222-35
50. Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA. Silencing of aphid genes by dsRNA feeding from plants. PloS one. 2011;6:e25709
51. Younis A, Siddique MI, Kim C-K, Lim K-B. RNA Interference (RNAi) Induced Gene Silencing: A Promising Approach of Hi-Tech Plant Breeding. International journal of biological sciences. 2014;10:1150
Author contact
![]() Corresponding author: jsxhzau.edu.cn; Fax: (+86) 27 8728 0016; Tel: (+86) 27 8728 0016
Corresponding author: jsxhzau.edu.cn; Fax: (+86) 27 8728 0016; Tel: (+86) 27 8728 0016
Received 2015-4-21
Accepted 2015-7-28
Published 2015-9-15
