10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(7):884-897. doi:10.7150/ijbs.15194 This issue Cite
Review
Interactions between Autophagy and Inhibitory Cytokines
1. Department of Hepatobiliary Surgery, the 309th Hospital of Chinese PLA, Beijing 100091, People's Republic of China
2. Trauma Research Center, First Hospital Affiliated to the Chinese PLA General Hospital, Beijing 100048, People's Republic of China
3. State Key Laboratory of Kidney Disease, the Chinese PLA General Hospital, Beijing 100853, People's Republic of China
Received 2016-2-3; Accepted 2016-5-12; Published 2016-6-7
Abstract
Autophagy is a degradative pathway that plays an essential role in maintaining cellular homeostasis. Most early studies of autophagy focused on its involvement in age-associated degeneration and nutrient deprivation. However, the immunological functions of autophagy have become more widely studied in recent years. Autophagy has been shown to be an intrinsic cellular defense mechanism in the innate and adaptive immune responses. Cytokines belong to a broad and loose category of proteins and are crucial for innate and adaptive immunity. Inhibitory cytokines have evolved to permit tolerance to self while also contributing to the eradication of invading pathogens. Interactions between inhibitory cytokines and autophagy have recently been reported, revealing a novel mechanism by which autophagy controls the immune response. In this review, we discuss interactions between autophagy and the regulatory cytokines IL-10, transforming growth factor-β, and IL-27. We also mention possible interactions between two newly discovered cytokines, IL-35 and IL-37, and autophagy.
Keywords: autophagy, inhibitory cytokines, adaptive immune response, innate immune response
Introduction
Autophagy is an essential cellular pathway that contributes to homeostasis by degrading a variety of intracellular components [1, 2]; in particular, toxic protein aggregates, dysfunctional organelles, and invading pathogens [3-7]. Three major autophagy pathways have been described that vary in the method of cargo delivery: macroautophagy, microautophagy, and chaperone-mediated autophagy [8]. Macroautophagy is characterized by formation of intracellular double-membrane structures known as autophagosomes. A large body of evidence has confirmed that macroautophagy is a selective process and requires a variety of autophagy-related proteins (ATG) for the initiation, maturation, and termination of the process [9, 10]. In this review, we will discuss only macroautophagy (hereafter referred to as autophagy). Most early studies of autophagy focused on its involvement in age-associated degeneration and nutrient deprivation. However, in recent years, there has been a dramatic increase in studies of the immunological functions of autophagy. Thus far, autophagy has been shown to be an intrinsic cellular defense mechanism that protects against invading pathogens, such as viruses and bacteria. Abnormal autophagy appears to be linked to a wide spectrum of human disorders, including autoimmune disease, infection, sterile inflammation, and even neoplasms.
Cytokines belong to a broad and loose category of proteins and are crucial for both innate and adaptive immune responses. Inhibitory cytokines have significant functions in immune defense, as they have evolved to permit tolerance to self while also contributing to the eradication of invading pathogens. Interactions between cytokines and autophagy have been described in several recent articles that shed light on a novel mechanism for controlling the immune response. In this review, we will mainly discuss the interactions between autophagy and three regulatory cytokines: the conventional inhibitory cytokines IL-10 and transforming growth factor (TGF)-β, the newcomer IL-27. We will also speculate on the possible interactions between IL-35 and IL-37 and autophagy, which to date remain only theoretical.
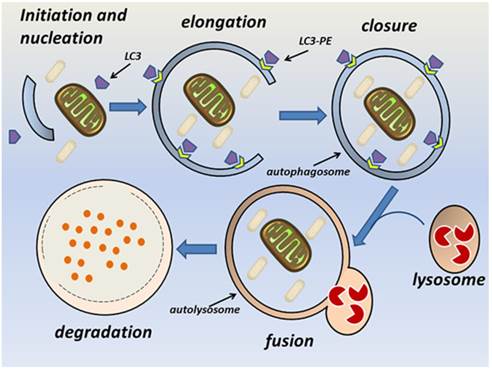
The autophagy pathway. Autophagy consists of sequential steps involving vacuole initiation, nucleation, elongation, closure, fusion, and degradation. Initiation occurs as a response to multiple potential autophagic stimuli. Nucleation is characterized by formation of an autophagic phagophore in the cytoplasm. During the elongation and closure stage, a double-membrane autophagosome develops into its mature form and fuses with a lysosome to become an autolysosome. This is followed by lysosomal degradation of the autophagic cargo.
The autophagy pathway
The autophagy pathway requires the orchestrated assembly of multiple ATG proteins into functional complexes [2]. Briefly, autophagy consists of sequential steps involving vacuole initiation, nucleation, elongation, closure, fusion, and degradation (Fig. 1). Initiation is triggered by the cellular response to multiple potential autophagic stimuli and is mediated mainly by the Unc-51-like kinase 1 (ULK1) complex. This complex is composed of ULK1, ATG13, FIP200, and ATG101 and is regulated by mammalian target of rapamycin (mTOR) [11-14]. At the nucleation stage, an isolated membrane structure known as an autophagic phagophore (autophagosome) is formed in the cytoplasm [9]. The key modulator of nucleation is the Beclin-1-class III phosphatidylinositol 3-kinase (PI3K) complex, which is composed of vacuolar protein sorting 34 (Vps34), ATG14, and Beclin-1. The principle function of this complex is to recruit lipids and proteins to the phagophore and thus promote membrane formation [18]. The UV irradiation resistance-associated gene (UVRAG) promotes trafficking of the Beclin-1-class III PI3K complex and ATG9 [15]. The endoplasmic reticulum, Golgi apparatus, and mitochondria can all be sources of the phagophore membrane [16-18]. At the elongation and closure stage, the phagophore can be visualized as a double-membrane structure that develops into a mature autophagosome [9]. Two ubiquitin-like conjugation systems facilitate elongation of the phagophore; namely, the Atg12-Atg5-ATG16L and Atg4B-Atg3-Atg7 complexes [19, 20]. Atg7 and Atg3 [21, 22] facilitate conjugation of the cytosolic form of LC3 to phosphatidylethanolamine (PE), thereby converting it into the membrane-binding form LC3-PE. The Atg12-Atg5-ATG16L complex specifies the site of LC3 binding to the membrane and facilitates recruitment of other components of the autophagy machinery to sites of autophagosome formation [23, 24]. P62 (also known as sequestosome or SQSTM1) is an ubiquitin-binding protein that participates in this process, and changes in p62 levels can be used as an autophagic flux marker [25]. Endophilin B1 interacts with Beclin-1 through UVRAG to positively regulate and facilitate autophagosome formation [26]. At the fusion stage, the mature autophagosome fuses with a lysosome to form a single membrane-bound autolysosome through the interaction of syntaxin 17 and the homotypic fusion and vacuole protein sorting complex [27-29]. The fusion stage is followed by acidification of the lysosome and degradation of the autophagosome contents [9]. Several studies have demonstrated that autophagy is regulated by multiple immunological signals, including pattern recognition receptors (PRRs) [30, 31], mTOR [32], AMP-activated protein kinase [9], and high mobility group box-1 protein [33]. Additionally, pro-inflammatory cytokines, including IL-1β [34], interferon (IFN)-γ [35-37], and tumor necrosis factor (TNF)-α [38], stimulate autophagy, whereas Th2 cytokines, such as IL-4 and IL-13, inhibit autophagy [35].
Autophagy in the immune response
The innate immune system induces an inflammatory response that protects the host against microbial invasion. Multiple classes of PRRs, including Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), and retinoic acid-inducible gene (RIG-1)-I-like receptors (RLRs), have been shown to induce autophagy at different stages of the immune response. For example, TLR2, TLR3, TLR4, TLR7, and TLR8 all activate innate immune cells [39], and TLR stimulation can induce autophagy, which contributes to the anti-microbial response [40]. Conversely, autophagy can stimulate TLRs by transporting microbe-derived cytoplasmic pathogen-associated molecular patterns (PAMPs) to the endosomal lumen, where they activate TLR signaling [41]. A relationship between NLRs and autophagy has been revealed in mammals. NOD1 and NOD2 recruit ATG16L1 to the pathogen-entering site at the plasma membrane [42] and thus induce defensive autophagy. NLRX1 promotes the formation of ATG5-ATG12-ATG16L1 complexes [43]. In contrast, NLRP4 negatively regulates autophagy through its interaction with Beclin-1; consistent with this, RNA interference-mediated blockade of NLRP4 enhances autophagy under physiological conditions and during invasive bacterial infections [44]. NLRP4 also inhibits maturation of the autophagosome by associating with the class C vacuolar protein sorting complex [44]. RLR signaling also interacts with autophagy. Atg5 deficiency augments RLR stimulation, most likely via increased intracellular levels of reactive oxygen species (ROS) [45]. The Atg5-Atg12 conjugate interacts with caspase recruitment domains of RIG-I and mitochondrial antiviral signaling and thus inhibits IFN induction [46]. However, a downstream component of RLR signaling, stimulator of IFN genes, has been reported to activate TANK-binding kinase 1 and type I IFN production, which induces autophagy [47-49]. Autophagy also supplements the anti-microbial barrier function of conventional PRRs by mediating clearance of cytoplasmic pathogens through sequestosome-1/p62-like receptors (SLRs) [50-52].
Accumulating evidence indicates that autophagy suppresses activation of inflammasomes [53-55], which are expressed in myeloid cells and are the components of the innate immune system. For example, a lack of functional ATG16L1 increases the secretion of IL-1β and IL-18 [56]. Damaged mitochondria are an important source of inflammasome agonists, particularly mitochondrial DNA and ROS [55, 57]. Mitophagy is a specialized form of autophagy that removes aging and dysfunctional mitochondria and thus limits inflammasome activation [53]. Upon recognition by p62, inflammasomes are selectively degraded through autophagy, decreasing the production of IL-18 and IL-1β. The latter cytokine can be directly degraded in autolysosomes [58]. Previous work has demonstrated that cytosolic lipopolysaccharide (LPS) activates non-canonical inflammasomes and induces the production of IL-1α and/or IL-1β and IL-18 [59]. Autophagy eliminates bacteria-containing vacuoles by lysosomal degradation and thus inhibits activation of such non-canonical inflammasomes [59].
Recent work has provided insight into the role of autophagy in antigen-specific immune responses. In antigen presenting cells (APCs), autophagy degrades pathogen-derived antigens into peptides and transports them from the cytoplasm to major histocompatibility complex (MHC) compartments where they are assembled with MHC class II molecules [60-62]. Induction of autophagy or association of antigens with autophagosomes enhances MHC class II-mediated presentation of intracellular antigens to CD4+ T cells [63-65]. In addition to endogenous antigens, exogenous antigen presentation by MHC class II is promoted by autophagy via PRR-stimulated LC3-associated phagocytosis [66], in which LC3 binds to phagosomal membranes conjugated to TLR2, TIM4, Fc receptors, or dectin-1-bound cargo [67-70]. In contrast, autophagy may play dual roles in MHC class I-mediated antigen presentation because although it can limit protein degradation by proteasomes [71], it may also contribute to antigen presentation during herpes simplex virus type 1 or human cytomegalovirus infections [72, 73]. Autophagy is required for the unconventional pathway of MHC class I-mediated antigen presentation; namely, cross-presentation [72, 74]. Autophagosomes process and degrade protein antigens into peptides, which are assembled with MHC class I molecules in the endoplasmic reticulum [72]. Changes in autophagosome maturation can result in autophagic exocytosis, which might contribute to antigen processing and facilitate cross-presentation [75, 76].
In addition to promoting antigen processing, autophagy can regulate adaptive immunity via effects on the development, homeostasis, and function of T cells. After leaving the thymus, naive T cells rely on autophagy and mitochondrial quality control for further maturation [77]. In activated T cells, not only is autophagy increased by CD3 and CD28 co-stimulation [78], it is also required for cell survival [79]. Furthermore, autophagy seems indispensable for the expansion of lymphocyte populations and maintenance of memory [80-82]. Autophagy is also necessary for the survival of B1 cells [83] and plays an important role in plasma cell antibody secretion, survival, and homeostasis [84].
Inhibitory cytokines and the immune response
IL-10
IL-10 is an immunomodulatory cytokine that plays an essential role in maintaining immune homeostasis [85]. Binding of IL-10 to its receptor activates Janus kinase 1 and Tyk2, which phosphorylate signal transducer and activation of transcription (STAT)1 and STAT3 [86-89]. The phosphorylated STATs then translocate to the nucleus where they regulate the expression of many key genes involved in cytokine production [90]. IL-10 inhibits both the innate and adaptive immune responses, and it decreases the ability of monocytes and macrophages to produce a variety of pro-inflammatory cytokines [91]. IL-10 affects the expression of matrix metalloproteinase-9, inducible nitric oxide synthase, and IFN, and induces production of miR-187, a microRNA that reduces the stability of pro-inflammatory cytokines, including TNF-α [92]. Moreover, IL-10 interferes with IFN-γ-induced activation of monocytes [93] and cathepsin S expression in macrophages [94], hampering MHC class II formation and antigen presentation [94]. It also suppresses macrophage function by inhibiting nitric oxide production and is essential for the immunosuppressive function of Tr1 cells [95]. In various subsets of CD4+ T cells, including Th17 and Th1 cells, IL-10 plays regulatory roles through self-regulation and through control of Tregs [96]. In the innate immune response, IL-10 directly suppresses TLR signaling and activates the PI3K-Akt-GSK signaling pathway, which is involved in suppression of inflammatory genes [97].
TGF-β
TGF-β plays a key role in regulating the inflammatory response and maintaining immune homeostasis. TGF-β binding to its receptors induces phosphorylation of Smad2 and Smad3, which elicit diverse cellular actions [98, 99]. Inhibitory Smad7 acts as a negative regulator of TGF-β signaling [100]. In different cell contexts, TGF-β activates a variety of Smad-independent signaling pathways, including PI3K, mitogen-activated protein kinase (MAPK), Rho family proteins, the epithelial polarity protein Par6, and PP2A phosphatase [101, 102]. TGF-β regulates the proliferation, differentiation, and survival of a number of immune cells; for example, it downregulates T cell activation at the level of T cell receptor signaling, and its suppression of Th1 polarization is important in preventing Th1-mediated colitis [103, 104]. TGF-β also downregulates CD8+ T cell proliferation and cytotoxic activity [105-108] and promotes generation of induced Tregs [109-115]. In the presence of retinoic acid, TGF-β converts CD4+ T cells into Tregs [116-121]. The effects of TGF-β on Th17 cell differentiation are controversial. Several reports using various methods of TGF-β signaling blockade have revealed important effects of TGF-β on Th17 differentiation [122-126]. However, deletion of Smad2, Smad3, or Smad4 in T cells failed to impair Th17 development [127-129]. TGF-β suppresses NK cell cytolytic activity by inhibiting NKp30 and NKG2D receptors [130, 131], and exogenous administration of TGF-β suppresses B cell proliferation and antibody secretion [132, 133]. TGF-β1 blocks nuclear factor (NF)-κB activation in TLR-stimulated macrophages by facilitating the proteasomal degradation of MyD88 [134]. In dendritic cells (DCs), TGF-β downregulates pro-inflammatory cytokines, including TNF-α, IL-12, and IFN-α, but upregulates regulatory cytokines, including TGF-β itself [135, 136].
IL-27
IL-27 is an immunomodulatory cytokine expressed mainly by APCs stimulated with microbial products or inflammatory mediators [137-139]. Tregs, IFN-γ, and statins can induce IL-27 production by APCs. Binding of IL-27 to its receptor (IL-27R) induces STAT1 and STAT3 activation [139, 140]. IL-27 plays a complex role in immunoregulation as it has pro-inflammatory and anti-inflammatory effects on many cell types, including T cells, B cells, macrophages, and DCs [141]. The effects of IL-27 on Th1 cells are controversial and may be context dependent. IL-27 was reported to promote the induction of Th1 differentiation [142, 143]. However, in Il27ra-/- mice, Th1 responses are sufficient to eliminate intracellular pathogens [144]. IL-27 suppresses expression of the transcription factors RORγt [145] and RORα [146], the granulocyte-macrophage colony-stimulating factor (GM-CSF) [147], and the Th2-specific transcription factor GATA3[148, 149]. IL-27Rα-deficient mice are more prone to intracellular pathogen infection than wild-type mice [143, 150, 151]. In the experimental autoimmune encephalitis mouse model of human multiple sclerosis, IL-27 downregulates the differentiation and development of Th17 cells and lowers the incidence and severity of the disease[152-154]. IL-27 promotes the differentiation and expansion of Tr1 cells by inducing the transcription factor c-Maf, IL-21, and costimulatory receptor ICOS [155], and thus enhances the production of inhibitory IL-10 [156, 157]. Finally, IL-27 reduces Fork head box 3 (Foxp3) expression in inducible Tregs [158] and antagonizes their generation by TGF-β and IL-2 [159, 160]. Further research will be necessary to investigate how the pro-inflammatory activity of IL-27 in limiting Tregs is balanced by its anti-inflammatory activity in inducing IL-10.
IL-35
IL-35 is an anti-inflammatory and immunosuppressive cytokine [161] expressed by thymus-derived nTregs, peripheral iTregs, and DCs [162-165]. Another important source of IL-35 is iTr35 cells, a population of CD4+ Tregs induced by IL-35 [166]. IL-35 has also been reported to be expressed by CD8+ Tregs in prostate cancer patients [167] and some tumor cells [168]. A broad range of cell types can express IL-35 in the context of inflammation [164]. IL-35 binding to its receptor induces phosphorylation of STAT1 and/or STAT4, which mediate many of the immunosuppressive effects of IL-35 [169-171]. The predominant regulatory property of IL-35 is its ability to suppress T cell proliferation and inhibit their effector functions. IL-35 is required for the maximal suppressive activity of Tregs [161], and its deficiency significantly decreases the ability of CD4+ Tregs to suppress T cell proliferation [161]. Administration of recombinant IL-35 decreases T cell proliferation and cytokine expression [161], and ectopic expression of IL-35 in conventional CD4+ T cells causes a switch of their phenotype from effector to regulator [161, 166]. Although IL-35 induces the proliferation of Tregs, it also promotes the expression of suppressive IL-10 [172, 173]. Additionally, IL-35 hampers the development and function of Th17 cells, and it mediates the function of regulatory B cells [166, 170, 172, 174]. As mentioned, IL-35 induces a specific Foxp3- Treg cell subset, iTr35, which suppresses immune responses via IL-35 rather than IL-10 or TGF-β [162, 166]. In response to IL-35, CD8+ CTLA-4+ Tregs suppress autologous T cell proliferation in a contact-independent fashion [167].
IL-37
IL-37 is a newly discovered member of the IL-1 family. As an anti-inflammatory cytokine [175], IL-37 is expressed at low levels in peripheral blood mononuclear cells (PBMCs), mainly in monocytes and DCs [176]. In general, IL-37 is upregulated by inflammatory stimuli [176], and it can be induced by various TLR agonists and pro-inflammatory cytokines, including IL-1β, TNF-α, and IFN-γ [177]. However, TGF-β is the most effective stimulus for IL-37 induction, whereas IL-4 and GM-CSF inhibit constitutive IL-37 expression [178]. IL-37 binds to IL-18Rα and exploits IL-1R8 (SIGIRR) to mediate its anti-inflammatory activities in mouse models [179, 180]. P38 MAPK and extracellular signal-regulated kinase (ERK)1/2 pathways may be involved in IL-37 production [181]. IL-37 is known to play a negative role in Th1/Th17 cell responses mediated by human PBMCs, M1 macrophages, or monocyte-induced DCs (moDCs). Human PBMCs treated with a neutralizing anti-IL-37 monoclonal antibody show increased secretion of pro-inflammatory cytokines, such as IL-6, IL-1β, and TNF-α [180]. Similarly, siRNA-mediated silencing of IL-37 elevates IL-6, IL-1β, and TNF-α secretion by PBMCs after stimulation with the TLR ligands LPS or Pam3CSK4 [178]. Conversely, treatment with IL-37 decreases expression of these cytokines in LPS-stimulated M1 macrophages, probably by reducing phosphorylation of p38 MAPK, ERK, and c-Jun N-terminal kinase (JNK) [180]. Moreover, IL-37 treatment significantly inhibits IL-6, IL-1β, TNF-α, and ROS production in LPS-stimulated moDCs [182]. When CD4+ T cells are cocultured with moDCs, administration of IL-37 decreases formation of IL-17 and IFN-γ [182]. Expression of IL-37b in a mouse macrophage cell line reduced the expression of a variety of inflammatory cytokines, and similar results were observed in human monocytic and epithelial cell lines [178]. Thus, IL-37 inhibits the production of pro-inflammatory components in response to inflammatory stimuli. A recent study reported that IL-37 might be involved in peripheral tolerance through generation of semi-mature tolerogenic DCs in a murine contact hypersensitivity model [183], suggesting another possible cellular mechanism by which IL-37 can regulate the adaptive immune response.
Interactions between autophagy and inhibitory cytokines
IL-10 and autophagy
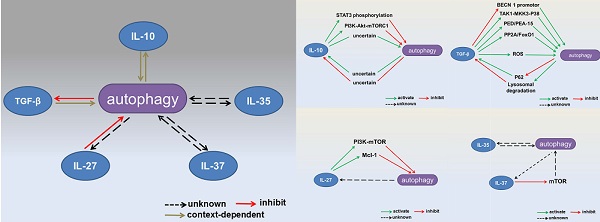
As an important regulator of the immune system, autophagy might be expected to affect IL-10 production by macrophages and DCs, and conversely, IL-10 is likely to have many effects on autophagy (Table 1, Figure 2 and Figure 3a). Indeed, there is evidence for both, although the precise regulatory mechanisms involved remain largely elusive.
Autophagy can enhance IL-10 production
Treatment of murine macrophages with globular adiponectin increases the abundance of autophagic vacuoles visible by transmission electron microscopy, indicative of autophagy induction, and elevates IL-10 mRNA levels [184]. However, the same treatment markedly downregulates IL-10 expression in Atg5-deficient macrophages [184], revealing a “direct” effect of autophagy on IL-10 expression. In mouse NG108-15 neural cells, addition of amyloid-β25-35 induces autophagosome formation and increases IL-10 levels, whereas IL-10 is significantly reduced by treatment with madecassoside, a triterpenoid saponin compound, or the autophagy inhibitor 3-methyladenine (3-MA) [185]. This report therefore indicates a positive effect of autophagy on IL-10 production. However, whether the reduction in IL-10 (determined by western blotting and ELISA) was the result of direct or indirect effects of autophagy inhibition could not be deduced. In DCs generated from the PBMCs of healthy volunteers, blockade of autophagy with ATG16L1 siRNA or 3-MA decreased IL-10 concentrations in the supernatant and increased T cell proliferation in an allogeneic mixed lymphocyte reaction [186]. Here too, the detailed mechanism by which this occurred was not reported.
Interactions between autophagy and IL-10
| Interactions | Authors | Year | Summary | References |
|---|---|---|---|---|
| Autophagy enhances IL-10 production | Qi, et al. | 2014 | In murine macrophages, autophagy increased the expression of IL-10. | [184] |
| Du, et al. | 2014 | In mouse neural cells, amyloid-β25-35 induced autophagosome formation accompanied by an elevated level of IL-10 and 3-MA resulted in significantly reduction of IL-10. | [185] | |
| Strisciugli, et al. | 2013 | In dendrite cells, blockade of autophagy resulted in decreased IL-10 production. | [186] | |
| Autophagy may reduce IL-10 production | Wang, et al. | 2015 | In the medium of LPS-treated spinal neuron-glia co-culture system, VEGF165 upregulated autophagy and decreased IL-10, and this effect were partially blocked by autophagy inhibitor. | [187] |
| IL-10 inhibits autophagy | Wang, et al. | 2014 | IL-10 inhibited autophagic flux of the MRC5 cells in the context of starvation or human CMV infection. | [190] |
| Santarelli, et al. | 2014 | IL-10 maintained STAT3 phosphorylation, which correlated to a blockade of autophagy. | [194] | |
| Buchser, et al. | 2012 | IL-10 attenuated the target tumor cell autophagy promoted by human peripheral blood lymphocytes. | [191] | |
| Park, et al. | 2011 | IL-10 signaling inhibited autophagy of murine macrophage. | [192] | |
| Van Grol, et al. | 2010 | Neutralization of IL-10 restored autophagy in by-standing macrophages/monocytes during HIV-1 infection. | [193] | |
| IL-10 may promote autophagy | Martinez-Outschoorn, et al. | 2011 | IL-10 induced autophagic response in fibroblasts. | [195] |
Autophagy may reduce IL-10 production
In contrast to the preceding reports, recent investigations have indicated possible negative effects of autophagy on IL-10 production. The growth factor VEGF165 upregulates Beclin-1 and LC3 expression in LPS-treated spinal neuron-glia co-cultures but decreases IL-1β, IL-10, and TNF-α expression in a manner that was partially blocked by 3-MA [187]. A coding polymorphism in ATG16L1 (threonine 300 to alanine) increases IL-10 and IL-6 expression in monocytes treated with Mycobacterium avium subspecies paratuberculosis [188], and systemic lupus erythematosus patients carrying an Atg5 gene mutation appear to have an increased ability to produce IL-10 [189]. The latter two reports suggest that impaired autophagy may correlate with elevated IL-10 production and that autophagy may thus negatively regulate IL-10 production. Nevertheless, a solid mechanistic explanation for this is lacking and must await further investigation.
IL-10 can inhibit autophagy
In MRC5 human lung fibroblasts subjected to nutrient deprivation or infected with CMV, IL-10 inhibits the autophagic flux via the PI3K/Akt signaling pathway, and this is reversed by induction of autophagy via rapamycin treatment or upregulation of BECN1 gene expression[190]. IL-10 also attenuates autophagy in tumor cells targeted by human peripheral blood lymphocytes (also called lymphocyte-mediated autophagy) [191]. As reported by Park [192], IL-10 signaling in murine macrophages inhibits autophagy and activates the class I PI3K pathway, which in turn leads to p70S6K phosphorylation through activation of Akt and mTORC1. Infection with HIV-1 blocks rapamycin-induced autophagy in bystander (uninfected) macrophages/monocytes, and this is reversed by neutralization of IL-10 [193]. Exposure of DCs to active or UV-inactivated Kaposi's sarcoma associated herpes virus stimulates STAT3 phosphorylation and increases secretion of IL-10 [194]. The subsequent maintenance of STAT3 phosphorylation by IL-10 correlates with a blockade in autophagy [194]. These reports document the inhibitory effect of IL-10 on autophagy. However, the underlying mechanisms by which the autophagy flux is altered remain to be elucidated. Exploration of this issue may shed new light on immunomodulation.
IL-10 may promote autophagy
While the majority of reports to date have demonstrated a negative effect of IL-10 on autophagy, Martinez-Outschoorn et al. showed that co-culture of fibroblasts with MCF7 breast cancer cells upregulates IL-10 expression, and further, that IL-10 induces the autophagic response in these cells [195]. Thus, the regulatory effect of IL-10 on autophagy appears to be context dependent.
TGF-β and autophagy
Interactions between TGF-β signaling and autophagy have been reported recently. As an intracellular degradation mechanism, autophagy controls TGF-β signaling in different ways, while TGF-β has been shown to induce autophagy in various cellular contexts. Several recent studies have also indicated that TGF-β can have a negative effect on autophagy (Table 2, Figure 2 and Figure 3b).
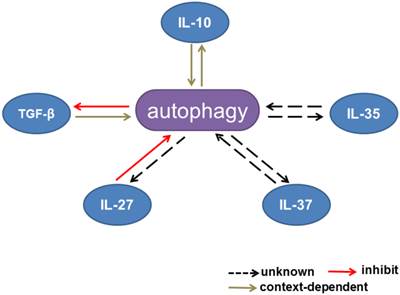
Interactions between autophagy and inhibitory cytokines. Interactions between autophagy and IL-10 production are context dependent. Autophagy inhibits TGF-β signaling, whereas TGF-β has dual effects on the autophagic flux. IL-27 has been reported to inhibit autophagy. The effects of IL-35 and IL-37 on autophagy remain unknown.
Interactions between autophagy and TGF-β
| Interactions | Authords | Year | Studied Cell Types | References |
|---|---|---|---|---|
| Autophagy inhibits TGF-β signaling | Araki, et al. | 2015 | cardiac fibroblasts | [197] |
| Ding, et al. | 2014 | primary renal proximal tubular epithelial cells | [196] | |
| TGF-β promotes autophagy | Men, et al. | 2015 | rat hepatic stellate cell line. | [205] |
| Ghavami, et al. | 2015 | human atrial myofibroblasts | [206] | |
| Wang, et al. | 2014 | human hepatocellular carcinoma cells | [201] | |
| Han, et al. | 2014 | human hepatocellular carcinoma cells | [202] | |
| Fu, et al. | 2014 | rat hepatic stellate cell line. | [204] | |
| Ding, et al. | 2014 | primary renal proximal tubular epithelial cells | [178] | |
| Xu, et al. | 2012 | primary renal proximal tubular epithelial cells | [189] | |
| Guido, et al. | 2012 | breast cancer tumor stromal cells | [208] | |
| Patel, et al. | 2012 | human lung fibroblasts | [209] | |
| Iovino, et al. | 2012 | mouse L6 and C2C12 myoblasts | [192] | |
| Lee, et al. | 2011 | mouse L6 and C2C12 myoblasts | [193] | |
| Tra, et al. | 2011 | human embryonic stem cells | [212] | |
| Ding, et al. | 2010 | mouse mesangial cells | [203] | |
| Kiyono, et al. | 2009 | human hepatocellular carcinoma cell lines | [200] | |
| Gajewska, et al. | 2005 | bovine mammary epithelial cell line. | [199] | |
| TGF-β down-regulates autophagy | Patschan, et al. | 2015 | murine early endothelial progenitor cells | [215] |
| Pan, et al. | 2015 | endothelial cells | [216] |
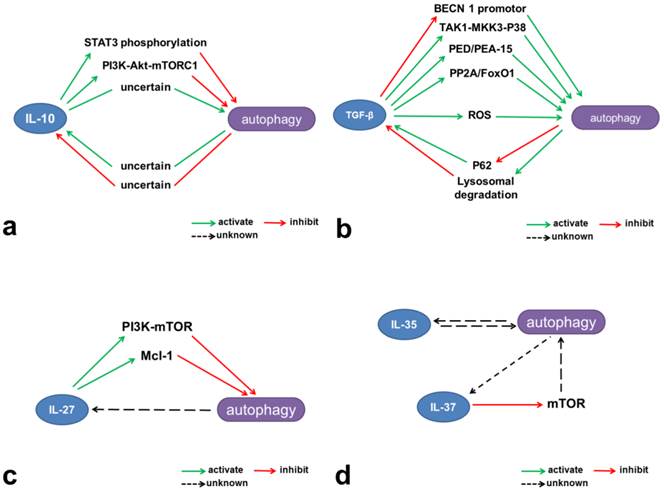
Mechanisms involved in the interactions between autophagy and inhibitory cytokines. (a) IL-10 contributes to activation of the JAK-STAT3 and PI3K-Akt-mTORC1 pathways and thus inhibits autophagy. (b) TGF-β induces autophagy through multiple mechanisms, including intervention in the TAK1-MKK3-p38 MAPK signaling pathway and regulation of PED/PEA-15, ROS, and PP2A/FoxO1. However, TGF-β can also inhibit the BECN1 promotor and thus reduce autophagy. Autophagy limits TGF-β production through lysosomal degradation and may negatively regulate TGF-β signaling through p62 degradation. (c) IL-27 activates mTOR and Mcl-1 and negatively regulates autophagy. (d) IL-37 inhibits mTOR and may have potential negative effects on autophagy.
Autophagy inhibits TGF-β signaling
Autophagic degradation may play an important role in TGF-β clearance. Ding et al. found that LC3 deficiency resulted in elevation of TGF-β levels in obstructed kidneys [196]. LC3 deficiency also increased TGF-β in human and mouse primary renal proximal tubular epithelial cells, as did bafilomycin A1, an autolysosomal inhibitor [196]. Sirt7 is a member of the mammalian sirtuin family and promotes oncogenic transformation. In cardiac fibroblasts, loss of Sirt7 activates autophagy and downregulates TβRI[197]. Treatment with chloroquine blocked TβRI downregulation [197]. In TGF-β-treated immortalized and tumor-derived epithelial cell lines, the autophagic signaling adaptor p62 was observed to accumulate and stabilize the TGF-β/Smad signaling co-activator Smad4; in this case, p62 accumulation was mainly due to induction of transcription [198]. Though the effects of autophagy on p62 accumulation and Smad4 stabilization were not investigated in this study, it could be hypothesized that autophagy might negatively affect TGF-β signaling through reduction of p62.
TGF-β can promote autophagy
As a pleiotropic cytokine, TGF-β could regulate autophagy through many different pathways. In 2005, Gajewska et al. [199] reported that treatment of a bovine mammary epithelial cell line with TGF-β1 significantly increased cytoplasmic levels of LC3 and Beclin-1. This was the first report of a potential effect of TGF-β on autophagy. Subsequent reports demonstrated positive effects of TGF-β signaling on autophagy in a variety of cellular contexts. For example, in human hepatocellular carcinoma cell (HCC) lines, TGF-β increased autophagosome formation, LC3 conversion, and Beclin-1, ATG5, ATG7, and death-associated protein kinase (DAPK) mRNA expression [200]. Blockade of TGF-β signaling by knockdown of Smad2, Smad3, or Smad4 attenuated the TGF-β-induced autophagy [200]. Likewise, knockdown of DAPK or inhibition of JNK inhibited autophagy, indicating that both Smad and non-Smad pathways are involved in TGF-β-induced autophagy [200]. In HCC cells, galangin-induced autophagy requires TGF-β signaling, and it is blocked by either downregulation of Smad4 or inhibition of TGF-β receptor activation [201]. In a study of mesenchymal stem cell (MSC)-induced chemoresistance of HCC cells, IFN-γ in combination with TNF-α induced expression of TGF-β in the MSCs and induced autophagy in the HCC cells[202]. Knockdown of TGF-β attenuated the MSC-induced autophagy and chemoresistance in HCC cells [202]. TGF-β1 treatment of mouse mesangial cells enhances the expression of LC3 and suppresses caspase-3 activation, representing an adaptive mechanism to glomerular injury [203]. Serum deprivation causes apoptosis of the rat HSC cell line, and addition of TGF-β1 reduces this through an autophagy-dependent mechanism[204]. Thus, TGF-β1 provides protection against serum deprivation via autophagy induction. Nogo-B deficiency in the rat HSC cell line decreases ER stress and autophagy, and conversely, TGF-β treatment increases Nogo-B expression in a time- and dose-dependent manner [205]. TGF-β has also been reported to induce autophagy in human atrial myofibroblasts [206], primary mouse and human renal proximal tubular epithelial cells [196, 207], breast cancer tumor stromal cells [208], human lung fibroblasts [209], mouse L6 and C2C12 myoblasts [210, 211], human embryonic stem cells [212], and renal tubules [213]. Multiple mechanisms might be involved the induction of autophagy by TGF-β, including the TAK1-MKK3-p38 MAPK signaling pathway [214], ROS [207], PP2A/FoxO1, and PED/PEA15 [210].
TGF-β downregulates autophagy
Recent studies have shown that TGF-β signaling can inhibit autophagy. In cultured murine early endothelial progenitor cells, TGF-β treatment decreased the density of LC3-II-positive autophagosomes, and this was countered by treatment with the autophagy inducer suberoylanilide hydroxamic acid [215]. Smad2, a key component of TGF-β signaling, was found to target Beclin-1 gene expression and suppress the BECN1 promotor in endothelial cells [216].
IL-27 and autophagy
At present, the connections between IL-27 and autophagy are largely unclear. Sharma and colleagues were the first to show that IL-27 inhibits IFN-induced autophagy [217]. They showed that IL-27 downregulated IFN-induced autophagosome generation and phagosome maturation in M. tuberculosis H37Rv-infected macrophages, and suppressed nutrient deprivation-induced autophagy and phagosomal acidification in these cells. As a result, IL-27 effectively promoted survival of the intracellular mycobacteria. Several mechanisms might be involved in this process, including activation of the JAK-PI3K-mTOR pathway and upregulation of the anti-apoptotic protein Mcl-1 (Figure 2 and Figure 3c). However, of all the steps involved in autophagy, IL-27 is perhaps most likely to affect autophagosome formation, and its effects on lysosomal degradation remain unclear. Given that IL-27 activates STAT1 and STAT3, the potential role of these signaling proteins in the effect of IL-27 on autophagy needs further investigation.
IL-35, IL-37 and autophagy
The molecular mechanisms by which IL-35 exerts its immunosuppressive effects are not well understood, although promotion of Tregs, inhibition of effector T cells, induction of iTr35 cells, and augmentation of IL-10 secretion have all been implicated. Autophagy is a critical process and, under certain circumstances, it may even dictate cell survival or death. Given the increasing evidence that autophagy has substantial effects on the immune system, it would not be surprising if this process were involved in IL-35-mediated immunosuppression at the molecular and subcellular levels.
Although there is currently no evidence that IL-37 and autophagy do interact, some clues about this can be gleaned from the literature. Transfection of the human monocytic cell line THP-1 with IL-37b reduces expression of mTOR [178], and IL-37 plus IL-1R8 induce a starvation-like effect on mTOR [179] (Figure 2 and Figure 3d). mTOR is a metabolic checkpoint kinase and upstream regulator of autophagy, and has direct effects on ULK1 and the initiation of autophagy. Thus, it will be interesting to investigate the effects of IL-37 on autophagy, particularly with respect to the mTOR/ULK1 complex pathway.
Concluding remarks
Recent work has demonstrated that autophagy plays an important role in the immune response. As mentioned, autophagy serves as the effector mechanism for PPRs, inhibits inflammasome formation, and promotes MHC class II-mediated antigen presentation and cross-presentation. In addition, autophagy is induced by several pro-inflammatory cytokines and is reduced by Th2-associated cytokines. Reciprocally, autophagy is involved in regulating the production and degradation of cytokines. The interaction between autophagy and cytokines is an emerging topic in immunology. Since inhibitory cytokines are crucial for maintaining immune homeostasis, the interactions between inhibitory cytokines and autophagy may have profound implications for the immune system, and could suggest new strategies for the treatment of autoimmune diseases, infection, tumors, and even severe trauma. However, it is clear from the literature that there are still large gaps in our understanding of this field.
First, theoretically, autophagy may play a dual role in the regulation of cytokines. Enhanced production of cytokines is usually accompanied by an increasing need for energy and subcellular material for cytokine synthesis, both of which may intrinsically stimulate autophagy. Under normal circumstances, autophagy contributes to energy production and recycling of cellular material, and a deficit in autophagy may result in ATP depletion. However, autophagy is also an important mechanism for cytokine degradation, so increasing the process may promote a reduction in cytokines. The overall effect will thus depend on the equilibrium between cytokine production and degradation, and it may be controlled by a highly complex and mostly unknown signal transduction network. Second, to date, studies of autophagy have mainly focused on the formation of autophagosomes, which is the characteristic structure of autophagy. However, autophagosome formation is only one part of the autophagic flux. Degradation in autolysosomes is the ultimate fate of autophagic cargo and this step demands more attention. Third, the spectrum of cell types in which autophagy-inhibitory cytokine interactions have been investigated needs to be expanded. For example, whether IL-10 and autophagy interact in T cells is unknown, and the effects of TGF-β on autophagy in lymphocytes need further investigation. In this regard, more effort should be devoted to the newcomers in this area, IL-35 and IL-37.
From a clinical perspective, autophagy modulators such as rapamycin, resveratrol, and chloroquine have been widely administered to patients for treatment of various disorders, including cancer, transplantation rejection, and infection. Activation or inhibition of autophagy will endow novel therapeutic applications to these conventional pharmaceutical agents and may provide benefits with relatively few risks. Therefore, therapeutic manipulation of the interactions between autophagy and inhibitory cytokines might represent a novel method of regulating the immune response and a more facilitated clinical translation.
Abbreviations
ATG: autophagy-related proteins; TGF: transforming growth factor; ULK1: Unc-51-like kinase 1; mTOR: mammalian target of rapamycin; PI3K: phosphatidylinositol 3-kinase; UVRAG: UV irradiation resistance-associated gene; PE: phosphatidylethanolamine; PRR: pattern recognition receptor; IFN: interferon; TNF: tumor necrosis factor; TLR: Toll-like receptor; NLR: (NOD)-like receptor; RLR: (RIG-1)-I-like receptor; PAMPs: pathogen-associated molecular patterns; ROS: reactive oxygen species; SLRs: sequestosome-1/p62-like receptors; LPS: lipopolysaccharide; APC: antigen presentation cell; MHC: major histocompatibility complex; STAT: signal transducer and activation of transcription; MAPK: mitogen-activated protein kinase; DC: dendritic cell; GM-CSF: granulocyte-macrophage colony-stimulating factor; Foxp3: Fork head box 3; PBMC: peripheral blood mononuclear cells; ERK: extracellular signal-regulated kinase; moDC: monocyte-induced dendritic cell; JNK: c-jun amino-terminal kinase; HCC: hepatocellular carcinoma cell; DAPK: death-associated protein kinase; MSC: mesenchymal stem cell.
Acknowledgements
This study was supported, in part, by grants from the National Natural Science Foundation of China (Nos. 81130035, 81372054) and the National Basic Research Program of China (No. 2012CB518102).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Developmental cell. 2004;6:463-77
2. Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nature reviews Molecular cell biology. 2012;13:7-12
3. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nature cell biology. 2013;15:713-20
4. Cuervo AM, Macian F. Autophagy, nutrition and immunology. Molecular aspects of medicine. 2012;33:2-13
5. Kaushik S, Singh R, Cuervo AM. Autophagic pathways and metabolic stress. Diabetes, obesity & metabolism. 2010;12(Suppl 2):4-14
6. Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell metabolism. 2011;13:495-504
7. Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. The Journal of pathology. 2012;226:255-73
8. Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. The Journal of cell biology. 2010;189:925-35
9. Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107-32
10. Yoshii SR, Mizushima N. Autophagy machinery in the context of mammalian mitophagy. Biochimica et biophysica acta. 2015;1853:2797-801
11. Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. The Journal of biological chemistry. 2009;284:12297-305
12. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y. et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Molecular biology of the cell. 2009;20:1981-91
13. Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649-62
14. Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J. et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Molecular biology of the cell. 2009;20:1992-2003
15. He S, Ni D, Ma B, Lee JH, Zhang T, Ghozalli I. et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nature cell biology. 2013;15:1206-19
16. Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature cell biology. 2009;11:1433-7
17. Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK. et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656-67
18. Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Molecular biology of the cell. 2010;21:2257-69
19. Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T. et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. The Journal of biological chemistry. 2007;282:37298-302
20. Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9:424-5
21. Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S, Singh R. Autophagy proteins regulate ERK phosphorylation. Nature communications. 2013;4:2799
22. Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N. et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488-92
23. Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Current opinion in cell biology. 2010;22:124-31
24. Virgin HW, Levine B. Autophagy genes in immunity. Nature immunology. 2009;10:461-70
25. Lee HS, Daniels BH, Salas E, Bollen AW, Debnath J, Margeta M. Clinical utility of LC3 and p62 immunohistochemistry in diagnosis of drug-induced autophagic vacuolar myopathies: a case-control study. PloS one. 2012;7:e36221
26. Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y. et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nature cell biology. 2007;9:1142-51
27. Hegedus K, Takats S, Kovacs AL, Juhasz G. Evolutionarily conserved role and physiological relevance of a STX17/Syx17 (syntaxin 17)-containing SNARE complex in autophagosome fusion with endosomes and lysosomes. Autophagy. 2013;9:1642-6
28. Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T. et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Molecular biology of the cell. 2014;25:1327-37
29. Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell death and differentiation. 2014;21:348-58
30. Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M. et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nature cell biology. 2013;15:406-16
31. Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Science signaling. 2010;3:ra42
32. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274-93
33. Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P. et al. Endogenous HMGB1 regulates autophagy. The Journal of cell biology. 2010;190:881-92
34. Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA. et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223-34
35. Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M. et al. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505-17
36. Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753-66
37. Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E. et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nature cell biology. 2010;12:1154-65
38. Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T. et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. The Journal of biological chemistry. 2011;286:26987-95
39. Zeng R, Cui Y, Hai Y, Liu Y. Pattern recognition receptors for respiratory syncytial virus infection and design of vaccines. Virus research. 2012;167:138-45
40. Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135-44
41. Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398-401
42. Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature immunology. 2010;11:55-62
43. Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG. et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933-46
44. Jounai N, Kobiyama K, Shiina M, Ogata K, Ishii KJ, Takeshita F. NLRP4 negatively regulates autophagic processes through an association with beclin1. J Immunol. 2011;186:1646-55
45. Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2770-5
46. Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ. et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14050-5
47. Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK. et al. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol. 2011;187:5268-76
48. McFarlane S, Aitken J, Sutherland JS, Nicholl MJ, Preston VG, Preston CM. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. Journal of virology. 2011;85:4212-21
49. Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803-15
50. Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nature immunology. 2009;10:1215-21
51. Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228-33
52. Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M. et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nature cell biology. 2009;11:1233-40
53. Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J. et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nature immunology. 2013;14:480-8
54. Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA. et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nature immunology. 2012;13:255-63
55. Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221-5
56. Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264-8
57. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC. et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2011;12:222-30
58. Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA. et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. The Journal of biological chemistry. 2011;286:9587-97
59. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013-22
60. Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R. et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654-69
61. Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T. et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593-6
62. Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396-400
63. Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nature medicine. 2009;15:267-76
64. Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79-92
65. Jin Y, Sun C, Feng L, Li P, Xiao L, Ren Y. et al. Regulation of SIV antigen-specific CD4+ T cellular immunity via autophagosome-mediated MHC II molecule-targeting antigen presentation in mice. PloS one. 2014;9:e93143
66. Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A. et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227-39
67. Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S. et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253-7
68. Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nature cell biology. 2011;13:1335-43
69. Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D. et al. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. The Journal of cell biology. 2013;203:757-66
70. Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. The Journal of biological chemistry. 2012;287:34149-56
71. Wenger T, Terawaki S, Camosseto V, Abdelrassoul R, Mies A, Catalan N. et al. Autophagy inhibition promotes defective neosynthesized proteins storage in ALIS, and induces redirection toward proteasome processing and MHCI-restricted presentation. Autophagy. 2012;8:350-63
72. English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D. et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nature immunology. 2009;10:480-7
73. Tey SK, Khanna R. Autophagy mediates transporter associated with antigen processing-independent presentation of viral epitopes through MHC class I pathway. Blood. 2012;120:994-1004
74. Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS. et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343:313-7
75. Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell death and differentiation. 2009;16:991-1005
76. Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer research. 2008;68:6889-95
77. Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046-55
78. Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349-57
79. Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8670-5
80. Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I. et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. eLife. 2014:3
81. Henson SM, Lanna A, Riddell NE, Franzese O, Macaulay R, Griffiths SJ. et al. p38 signaling inhibits mTORC1-independent autophagy in senescent human CD8(+) T cells. The Journal of clinical investigation. 2014;124:4004-16
82. Xu X, Araki K, Li S, Han JH, Ye L, Tan WG. et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nature immunology. 2014;15:1152-61
83. Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK. et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309-14
84. Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A. et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nature immunology. 2013;14:298-305
85. Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546-58
86. Gemelli C, Zanocco Marani T, Bicciato S, Mazza EM, Boraschi D, Salsi V. et al. MafB is a downstream target of the IL-10/STAT3 signaling pathway, involved in the regulation of macrophage de-activation. Biochimica et biophysica acta. 2014;1843:955-64
87. Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. The EMBO journal. 1997;16:5894-903
88. Wang Y, Li T, Wu B, Liu H, Luo J, Feng D. et al. STAT1 regulates MD-2 expression in monocytes of sepsis via miR-30a. Inflammation. 2014;37:1903-11
89. Liu WH, Liu JJ, Wu J, Zhang LL, Liu F, Yin L. et al. Novel mechanism of inhibition of dendritic cells maturation by mesenchymal stem cells via interleukin-10 and the JAK1/STAT3 signaling pathway. PloS one. 2013;8:e55487
90. Redford PS, Murray PJ, O'Garra A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal immunology. 2011;4:261-70
91. Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O'Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815-22
92. Rossato M, Curtale G, Tamassia N, Castellucci M, Mori L, Gasperini S. et al. IL-10-induced microRNA-187 negatively regulates TNF-alpha, IL-6, and IL-12p40 production in TLR4-stimulated monocytes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3101-10
93. Avdic S, Cao JZ, McSharry BP, Clancy LE, Brown R, Steain M. et al. Human cytomegalovirus interleukin-10 polarizes monocytes toward a deactivated M2c phenotype to repress host immune responses. Journal of virology. 2013;87:10273-82
94. Chan LL, Cheung BK, Li JC, Lau AS. A role for STAT3 and cathepsin S in IL-10 down-regulation of IFN-gamma-induced MHC class II molecule on primary human blood macrophages. Journal of leukocyte biology. 2010;88:303-11
95. Astier AL, Meiffren G, Freeman S, Hafler DA. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. The Journal of clinical investigation. 2006;116:3252-7
96. Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM. et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566-78
97. Antoniv TT, Ivashkiv LB. Interleukin-10-induced gene expression and suppressive function are selectively modulated by the PI3K-Akt-GSK3 pathway. Immunology. 2011;132:567-77
98. Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS letters. 2006;580:2811-20
99. Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685-700
100. Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R. et al. Identification of Smad7, a TGF beta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631-5
101. Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577-84
102. Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603-9
103. Li MO, Wan YSY, Flavell RA. T cell-produced transforming growth factor-beta 1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579-91
104. Di Sabatino A, Pickard KM, Rampton D, Kruidenier L, Rovedatti L, Leakey NAB. et al. Blockade of transforming growth factor beta upregulates T-box transcription factor T-bet, and increases T helper cell type 1 cytokine and matrix metalloproteinase-3 production in the human gut mucosa. Gut. 2008;57:605-12
105. Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99-146
106. Worthington JJ, Fenton TM, Czajkowska BI, Klementowicz JE, Travis MA. Regulation of TGFbeta in the immune system: an emerging role for integrins and dendritic cells. Immunobiology. 2012;217:1259-65
107. Gorelik L, Fields PE, Flavell RA. Cutting edge: TGF-beta inhibits Th type 2 development through inhibition of GATA-3 expression. J Immunol. 2000;165:4773-7
108. Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H. et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. The Journal of experimental medicine. 2002;195:1129-43
109. Trapani JA. The dual adverse effects of TGF-beta secretion on tumor progression. Cancer cell. 2005;8:349-50
110. Nakamura K, Kitani A, Strober W. Cell contact-dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface-bound transforming growth factor beta. The Journal of experimental medicine. 2001;194:629-44
111. Ghiringhelli F, Menard C, Terme M, Flament C, Taieb J, Chaput N. et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. The Journal of experimental medicine. 2005;202:1075-85
112. Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636-45
113. Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N. et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine. 2003;198:1875-86
114. Chen WJ, Jin WW, Hardegen N, Lei KJ, Li L, Marinos N. et al. Conversion of peripheral CD4(+)CD25(-) naive T cells to CD4(+)CD25(+) regulatory T cells by TGF-beta induction of transcription factor Foxp3. Journal of Experimental Medicine. 2003;198:1875-86
115. Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, Neurath MF. Cutting edge: TGF-beta induces a regulatory phenotype in CD4(+)CD25(-) T cells through Foxp3 induction and down-regulation of Smad7. Journal of Immunology. 2004;172:5149-53
116. Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR. et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. The Journal of experimental medicine. 2007;204:1775-85
117. Xiao S, Jin H, Korn T, Liu SM, Oukka M, Lim B. et al. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J Immunol. 2008;181:2277-84
118. Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. The Journal of experimental medicine. 2007;204:1765-74
119. Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR. et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. Journal of Experimental Medicine. 2007;204:1775-85
120. Coombes JL, Siddiqui KRR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y. et al. A functionally specialized population of mucosal CD103(+) DCs induces Foxp3(+) regulatory T cells via a TGF-beta- and retinoic acid-dependent mechanism. Journal of Experimental Medicine. 2007;204:1757-64
121. Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M. et al. Reciprocal T(H)17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256-60
122. Gutcher I, Donkor MK, Ma Q, Rudensky AY, Flavell RA, Li MO. Autocrine transforming growth factor-beta1 promotes in vivo Th17 cell differentiation. Immunity. 2011;34:396-408
123. Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO. et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231-4
124. Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235-8
125. Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392-404
126. Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE. et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967-71
127. Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP. et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44-56
128. Martinez GJ, Zhang Z, Reynolds JM, Tanaka S, Chung Y, Liu T. et al. Smad2 positively regulates the generation of Th17 cells. The Journal of biological chemistry. 2010;285:29039-43
129. Takimoto T, Wakabayashi Y, Sekiya T, Inoue N, Morita R, Ichiyama K. et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J Immunol. 2010;185:842-55
130. Castriconi R, Cantoni C, Della Chiesa M, Vitale M, Marcenaro E, Conte R. et al. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4120-5
131. Lee JC, Lee KM, Kim DW, Heo DS. Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J Immunol. 2004;172:7335-40
132. Kehrl JH, Thevenin C, Rieckmann P, Fauci AS. Transforming growth factor-beta suppresses human B lymphocyte Ig production by inhibiting synthesis and the switch from the membrane form to the secreted form of Ig mRNA. J Immunol. 1991;146:4016-23
133. Cazac BB, Roes J. TGF-beta receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13:443-51
134. Naiki Y, Michelsent KS, Zhang WX, Chen SA, Doherty TM, Arditi M. Transforming growth factor-beta differentially inhibits MyD88-dependent, but not TRAM- and TRIF-dependent, lipopolysaccharide-induced TLR4 signaling. Journal of Biological Chemistry. 2005;280:5491-5
135. Lievens D, Habets KL, Robertson AK, Laouar Y, Winkels H, Rademakers T. et al. Abrogated transforming growth factor beta receptor II (TGFbetaRII) signalling in dendritic cells promotes immune reactivity of T cells resulting in enhanced atherosclerosis. European heart journal. 2013;34:3717-27
136. Bonnefoy F, Couturier M, Clauzon A, Remy-Martin JP, Gaugler B, Tiberghien P. et al. TGF-beta-exposed plasmacytoid dendritic cells participate in Th17 commitment. J Immunol. 2011;186:6157-64
137. Molle C, Goldman M, Goriely S. Critical role of the IFN-stimulated gene factor 3 complex in TLR-mediated IL-27p28 gene expression revealing a two-step activation process. J Immunol. 2010;184:1784-92
138. Molle C, Nguyen M, Flamand V, Renneson J, Trottein F, De Wit D. et al. IL-27 synthesis induced by TLR ligation critically depends on IFN regulatory factor 3. J Immunol. 2007;178:7607-15
139. Pflanz S, Timans JC, Cheung J, Rosales R, Kanzler H, Gilbert J. et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4+ T cells. Immunity. 2002;16:779-90
140. Pflanz S, Hibbert L, Mattson J, Rosales R, Vaisberg E, Bazan JF. et al. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J Immunol. 2004;172:2225-31
141. Villarino AV, Huang E, Hunter CA. Understanding the pro- and anti-inflammatory properties of IL-27. J Immunol. 2004;173:715-20
142. Takeda A, Hamano S, Yamanaka A, Hanada T, Ishibashi T, Mak TW. et al. Cutting edge: role of IL-27/WSX-1 signaling for induction of T-bet through activation of STAT1 during initial Th1 commitment. J Immunol. 2003;170:4886-90
143. Yoshida H, Hamano S, Senaldi G, Covey T, Faggioni R, Mu S. et al. WSX-1 is required for the initiation of Th1 responses and resistance to L. major infection. Immunity. 2001;15:569-78
144. Owaki T, Asakawa M, Morishima N, Hata K, Fukai F, Matsui M. et al. A role for IL-27 in early regulation of Th1 differentiation. J Immunol. 2005;175:2191-200
145. Diveu C, McGeachy MJ, Boniface K, Stumhofer JS, Sathe M, Joyce-Shaikh B. et al. IL-27 blocks RORc expression to inhibit lineage commitment of Th17 cells. J Immunol. 2009;182:5748-56
146. El-behi M, Ciric B, Yu S, Zhang GX, Fitzgerald DC, Rostami A. Differential effect of IL-27 on developing versus committed Th17 cells. J Immunol. 2009;183:4957-67
147. Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L. et al. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nature immunology. 2011;12:560-7
148. Lucas S, Ghilardi N, Li J, de Sauvage FJ. IL-27 regulates IL-12 responsiveness of naive CD4+ T cells through Stat1-dependent and -independent mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15047-52
149. Yoshimoto T, Yasuda K, Mizuguchi J, Nakanishi K. IL-27 suppresses Th2 cell development and Th2 cytokines production from polarized Th2 cells: a novel therapeutic way for Th2-mediated allergic inflammation. J Immunol. 2007;179:4415-23
150. Chen Q, Ghilardi N, Wang H, Baker T, Xie MH, Gurney A. et al. Development of Th1-type immune responses requires the type I cytokine receptor TCCR. Nature. 2000;407:916-20
151. Artis D, Johnson LM, Joyce K, Saris C, Villarino A, Hunter CA. et al. Cutting edge: early IL-4 production governs the requirement for IL-27-WSX-1 signaling in the development of protective Th1 cytokine responses following Leishmania major infection. J Immunol. 2004;172:4672-5
152. Stumhofer JS, Laurence A, Wilson EH, Huang E, Tato CM, Johnson LM. et al. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nature immunology. 2006;7:937-45
153. Batten M, Li J, Yi S, Kljavin NM, Danilenko DM, Lucas S. et al. Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nature immunology. 2006;7:929-36
154. Fitzgerald DC, Ciric B, Touil T, Harle H, Grammatikopolou J, Das Sarma J. et al. Suppressive effect of IL-27 on encephalitogenic Th17 cells and the effector phase of experimental autoimmune encephalomyelitis. J Immunol. 2007;179:3268-75
155. Pot C, Jin H, Awasthi A, Liu SM, Lai CY, Madan R. et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J Immunol. 2009;183:797-801
156. Fitzgerald DC, Zhang GX, El-Behi M, Fonseca-Kelly Z, Li H, Yu S. et al. Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nature immunology. 2007;8:1372-9
157. Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA. et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nature immunology. 2007;8:1363-71
158. Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D. et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nature immunology. 2010;11:854-61
159. Huber M, Steinwald V, Guralnik A, Brustle A, Kleemann P, Rosenplanter C. et al. IL-27 inhibits the development of regulatory T cells via STAT3. International immunology. 2008;20:223-34
160. Neufert C, Becker C, Wirtz S, Fantini MC, Weigmann B, Galle PR. et al. IL-27 controls the development of inducible regulatory T cells and Th17 cells via differential effects on STAT1. European journal of immunology. 2007;37:1809-16
161. Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM. et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566-9
162. Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nature reviews Immunology. 2008;8:523-32
163. Seyerl M, Kirchberger S, Majdic O, Seipelt J, Jindra C, Schrauf C. et al. Human rhinoviruses induce IL-35-producing Treg via induction of B7-H1 (CD274) and sialoadhesin (CD169) on DC. European journal of immunology. 2010;40:321-9
164. Li X, Mai J, Virtue A, Yin Y, Gong R, Sha X. et al. IL-35 is a novel responsive anti-inflammatory cytokine-a new system of categorizing anti-inflammatory cytokines. PloS one. 2012;7:e33628
165. Guttek K, Reinhold D. Stimulated human peripheral T cells produce high amounts of IL-35 protein in a proliferation-dependent manner. Cytokine. 2013;64:46-50
166. Collison LW, Chaturvedi V, Henderson AL, Giacomin PR, Guy C, Bankoti J. et al. IL-35-mediated induction of a potent regulatory T cell population. Nature immunology. 2010;11:1093-101
167. Olson BM, Jankowska-Gan E, Becker JT, Vignali DA, Burlingham WJ, McNeel DG. Human prostate tumor antigen-specific CD8+ regulatory T cells are inhibited by CTLA-4 or IL-35 blockade. J Immunol. 2012;189:5590-601
168. Wang Z, Liu JQ, Liu Z, Shen R, Zhang G, Xu J. et al. Tumor-derived IL-35 promotes tumor growth by enhancing myeloid cell accumulation and angiogenesis. J Immunol. 2013;190:2415-23
169. Delgoffe GM, Murray PJ, Vignali DA. Interpreting mixed signals: the cell's cytokine conundrum. Current opinion in immunology. 2011;23:632-8
170. Collison LW, Delgoffe GM, Guy CS, Vignali KM, Chaturvedi V, Fairweather D. et al. The composition and signaling of the IL-35 receptor are unconventional. Nature immunology. 2012;13:290-9
171. Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nature immunology. 2012;13:722-8
172. Niedbala W, Wei XQ, Cai B, Hueber AJ, Leung BP, McInnes IB. et al. IL-35 is a novel cytokine with therapeutic effects against collagen-induced arthritis through the expansion of regulatory T cells and suppression of Th17 cells. European journal of immunology. 2007;37:3021-9
173. Kochetkova I, Golden S, Holderness K, Callis G, Pascual DW. IL-35 stimulation of CD39+ regulatory T cells confers protection against collagen II-induced arthritis via the production of IL-10. J Immunol. 2010;184:7144-53
174. Wang RX, Yu CR, Dambuza IM, Mahdi RM, Dolinska MB, Sergeev YV. et al. Interleukin-35 induces regulatory B cells that suppress autoimmune disease. Nature medicine. 2014;20:633-41
175. Kumar S, McDonnell PC, Lehr R, Tierney L, Tzimas MN, Griswold DE. et al. Identification and initial characterization of four novel members of the interleukin-1 family. The Journal of biological chemistry. 2000;275:10308-14
176. Ye L, Ji L, Wen Z, Zhou Y, Hu D, Li Y. et al. IL-37 inhibits the production of inflammatory cytokines in peripheral blood mononuclear cells of patients with systemic lupus erythematosus: its correlation with disease activity. Journal of translational medicine. 2014;12:69
177. Bufler P, Gamboni-Robertson F, Azam T, Kim SH, Dinarello CA. Interleukin-1 homologues IL-1F7b and IL-18 contain functional mRNA instability elements within the coding region responsive to lipopolysaccharide. The Biochemical journal. 2004;381:503-10
178. Nold MF, Nold-Petry CA, Zepp JA, Palmer BE, Bufler P, Dinarello CA. IL-37 is a fundamental inhibitor of innate immunity. Nature immunology. 2010;11:1014-22
179. Nold-Petry CA, Lo CY, Rudloff I, Elgass KD, Li S, Gantier MP. et al. IL-37 requires the receptors IL-18Ralpha and IL-1R8 (SIGIRR) to carry out its multifaceted anti-inflammatory program upon innate signal transduction. Nature immunology. 2015;16:354-65
180. Li S, Neff CP, Barber K, Hong J, Luo Y, Azam T. et al. Extracellular forms of IL-37 inhibit innate inflammation in vitro and in vivo but require the IL-1 family decoy receptor IL-1R8. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2497-502
181. He L, Liang Z, Zhao F, Peng L, Chen Z. Modulation of IL-37 expression by triptolide and triptonide in THP-1 cells. Cellular & molecular immunology. 2015;12:515-8
182. Ye Z, Wang C, Kijlstra A, Zhou X, Yang P. A possible role for interleukin 37 in the pathogenesis of Behcet's disease. Current molecular medicine. 2014;14:535-42
183. Luo Y, Cai X, Liu S, Wang S, Nold-Petry CA, Nold MF. et al. Suppression of antigen-specific adaptive immunity by IL-37 via induction of tolerogenic dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15178-83
184. Qi GM, Jia LX, Li YL, Li HH, Du J. Adiponectin suppresses angiotensin II-induced inflammation and cardiac fibrosis through activation of macrophage autophagy. Endocrinology. 2014;155:2254-65
185. Du B, Zhang Z, Li N. Madecassoside prevents Abeta(25-35)-induced inflammatory responses and autophagy in neuronal cells through the class III PI3K/Beclin-1/Bcl-2 pathway. International immunopharmacology. 2014;20:221-8
186. Strisciuglio C, Duijvestein M, Verhaar AP, Vos AC, van den Brink GR, Hommes DW. et al. Impaired autophagy leads to abnormal dendritic cell-epithelial cell interactions. Journal of Crohn's & colitis. 2013;7:534-41
187. Wang H, Wang Y, Li D, Liu Z, Zhao Z, Han D. et al. VEGF inhibits the inflammation in spinal cord injury through activation of autophagy. Biochemical and biophysical research communications. 2015;464:453-8
188. Glubb DM, Gearry RB, Barclay ML, Roberts RL, Pearson J, Keenan JI. et al. NOD2 and ATG16L1 polymorphisms affect monocyte responses in Crohn's disease. World journal of gastroenterology: WJG. 2011;17:2829-37
189. Lopez P, Alonso-Perez E, Rodriguez-Carrio J, Suarez A. Influence of Atg5 mutation in SLE depends on functional IL-10 genotype. PloS one. 2013;8:e78756
190. Wang L, Zhang H, Qian J, Wang K, Zhu J. Interleukin-10 blocks in vitro replication of human cytomegalovirus by inhibiting the virus-induced autophagy in MRC5 cells. Biochemical and biophysical research communications. 2014;448:448-53
191. Buchser WJ, Laskow TC, Pavlik PJ, Lin HM, Lotze MT. Cell-mediated autophagy promotes cancer cell survival. Cancer research. 2012;72:2970-9
192. Park HJ, Lee SJ, Kim SH, Han J, Bae J, Kim SJ. et al. IL-10 inhibits the starvation induced autophagy in macrophages via class I phosphatidylinositol 3-kinase (PI3K) pathway. Molecular immunology. 2011;48:720-7
193. Van Grol J, Subauste C, Andrade RM, Fujinaga K, Nelson J, Subauste CS. HIV-1 inhibits autophagy in bystander macrophage/monocytic cells through Src-Akt and STAT3. PloS one. 2010;5:e11733
194. Santarelli R, Gonnella R, Di Giovenale G, Cuomo L, Capobianchi A, Granato M. et al. STAT3 activation by KSHV correlates with IL-10, IL-6 and IL-23 release and an autophagic block in dendritic cells. Scientific reports. 2014;4:4241
195. Martinez-Outschoorn UE, Whitaker-Menezes D, Lin Z, Flomenberg N, Howell A, Pestell RG. et al. Cytokine production and inflammation drive autophagy in the tumor microenvironment: role of stromal caveolin-1 as a key regulator. Cell Cycle. 2011;10:1784-93
196. Ding Y, Kim SL, Lee SY, Koo JK, Wang Z, Choi ME. Autophagy regulates TGF-beta expression and suppresses kidney fibrosis induced by unilateral ureteral obstruction. J Am Soc Nephrol. 2014;25:2835-46
197. Araki S, Izumiya Y, Rokutanda T, Ianni A, Hanatani S, Kimura Y. et al. Sirt7 Contributes to Myocardial Tissue Repair by Maintaining TGF-beta Signaling Pathway. Circulation. 2015
198. Bertrand M, Petit V, Jain A, Amsellem R, Johansen T, Larue L. et al. SQSTM1/p62 regulates the expression of junctional proteins through epithelial-mesenchymal transition factors. Cell Cycle. 2015;14:364-74
199. Gajewska M, Gajkowska B, Motyl T. Apoptosis and autophagy induced by TGF-B1 in bovine mammary epithelial BME-UV1 cells. Journal of physiology and pharmacology: an official journal of the Polish Physiological Society. 2005;56(Suppl 3):143-57
200. Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR. et al. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer research. 2009;69:8844-52
201. Wang Y, Wu J, Lin B, Li X, Zhang H, Ding H. et al. Galangin suppresses HepG2 cell proliferation by activating the TGF-beta receptor/Smad pathway. Toxicology. 2014;326:9-17
202. Han Z, Jing Y, Xia Y, Zhang S, Hou J, Meng Y. et al. Mesenchymal stem cells contribute to the chemoresistance of hepatocellular carcinoma cells in inflammatory environment by inducing autophagy. Cell & bioscience. 2014;4:22
203. Ding Y, Kim JK, Kim SI, Na HJ, Jun SY, Lee SJ. et al. TGF-{beta}1 protects against mesangial cell apoptosis via induction of autophagy. The Journal of biological chemistry. 2010;285:37909-19
204. Fu MY, He YJ, Lv X, Liu ZH, Shen Y, Ye GR. et al. Transforming growth factorbeta1 reduces apoptosis via autophagy activation in hepatic stellate cells. Molecular medicine reports. 2014;10:1282-8
205. Men R, Wen M, Dan X, Zhu Y, Wang W, Li J. et al. Nogo-B: A potential indicator for hepatic cirrhosis and regulator in hepatic stellate cell activation. Hepatology research: the official journal of the Japan Society of Hepatology. 2015;45:113-22
206. Ghavami S, Cunnington RH, Gupta S, Yeganeh B, Filomeno KL, Freed DH. et al. Autophagy is a regulator of TGF-beta1-induced fibrogenesis in primary human atrial myofibroblasts. Cell death & disease. 2015;6:e1696
207. Xu Y, Yang S, Huang J, Ruan S, Zheng Z, Lin J. Tgf-beta1 induces autophagy and promotes apoptosis in renal tubular epithelial cells. International journal of molecular medicine. 2012;29:781-90
208. Guido C, Whitaker-Menezes D, Capparelli C, Balliet R, Lin Z, Pestell RG. et al. Metabolic reprogramming of cancer-associated fibroblasts by TGF-beta drives tumor growth: connecting TGF-beta signaling with "Warburg-like" cancer metabolism and L-lactate production. Cell Cycle. 2012;11:3019-35
209. Patel AS, Lin L, Geyer A, Haspel JA, An CH, Cao J. et al. Autophagy in idiopathic pulmonary fibrosis. PloS one. 2012;7:e41394
210. Iovino S, Oriente F, Botta G, Cabaro S, Iovane V, Paciello O. et al. PED/PEA-15 induces autophagy and mediates TGF-beta1 effect on muscle cell differentiation. Cell death and differentiation. 2012;19:1127-38
211. Lee JY, Hopkinson NS, Kemp PR. Myostatin induces autophagy in skeletal muscle in vitro. Biochemical and biophysical research communications. 2011;415:632-6
212. Tra T, Gong L, Kao LP, Li XL, Grandela C, Devenish RJ. et al. Autophagy in human embryonic stem cells. PloS one. 2011;6:e27485
213. Koesters R, Kaissling B, Lehir M, Picard N, Theilig F, Gebhardt R. et al. Tubular overexpression of transforming growth factor-beta1 induces autophagy and fibrosis but not mesenchymal transition of renal epithelial cells. Am J Pathol. 2010;177:632-43
214. Kim SI, Na HJ, Ding Y, Wang Z, Lee SJ, Choi ME. Autophagy promotes intracellular degradation of type I collagen induced by transforming growth factor (TGF)-beta1. J Biol Chem. 2012;287:11677-88
215. Patschan D, Schwarze K, Henze E, Patschan S, Muller GA. Endothelial autophagy and Endothelial-to-Mesenchymal Transition (EndoMT) in eEPC treatment of ischemic AKI. Journal of nephrology. 2015
216. Pan CC, Kumar S, Shah N, Bloodworth JC, Hawinkels LJ, Mythreye K. et al. Endoglin Regulation of Smad2 Function Mediates Beclin1 Expression and Endothelial Autophagy. The Journal of biological chemistry. 2015;290:14884-92
217. Sharma G, Dutta RK, Khan MA, Ishaq M, Sharma K, Malhotra H. et al. IL-27 inhibits IFN-gamma induced autophagy by concomitant induction of JAK/PI3 K/Akt/mTOR cascade and up-regulation of Mcl-1 in Mycobacterium tuberculosis H37Rv infected macrophages. The international journal of biochemistry & cell biology. 2014;55:335-47
Author contact
![]() Corresponding author: Yong-Ming Yao, MD, PhD, Trauma Research Center, First Hospital Affiliated to the Chinese PLA General Hospital, No. 51 Fucheng Road, Haidian District, Beijing 100048, China. Tel: +86 1066867394; FAX: +86 1068989955; E-mail: yaoyongmingcom.cn
Corresponding author: Yong-Ming Yao, MD, PhD, Trauma Research Center, First Hospital Affiliated to the Chinese PLA General Hospital, No. 51 Fucheng Road, Haidian District, Beijing 100048, China. Tel: +86 1066867394; FAX: +86 1068989955; E-mail: yaoyongmingcom.cn

 Global reach, higher impact
Global reach, higher impact