10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(9):1083-1092. doi:10.7150/ijbs.15633 This issue Cite
Review
Cytoskeleton Molecular Motors: Structures and Their Functions in Neuron
1. Faculty of Health Sciences, University of Macau, Taipa, Macau, China
2. Department of Biology, Southern University of Science and Technology, Shenzhen, 518055, China
Received 2016-3-24; Accepted 2016-5-28; Published 2016-7-18
Abstract
Cells make use of molecular motors to transport small molecules, macromolecules and cellular organelles to target region to execute biological functions, which is utmost important for polarized cells, such as neurons. In particular, cytoskeleton motors play fundamental roles in neuron polarization, extension, shape and neurotransmission. Cytoskeleton motors comprise of myosin, kinesin and cytoplasmic dynein. F-actin filaments act as myosin track, while kinesin and cytoplasmic dynein move on microtubules. Cytoskeleton motors work together to build a highly polarized and regulated system in neuronal cells via different molecular mechanisms and functional regulations. This review discusses the structures and working mechanisms of the cytoskeleton motors in neurons.
Keywords: Cytoskeleton motors, Myosin, Kinesin, Cytoplasmic dynein, Neuron
Introduction
A neuron cell is highly polarized, composing of a cell body, an axon and dendrites. Macromolecules, such as proteins and RNA, are synthesized in neuronal cell body, and are then transported to the axon or dendrites. Cytoskeleton motors are vital to transport cargos in the biological processes by using cytoskeleton as walking tracks and hydrolyze ATP to provide mechanical energy. Myosin is one superfamily of cytoskeletal motors, walking along the F-actin filaments. Kinesin and Dynein are both microtubule motors, walking towards microtubule's plus-end and minus-end directions respectively. The dysfunction of these motor proteins in neuron system is related to many human neuronal diseases. In this work, we review the structural and functional characteristics of cytoskeleton motors and discuss the roles of cytoskeleton motors in neuronal polarization, development and neurotransmission processes. Finally, some possible future directions of cytoskeleton motors study will be highlighted.
Actin-based motors
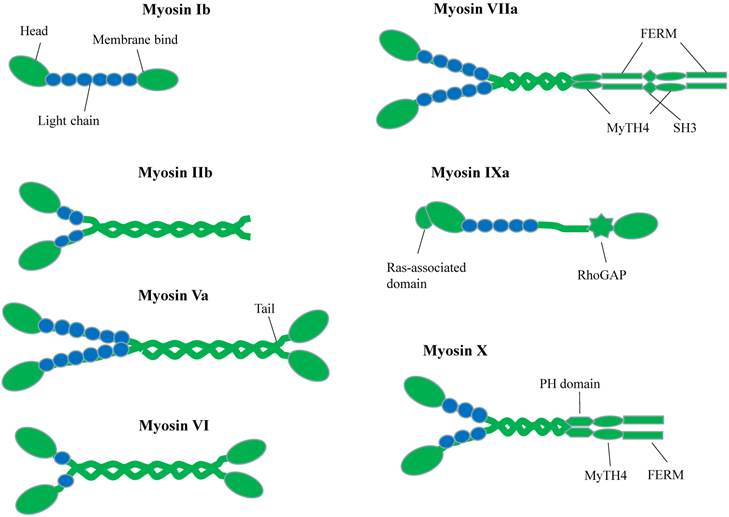
According to their gene sequence homology, myosin can be classified into 35 classes in eukaryote [1]. Mammals have 40 myosin genes, whose expression products fall into 13 classes [2]. Figure 1 shows the selected myosin families, which have been reported and expressed in neuronal cells [3,4]. Myosins compose of three common domains [5]: (1) the N-terminal motor domains that interact with actin and hydrolyze ATP to generate force [6], (2) the neck domains or lever arms that transduce and amplify the force generated by motor domain [7,8] and (3) the tail domains offer specific binding sites for different cargos [9].
The motor domain of myosin has an actin-binding site and a nucleotide-binding site, which are mutually exclusive. Following the motor domain is lever arm, which usually contains some IQ motifs (consensus sequence IQxxxRGxxR) for calmodulin (CaM) or calmodulin-like proteins binding [10,11]. The numbers of IQ motifs vary among different myosin classes. The length of lever arm determines the step size along actin. The most essential difference among myosin motor proteins lies in their C-terminal globular tail domain (GTD). This domain recognizes various cargos through direct interactions or via adaptor proteins. Usually, GTD binds to the adaptor protein, which can interact with organelle-specific Rab family GTPase protein and therefore link to the specific organelle or vesicle [12,13].
Schematic Diagrams of Myosin superfamily representations in neuron. Different myosins possess conserved N-terminal motor domains and diverse C-terminal tail regions. The neck domain binds to calmodulin or calmodulin-like light chains.
Microtubule-based motors
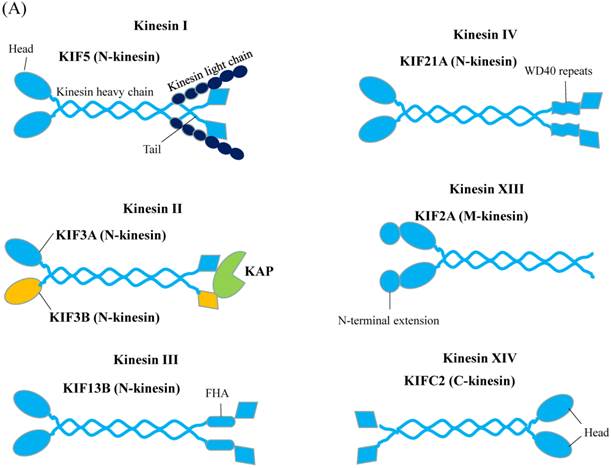
Kinesins (also referred to as KIFs) propel cargos along the microtubule. There are about 45 KIF genes in mammal genome [14]. A standard kinesin nomenclature system had been developed for the kinesin family members based on phylogenic analysis [15]. Accordingly, the kinesin proteins can be classified into 14 subfamilies, namely kinesin 1 to kinesin 14. Figure 2A gives representative kinesin of each family found in neuron [16]. All kinesins contain a highly conserved globular motor domain comprising a microtubule-binding site and an ATP-binding site as well as a diverse tail domain, which is responsible for cargo recognition and binding via the adaptor or scaffolding proteins [17]. Unlike myosin whose motor domain is always located in N-terminus, the motor domain of kinesins can be found in their N-terminal (N-type), middle (M-type), or C-terminal (C-type) region [18]. On microtubule, N-type kinesin moves to the plus end of microtubule, C-type kinesin moves to minus end of microtubule and M-type kinesin can destabilize microtubule tracks [19]. The energy supplying for kinesin processive motility comes from hydrolyzing ATP in the motor domain.
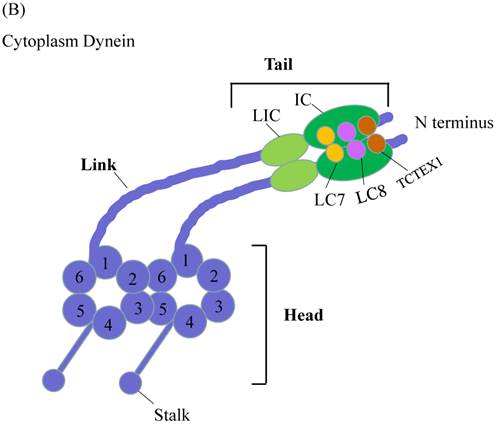
Another microtubule-based motor is cytoplasmic dynein (Figure 2B). Compared to myosin and kinesin, dynein is larger in size with more complicated composition and its working mechanisms are not well understood. The motor domain of dynein is located in C-terminal region, while tail domain is situated in the N-terminal region. The motor domain composes of six ATPases associated with diverse cellular activities (AAA) domains. The six AAA domains arrange in a large doughnut-shape ring [20,21]. AAA1 and AAA3 are responsible for ATP binding and hydrolysis, AAA2 and AAA4 play essential regulatory roles [22,23]. The function of AAA5 and AAA6 is not clear, because they lacked the required ATP-binding residues and exhibited other functions [21]. Extended from AAA4 domain is an antiparallel coiled-coil region, at the tip of which is the microtubule-binding domain. Microtubule-binding domain, coupled with ATP-hydrolysis cycle, moves the protein towards minus-end microtubule cytoskeleton. The dynein tail domain is responsible for dynein homodimerization and serves as binding sites with other subunits of dynein, including intermediate chain (IC), light intermediate chain (LIC), light chain 8 (LC8), roadblock (also known as LC7) and T-complex testis-specific protein 1 (TCTEX1). These dynein subunits can interact with adaptor proteins to form bridges [24-27]. With these bridges, dynein tail domain can thus carry the target cargos.
Schematic Diagrams of Microtubule-based motor proteins. (A) Kinesin superfamily representations in neuron. Like myosins, kinesins have conserved motor (head) domains and diverse tails. According to the positions of the motor domain, kinesins can be grouped into N-type, M-type and C-type kinesins. (B) Cytoplasmic dynein. The cytoplasmic dynein consists of C-terminal motor domain and N-terminal tail domain. The tail domains not only homodimerize the dynein heavy chains (purple color), but also recruit five other subunits (LIC, IC, LC8, LC7, TCTEX1) to accomplish dynein assembly.
Structural characteristics of cytoskeleton motors. The graphics of 3D views were drawn by PyMOL. (A) Complex structure of myosin VIIa/Sans. Cartoon representation indicates Sans-CEN1 domain binds to myosin VIIa-FERM domain which consists of F1, F2 and F3 lobes. (B) Structure of KIF13B-FHA domain bound to CENTA1. Cartoon diagram shows KIF13B-FHA domain interacts with dimerized CENTA1 PH1 and ArfGAP domains.
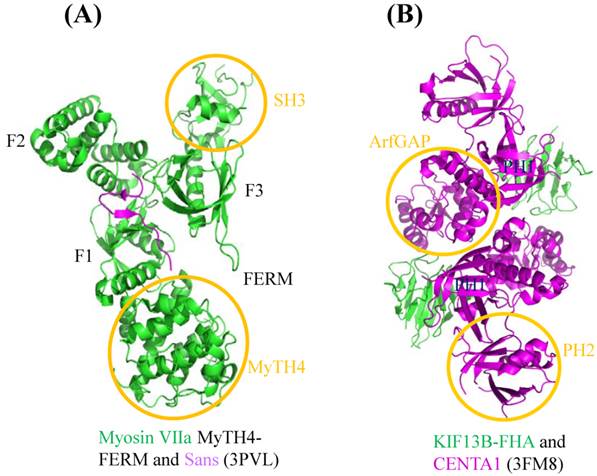
Structural biology and cytoskeleton motors in neuron
A large number of structural studies have greatly enhanced our understanding of neurological functions of many cytoskeleton motors. We summarize a few recent cases as examples to highlight the power and uniqueness of structural biology in uncovering the molecular mechanisms of cytoskeleton motors.
Many mutations in cytoskeleton motor proteins directly lead to diseases and aberrant developments. In neuron system, mutations found in myosin VIIa tail region result in the neuron disease named Usher syndrome I (USH1), which is characterized by deafness and visual impairment [28,29]. Crystallographic study shows that myosin VIIa tail region MyTH4-FERM domain binds to Sans central domain (Figure 3A) [30]. Sans is an adaptor protein interacting with both myosin VIIa and hormonin to form the ternary complex, which plays pivotal role in sensory neuroepithelia stereocilia formation [31]. In the complex structure, numerous amino acid mutations documented in Usher syndrome patients, such as E1327K, A1340P, E1349K, are located in the MyTH4-FERM domain of myosin VIIa and affect the interaction between the motor and Sans, leading to the disruption of the hormonin targeting to the neuroepithelia stereocilia of the inner ear. Based on the current study, we can further explore how these mutations individually impact on these interactions. For example, some mutations may function to weaken the interaction, while others may involve in regulation of other partners or signal pathways.
During axonal development, Phosphatidylinositol (3,4,5)-trisphosphate (PIP3)-containing vesicles function as the polarization factor, and need to be transported away from the soma to the tip of axon (more details will be given later). Kinesin III family KIF13B binds to PIP3 by using Centaurin-α1 (CENTA1) as adaptor protein to undertake the anterograde transport in vivo [32,33]. Structure of KIF13B-CENTA1 complex had been solved in the presence of phosphatidylinositol (Figure 3B) [34]. KIF13B binds to the first pleckstrin homology (PH1) domain and ArfGAP domain of CENTA1 through forkhead-associated (FHA) domain. According to the study, PH1 and PH2 domain of CENTA1 can bind simultaneously to PIP3 head group (IP4) [34]. Because of homodimerization of full-length KIF13B, it is expected to form a supercomplex with CENTA1 and PIP3-containing vesicle and move along microtubule towards the axonal terminus. This complex structure not only built the first molecular model of kinesin-adaptor interaction, but confirmed the long-speculated phosphorylation-independent interaction between CENTA1 and KIF13B-FHA, which represents the only known phosphothreonine-specific recognition domain but is unique among kinesin FHA domain [35].
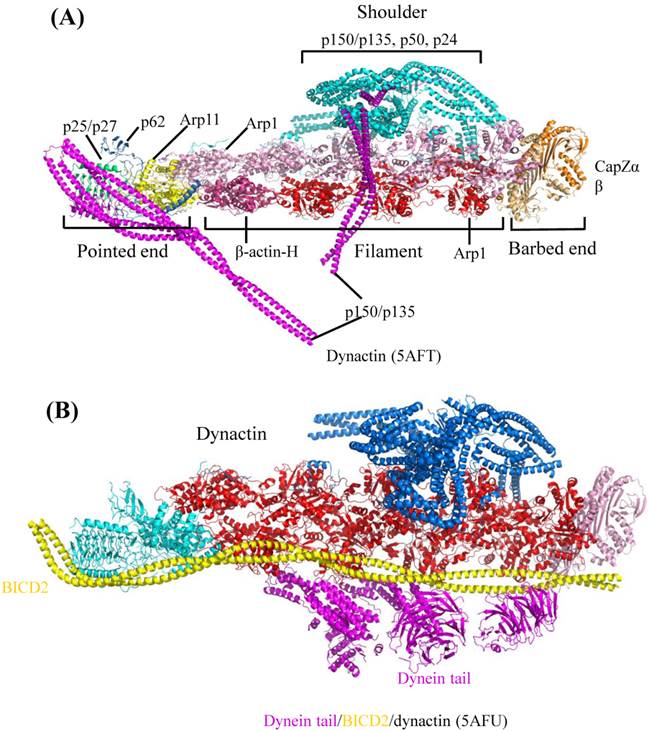
Dynactin (dynein activator) is one of the most important dynein's adaptors, which is a very complicated molecule, comprising of 11 different subunits [36]. Dynactin can bind to dynein N-terminal tail region and also microtubule [36,37]. In neuron cells, the ternary complex, consisting of dynein, dynactin and microtubule, is necessary for dendritic polarization formation and axonal uniformed microtubule development [38]. According to recently published Cryo-electron microscopy (EM) structures [39], dynactin consists of four parts: pointed end, filament, barbed end and shoulder (Figure 4A). Pointed end is composed of pointed-end proteins (p25, p27, p62) and actin-related protein (Arp11). Filament contains Arp1 and β-actin. Barbed end consists of actin capping proteins (CapZα and CapZβ). The shoulder contains p150Glued, p50 and p24. The pointed end interacts with cargo, while the filament binds with dynein tail domain and conserved motor adaptor Bicaudal D2 (BICD2) (Figure 4B), which can stabilize the interaction between dynein tail and dynactin by forming stable ternary complex. These complex structures by Cryo-EM showed us the assembly of dynactin and the detail of its interaction with dynein. Considering the facts that human dynein mutations are related with nervous diseases, such as sensory, motor and cognitive deficits [40], the structural elucidation of dynein and its adaptors may provide fundamental knowledge to help the rationalization of the prognosis of these diseases. Clearly, the above-mentioned work highlighted the power of cryo-EM in the elucidation of complicated macromolecule interaction involved in large cytoskeleton motors, such as dynein.
Cryo-EM Structure of dynactin and its complex with dynein tail. The graphics of 3D views were drawn by PyMOL. (A) Cryo-EM structure of dynactin. According to the molecular model, complicated dynactin is composed of four parts (shoulder, pointed end, filament and barbed end), each of these parts contains several subunits. (B) Complex structure of dynactin interacting with BICD2 and dynein tail. Structural architecture shows dynactin interacting with dynein tail region. The other subunits of dynactin can interact with other proteins or cargos, which facilitates the dynein-dependent cargo trafficking intracellular.
Roles in neuron
The processes of neuron development and maturation are very complicated. The polarizing factors and neurotrophin factors play essential roles in neuronal development and maturation processes. Notably, the neurotransmission is the most important function for neuron system. However, it remains ambiguous about what roles cytoskeleton motors play during these processes and what the structure-based mechanisms lie behind these roles. Here, we seek to summarize the current understanding about cytoskeleton motors trafficking in neuron function.
Polarizing-related factors transport
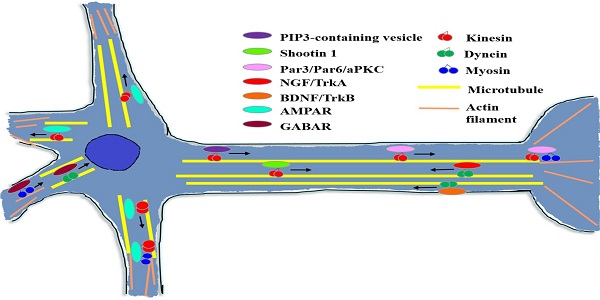
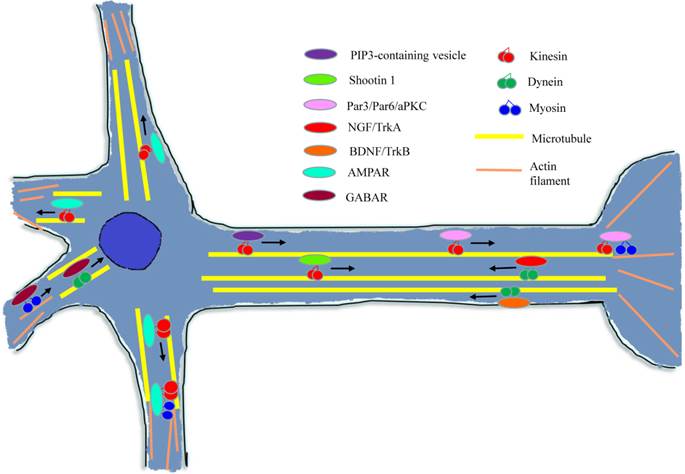
During the early stage of neuron development, polarization-related factors are essential for axon/dendrite determination and formation [41]. As shown in Figure 5, many of these factors need to be transported to the target region and then affect the neuron polarization through a variety of signal pathways or mechanisms.
Diagram of cytoskeleton motors in neuron. During neuron growth and development processes, cytoskeleton molecular motors get involved in signal molecules and growth factors transport in neurons. When conducting neurotransmission, cytoskeleton motors are responsible for kinds of neurotransmitter receptors trafficking.
Transport of PIP3 by KIF13B
It has been well-established that PIP3 can stimulate self-generation by activating Rho GTPases, which can subsequently activate PI3K to produce more PIP3 [42]. When treated by PI3K inhibitors to disrupt this feedback circuit, the asymmetrical polarization of neuron cells was reduced dramatically. This observation established that PIP3 is critical for neuronal polarization. In cultured hippocampal neuron cells, PIP3 has been found to accumulate in the tip of the longest neurite, which developed into an axon [43]. Later, KIF13B was found to bind to PIP3 by using Centaurin-α1 as adaptor protein and carried them away from the soma to the tip of axon [32,33]. As mentioned before, molecular mechanism of KIF13B-mediated phosphoinositide trafficking has been revealed by crystallographic study [34]. It has been reported that KIF13B can be phosphorylated by Par1, leading to a reduction in PIP3 transportation. However, Par1 can be phosphorylated and inactivated by Par3/Par6/atypical PKC (aPKC) [44]. This regulation mechanism indicates that there may exist some unknown connections between cellular signaling (not limited to phosphorylation cascade) and the motor trafficking processes. These deserve further investigation.
Transport of shootin 1 by KIF20B
Shootin 1 was discovered through proteomic analysis of cultured hippocampal neurons [45]. Shootin 1 showed fluctuating asymmetrical location in multiple neurites, but eventually, it anchored on a single neurite and accumulated in this neurite, which subsequently grew and developed into the axon [45]. Shootin 1 overexpression disrupted its asymmetrical accumulation and resulted in the surplus axons formation. Conversely, downregulated shootin 1 expression led to the neuronal polarization inhibition. It has been demonstrated that shootin 1 could regulate the asymmetrical accumulation of PIP3, thus the PI3K activity in the growth cone, but the reverse was not true. Conclusively, shootin 1 and PI3K are involved in the cell polarity pathway, during which shootin 1 is located in the upstream of PI3K and affects asymmetrical localization of PI3K [45]. During the axon determination, shootin 1 was observed to be actively transported by KIF20B from the soma cell body to the growth cone [46]. Disrupting shootin 1 anterograde transport decreases shootin 1 mobilizing to the growth cone, affecting asymmetrical distribution of PI3K activity in the neurite. However, the molecular mechanism of shootin 1 interacting with KIF20B requires further investigation.
Transport of Par3/Par6/atypical PKC (aPKC) polarity complex by KIF3A
Partitioning defective 3 homolog (Par3), Partitioning defective 6 homolog (Par6) and atypical protein kinase C (aPKC) form a polarity complex in epithelial or neuronal cell development [47]. In cultured hippocampal neurons, the axonal determination requires the localized activity of PI3K and aPKC [48]. The abnormal expression of Par3 or Par6 disrupts the axon specification and neuronal polarity. The polarity complex uses Par6 to bind to the small GTPase Cdc42 and uses Par3 to interact with Rac-specific guanine nucleotide-exchange factors (Rac GEF). Therefore, Par3/Par6/aPKC mediates Cdc42 induced Rac activation, which can subsequently induce PI3K activity to produce PIP3 [41,49]. In rat hippocampal neurons, Par3 was found to accumulate at the tip of developing axon. Assays show that KIF3A can anterograde transport the polarity complex to the tip of axonal neurite through directly binding to Par3 [50]. The localization of the polarity complex is essential to neuron polarity. Inhibition of Par3 and KIF3A interaction or disruption of Par3/Par6/aPKC transport to the growing axon can abolish the neuronal polarity development.
Neurotrophin-related factors transport
Neurotropins are the family of proteins implicated to induce neuron survival, development and modulating neuron plasticity [51]. Neurotrophin family contains four members: nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3) and neurotrophin-4 (NT-4) [52]. After their formation, these neurotrophins interact with the plasma membrane neurotrophin receptors, Trk and p75NTR, to regulate the neuronal survival [53,54]. Neurotrophin receptors bind with neurotrophin factors with different specificities [55]: TrkA binds tendentiously to NGF, TrkB binds preferentially to BDNF and NT-4, and TrkC to NT-3; while p75NTR can bind to all neurotrophin factors and their precursors. The activated neurotrophin and receptor complex can promote neuron growth through regulating gene expression, only when the complex has been transported to neuron soma from the terminal of axon or presynapse (Figure 5). [56,57]. It appeared that neurotrophin and its receptor are firstly endocytosed into cells and then transported to the cell nucleus periphery by interacting with cytoplasmic dynein in some unknown manner, which allows extracellular signals to be sent to the cell nucleus [58].
NGF/TrkA signaling endosome
NGF binding to TrkA extracellular domain leads to a conformation change in its cytoplasmic domain and autophosphorylation its tyrosine residues, which enhance TrkA kinase activity [59]. With surrounding plasma membrane, the activated NGF/TrkA complex forms the signaling endosome, which is endocytosed or internalized into the cell and activates the downstream signaling cascades [60]. NGF/TrkA signaling endosomes has been shown to require dynein motor protein for retrograde transport and for survival signaling propagation [61]. Disrupting the dynein trafficking through overexpression of dynamitin (p50) suppressed the perinuclear MAPK signaling pathway, which is activated by NGF/Trk, resulting in neuron cell atrophy and death [62].
BDNF/TrkB signaling endosome
BDNF has been demonstrated to regulate dendritic arborization and outgrowth during neuronal development [63]. BDNF binds to receptor TrkB, which also distributes on the axonal terminal membrane surface. Initiated by BDNF binding to TrkB, TrkB becomes phosphorylated, triggering internalization of BDNF/TrkB signaling endosome [58,64,65]. The signaling endosomes then associate with dynein motor to undergo axonal retrograde transport to cell body to induce nuclear signal pathway [66]. It has been shown that Snapin acts as a dynein adaptor, linking dynein and BDNF/TrkB complex and ensuring the signaling endosome axonal transport [67]. Disrupting Snapin and dynein interaction leads to aberrant BDNF/Trk motility and decreased dendrites growth.
Neurotransmission-related vesicles transport
Neurotransmission is the most important function of neuron system, a process of systematic communication between two neuronal cells. During the transmission, the neurotransmitters in the presynapse are released into the synaptic cleft and bind to the specific receptors on the postsynapse membrane. In presynapse, the synaptic vesicles containing neurotransmitters need to undergo a long axonal anterograde transport to reach the presynapse [68]. KIF1A and KIF1Bβ transport synaptic vesicles in axon, such as synaptotagmin, synaptophysin and Rab3A [68,69]. Additionally, myosin V has also been shown to associate with synaptic vesicles. It has been demonstrated that on the surface of synaptic vesicles, the synaptobrevin-synaptophysin complex binds to myosin V through the cytoplasmic domain of synaptobrevin/synaptophysin to form a multimeric complex [70,71]. In hippocampal neurons, myosin Va associates with the postsynaptic density-95 (PSD-95) through directly interacting with guanylate kinase domain-associated protein (GKAP), which binds to PSD-95 [72,73]. Simultaneously, myosin VI also binds to PSD-95 through its binding partner synapse-associated protein 97 (SAP97) interacting with GKAP [74,75]. As a result, both myosin V and myosin VI can interact with the postsynaptic density protein PSD-95 and may work in concert to transport PSD-95-containing vesicles. In postsynapse (Figure 5), the receptors need to be transported to and from the postsynaptic membrane [76]. Myosin VI has been reported to interact with alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor (AMPAR), a glutamate receptor, through binding directly to synapse-associated protein 97 (SAP97) [77]. Interestingly, KIF5 can also interact with AMPAR through adaptor glutamate receptor-interacting protein 1 (GRIP1) [78]. Hence, KIF5 carries AMPAR in soma and dendrites along microtubule; while myosin VI transports AMPAR on F-actin. It implies that the motors can recognize the cargo through specific interactions with adaptor proteins. Also, the different kinds of motors may act in a coherent manner during the cargo transportation process [79,80]. Besides AMPAR, both myosin VI and dynein can bind to muskelin that acts as an adaptor to link myosin VI and dynein to gamma-aminobutyric acid receptor (GABAR), and can regulate GABAR endocytosis process [80]. During this process, muskelin acts as the interconnector between actin-based retrograde trafficking and microtubule-based retrograde trafficking of GABAR. Another transmitter's receptor, N-methyl-D-aspartate receptor (NMDAR), can be transported by KIF17 through bridging by the complex LIN10-LIN2-LIN7 [81]. But the interaction between KIF17 and LIN10 can be suppressed by septin 9 (SEPT9) which directly binds to the tail region of KIF17 and competes with LIN10 [82].
Some cytoskeleton motors, including myosin Ic, myosin III and myosin IX, play other roles as mechanoforce sensor, tether and signaling regulator. Myosin Ic directly binds to cell membranes through the C-terminal pleckstrin homology (PH) motif, which allows it to crosslink the membranes and actin filaments and transduce the mechanical force from tail-binding membranes to head-binding filaments in inner ear stereocilia [83]. A recent structural study showed that myosin Ic undergoes CaM-mediated conformational transformation upon mechanical force transduction [84]. Myosin III was also found to stimulate the elongation of stereocilia in hair cells through tethering and activating Espin1 at the tips of stereocilia, the plus ends of actin filaments [85]. Gene mutations in encoding these proteins can lead to hear loss disease [86,87]. The recent solved myosin III/Espin complex structure provided a mechanistic explanation of myosin III-mediated stimulation of actin bundling formation [88]. Myosin IX contains a unique RhoGAP (GTPase-activating protein) domain in their tail region [89], which can switch active GTP-bound Rho to inactive GDP-bound Rho [90]. The structural study of myosin IXb RhoGAP domain indicates a regulation role of myosin IXb in SLIT/ROBO pathway [91]. In general, these diverse role manifests an integrated and complex function of myosin in cellular processes.
Perspectives
We have summarized and discussed the roles of cytoskeleton motors during neuronal development and transmission function. However, the structural basis for intracellular trafficking and regulations are not yet fully understood. Study on cytoskeleton motors' trafficking in neuron can help us to understand the cause of a variety of brain and nervous disorders. Although much progress has been made in past decades, structural study of neuronal-intracellular trafficking is still an attractive topic, especially in light of recent advance in macromolecule complex structure solvation technology, such as single particle cryo-EM [92].
To gain further mechanistic insights, we envisage that the following three aspects of intracellular trafficking pertaining to cytoskeleton motors deserve special attentions:
- What are the regulations between cargo transport and trafficking network? It has been an attractive and complex question. To address this issue calls for the structural and functional studies on motors inactive/active transition, motors competitive/cooperative coordinating during trafficking processes and cargos loading/unloading regulations.
- How do cytoskeleton motors and the asymmetrical localization of differentiation factors play out in specific cell development? During neuron differentiation process, motors are required to determine the asymmetrical localization of many development relevant factors, which may be also indispensable for epithelial cells or other dendritic cells maturation process. Many of these motor proteins have the characteristic of tissue-specific alternative splicing (also known as isoforms), which implies that the tissue-specific motor proteins expression may be related to the specific cell type development.
- What's the potential therapeutic implication of cytoskeleton motors for manipulating the vesicle transport processes? It has been shown that many human pathogens, including bacterium, virus, fungus and prion, can spread into host cells by motor trafficking, and move to nucleus periphery of host cells by moving along cytoskeletons. Therefore, further studies to understand the mechanisms and regulations may offer ideas for developing novel antipathogen therapies.
Acknowledgements
The authors thank the Science and Technology Development Fund, Macao S.A.R (FDCT) (project reference no.: 118/2013/A3) for the financial support. This work was supported partly by the startup of South University of Science and Technology of China and by Science, Technology and Innovation Commission of Shenzhen Municipality (Grant No. JCYJ20140417105742705) to Z.W.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196
2. Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci U S A. 2006;103:3681-6
3. Bridgman PC, Elkin LL. Axonal myosins. J Neurocytol. 2000;29:831-41
4. Wu X, Jung G, Hammer JA. Functions of unconventional myosins. Curr Opin Cell Biol. 2000;12:42-51
5. Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527-33
6. Sweeney HL, Houdusse A. Structural and functional insights into the myosin motor mechanism. Annu Rev Biophys. 2010;39:539-57
7. Sakamoto T, Wang F, Schmitz S, Xu Y, Xu Q, Molloy JE, Veigel C, Sellers JR. Neck length and processivity of myosin V. J Biol Chem. 2003;278:29201-7
8. Ruff C, Furch M, Brenner B, Manstein DJ, Meyhöfer E. Single-molecule tracking of myosins with genetically engineered amplifier domains. Nat Struct Biol. 2001;8:226-9
9. Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiology. 2005;20:239-51
10. Espindola FS, Suter DM, Partata LB, Cao T, Wolenski JS, Cheney RE, King SM, Mooseker MS. The light chain composition of chicken brain myosin-Va: calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell Motil Cytoskeleton. 2000;47:269-81
11. Koide H, Kinoshita T, Tanaka Y, Tanaka SI, Nagura N, Meyer zu Hörste G, Miyagi A, Ando T. Identification of the single specific IQ motif of myosin V from which calmodulin dissociates in the presence of Ca2+. Biochemistry. 2006;45:11598-604
12. Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA. Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002;4:271-8
13. Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and Myosin Va: implications of a tripartite protein complex for melanosome transport. J Biol Chem. 2002;277:12432-6
14. Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A. 2001;98:7004-11
15. Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19-22
16. Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39-71
17. Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201-14
18. Dagenbach EM, Endow SA. A new kinesin tree. J Cell Sci. 2004;117:3-7
19. Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519-26
20. Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467-80
21. Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27-43
22. Reck-Peterson SL, Vale RD. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein's AAA domains in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:1491-5
23. Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283:25839-45
24. Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681-96
25. Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507-16
26. Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol. 2004;6:427-35
27. Whyte J, Bader JR, Tauhata SB, Raycroft M, Hornick J, Pfister KK, Lane WS, Chan GK, Hinchcliffe EH, Vaughan PS, Vaughan KT. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J Cell Biol. 2008;183:819-34
28. Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62-4
29. Lefèvre G, Michel V, Weil D, Lepelletier L, Bizard E, Wolfrum U, Hardelin JP, Petit C. A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development. 2008;135:1427-37
30. Wu L, Pan L, Wei Z, Zhang M. Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science. 2011;331:757-60
31. Adato A, Michel V, Kikkawa Y, Reiners J, Alagramam KN, Weil D, Yonekawa H, Wolfrum U, El-Amraoui A, Petit C. Interactions in the network of Usher syndrome type 1 proteins. Hum Mol Genet. 2005;14:347-56
32. Venkateswarlu K, Hanada T, Chishti AH. Centaurin-α1 interacts directly with kinesin motor protein KIF13B. J Cell Sci. 2005;118:2471-84
33. Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174:425-36
34. Tong Y, Tempel W, Wang H, Yamada K, Shen L, Senisterra GA, MacKenzie F, Chishti AH, Park HW. Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin α1. Proc Natl Acad Sci U S A. 2010;107:20346-51
35. Tsai MD, Mahajan A, Yuan C, Lee H, Chen ES, Wu PY. Structure and function of the phosphothreonine-specific FHA domain. Sci Signal. 2009;2:er2
36. Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854-65
37. Waterman-Storer CM, Karki S, Holzbaur EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1). Proc Natl Acad Sci U S A. 1995;92:1634-8
38. Zheng Y, Wildonger J, Ye B, Zhang Y, Kita A, Younger SH, Zimmerman S, Jan LY, Jan YN. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat Cell Biol. 2008;10:1172-80
39. Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441-6
40. Willemsen MH, Vissers LE, Willemsen MA, van Bon BW, Kroes T, de Ligt J, de Vries BB, Schoots J, Lugtenberg D, Hamel BC, van Bokhoven H. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J Med Genet. 2012;49:179-83
41. Cheng PL, Poo MM. Early events in axon/dendrite polarization. Annu Rev Neurosci. 2012;35:181-201
42. Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, Bourne HR. Lipid products of PI (3) Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol. 2002;4:513-8
43. Ménager C, Arimura N, Fukata Y, Kaibuchi K. PIP3 is involved in neuronal polarization and axon formation. J Neurochem. 2004;89:109-18
44. Yoshimura Y, Terabayashi T, Miki H. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol Cell Biol. 2010;30:2206-19
45. Toriyama M, Shimada T, Kim KB, Mitsuba M, Nomura E, Katsuta K, Sakumura Y, Roepstorff P, Inagaki N. Shootin1: A protein involved in the organization of an asymmetric signal for neuronal polarization. J Cell Biol. 2006;175:147-57
46. Sapir T, Levy T, Sakakibara A, Rabinkov A, Miyata T, Reiner O. Shootin1 acts in concert with KIF20B to promote polarization of migrating neurons. J Neurosci. 2013;33:11932-48
47. Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr Opin Cell Biol. 2001;13:641-8
48. Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63-75
49. Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima YI, Ohno S, Hoshino M, Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270-7
50. Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol. 2004;6:328-34
51. Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545-64
52. Bibel M, Barde YA. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919-37
53. Radeke MJ, Misko TP, Hsu C, Herzenberg LA, Shooter EM. Gene transfer and molecular cloning of the rat nerve growth factor receptor. Nature. 1987;325:593-7
54. Hempstead BL, Martin-Zanca D, Kaplan DR, Parada LF, Chao MV. High-affinity NGF binding requires coexpression of the trk proto-oncogene and the low-affinity NGF receptor. Nature. 1991;350:678-83
55. Arevalo JC, Wu SH. Neurotrophin signaling: many exciting surprises!. Cell Mol Life Sci. 2006;63:1523-37
56. Gomes RA, Hampton C, El-Sabeawy F, Sabo SL, McAllister AK. The dynamic distribution of TrkB receptors before, during, and after synapse formation between cortical neurons. J Neurosci. 2006;26:11487-500
57. Deinhardt K, Salinas S, Verastegui C, Watson R, Worth D, Hanrahan S, Bucci C, Schiavo G. Rab5 and Rab7 control endocytic sorting along the axonal retrograde transport pathway. Neuron. 2006;52:293-305
58. Chowdary PD, Che DL, Cui B. Neurotrophin signaling via long-distance axonal transport. Annu Rev Phys Chem. 2012;63:571-94
59. Pérez P, Coll PM, Hempstead BL, Martı́n-Zanca D, Chao MV. NGF binding to the trk tyrosine kinase receptor requires the extracellular immunoglobulin-like domains. Mol Cell Neurosci. 1995;6:97-105
60. Howe CL, Valletta JS, Rusnak AS, Mobley WC. NGF signaling from clathrin-coated vesicles: evidence that signaling endosomes serve as a platform for the Ras-MAPK pathway. Neuron. 2001;32:801-14
61. Heerssen HM, Pazyra MF, Segal RA. Dynein motors transport activated Trks to promote survival of target-dependent neurons. Nat Neurosci. 2004;7:596-604
62. Wu C, Ramirez A, Cui B, Ding J, Delcroix JD, Valletta JS, Liu JJ, Yang Y, Chu S, Mobley WC. A functional dynein-microtubule network is required for NGF signaling through the Rap1/MAPK pathway. Traffic. 2007;8:1503-20
63. Cheung ZH, Chin WH, Chen Y, Ng YP, Ip NY. Cdk5 is involved in BDNF-stimulated dendritic growth in hippocampal neurons. PLoS Biol. 2007;5:e63
64. Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609-22
65. Zheng J, Shen WH, Lu TJ, Zhou Y, Chen Q, Wang Z, Xiang T, Zhu YC, Zhang C, Duan S, Xiong ZQ. Clathrin-dependent endocytosis is required for TrkB-dependent Akt-mediated neuronal protection and dendritic growth. J Biol Chem. 2008;283:13280-8
66. Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6:615-25
67. Zhou B, Cai Q, Xie Y, Sheng ZH. Snapin recruits dynein to BDNF-TrkB signaling endosomes for retrograde axonal transport and is essential for dendrite growth of cortical neurons. Cell Rep. 2012;2:42-51
68. Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769-80
69. Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, Saito M. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell. 2001;105:587-97
70. Prekeris R, Terrian DM. Brain myosin V is a synaptic vesicle-associated motor protein: evidence for a Ca2+-dependent interaction with the synaptobrevin-synaptophysin complex. J Cell Biol. 1997;137:1589-601
71. Ohyama A, Komiya Y, Igarashi M. Globular tail of myosin-V is bound to vamp/synaptobrevin. Biochem Biophys Res Commun. 2001;280:988-91
72. Naisbitt S, Valtschanoff J, Allison DW, Sala C, Kim E, Craig AM, Weinberg RJ, Sheng M. Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J Neurosci. 2000;20:4524-34
73. Walikonis RS, Jensen ON, Mann M, Provance DW, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20:4069-80
74. Wu H, Reissner C, Kuhlendahl S, Coblentz B, Reuver S, Kindler S, Gundelfinger ED, Garner CC. Intramolecular interactions regulate SAP97 binding to GKAP. EMBO J. 2000;19:5740-51
75. Bridgman PC. Myosin-dependent transport in neurons. J Neurobiol. 2004;58:164-74
76. Collingridge GL, Isaac JT, Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci. 2004;5:952-62
77. Wu H, Nash JE, Zamorano P, Garner CC. Interaction of SAP97 with minus-end-directed actin motor myosin VI. Implications for AMPA receptor trafficking. J Biol Chem. 2002;277:30928-34
78. Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417:83-7
79. Brown SS. Cooperation between microtubule- and actin-based motor proteins. Annu Rev Cell Dev Biol. 1999;15:63-80
80. Heisler FF, Loebrich S, Pechmann Y, Maier N, Zivkovic AR, Tokito M, Hausrat TJ, Schweizer M, Bähring R, Holzbaur EL, Schmitz D. Muskelin regulates actin filament- and microtubule-based GABA(A) receptor transport in neurons. Neuron. 2011;70:66-81
81. Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796-802
82. Bai X, Karasmanis EP, Spiliotis ET. Septin 9 interacts with kinesin KIF17 and interferes with the mechanism of NMDA receptor cargo binding and transport. Mol Biol Cell. 2016;27:897-906
83. McConnell RE, Tyska MJ. Leveraging the membrane-cytoskeleton interface with myosin-1. Trends Cell Biol. 2010;20:418-26
84. Lu Q, Li J, Ye F, Zhang M. Structure of myosin-1c tail bound to calmodulin provides insights into calcium-mediated conformational coupling. Nat Struct Mol Biol. 2015;22:81-8
85. Salles FT, Merritt RC, Manor U, Dougherty GW, Sousa AD, Moore JE, Yengo CM, Dosé AC, Kachar B. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat Cell Biol. 2009;11:443-50
86. Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies' eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci U S A. 2002;99:7518-23
87. Boulouiz R, Li Y, Soualhine H, Abidi O, Chafik A, Nürnberg G, Becker C, Nürnberg P, Kubisch C, Wollnik B, Barakat A. A novel mutation in the Espin gene causes autosomal recessive nonsyndromic hearing loss but no apparent vestibular dysfunction in a Moroccan family. Am J Med Genet A. 2008;146:3086-9
88. Liu H, Li J, Raval MH, Yao N, Deng X, Lu Q, Nie S, Feng W, Wan J, Yengo CM, Liu W. Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin. Elife. 2016;5:e12856
89. Reinhard J, Scheel AA, Diekmann D, Hall A, Ruppert C, Bähler M. A novel type of myosin implicated in signalling by rho family GTPases. EMBO J. 1995;14:697-704
90. Post PL, Bokoch GM, Mooseker MS. Human myosin-IXb is a mechanochemically active motor and a GAP for rho. J Cell Sci. 1998;111:941-50
91. Kong R, Yi F, Wen P, Liu J, Chen X, Ren J, Li X, Shang Y, Nie Y, Wu K, Fan D, Zhu L, Feng W, Wu JY. Myo9b is a key player in SLIT/ROBO-mediated lung tumor suppression. J Clin Invest. 2015;125:4407-20
92. Cheng Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell. 2015;161:450-7
Author contact
![]() Corresponding author: Tam, K.Y. (kintammo); Tel.: +853-88224988; fax: +853-88222314
Corresponding author: Tam, K.Y. (kintammo); Tel.: +853-88224988; fax: +853-88222314

 Global reach, higher impact
Global reach, higher impact