10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(8):976-984. doi:10.7150/ijbs.19191 This issue Cite
Research Paper
The Overexpression of CARM1 Promotes Human Osteosarcoma Cell Proliferation through the pGSK3β/β-Catenin/cyclinD1 Signaling Pathway
Department of Orthopedics, Shanghai Jiao Tong University Affiliated Sixth People's Hospital, No.600, Yishan Road, Shanghai, 200233, China.
† These authors contributed equally to this work.
Received 2017-1-15; Accepted 2017-4-16; Published 2017-7-18
Abstract
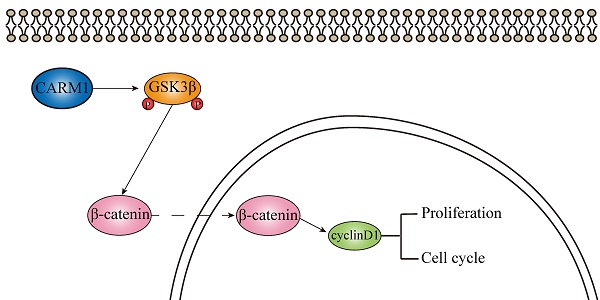
Osteosarcoma (OS) is a kind of malignant bone tumor that occurs frequently in the region surrounding the knee joint and poses a threat to the health of teenagers. Since the application of chemotherapy to treat OS, 5-year survival rate in patients has improved from 10% to 70%, but patient survival has not changed over the past four decades. Coactivator-associated arginine methyltransferase 1 (CARM1) is a member of the PRMT protein family; it acts as an oncogene in many cancers, but its function in OS is still unknown. In this study, we found that CARM1 is overexpressed in OS and its expression is correlated with the Enneking stage. CCK-8 and colony forming assays showed that proliferation in OS cell lines was downregulated when siRNA was used to knockdown CARM1 expression. The cell cycle was inhibited in S phase after si-CARM1 transfection in OS cell lines. An antibody array indicated that Erk1/2 (Thr202/Tyr204), PARS40 (Thr246), and GSK3β (Ser9) expression are affected by CARM1, and western blotting verified that CARM1 promotes OS cell proliferation via pGSK3β/β-catenin/cyclinD1 signaling. Accordingly, CARM1 is a crucial gene in OS and is a potential new treatment target.
Keywords: Osteosarcoma, CARM1, pGSK3β/β-catenin/cyclinD1
Introduction
Osteosarcoma (OS) is a kind of malignant bone tumor that occurs frequently in the region surrounding the knee joint [1]. With high rates of local invasion and early metastasis, osteosarcoma leads to disability in many adolescents as well as death. Cisplatin, cyclophosphamide, methotrexate, and Adriamycin have been used for chemotherapy in OS patients, improving the 5-year survival rate in patients from 10% to 70% [2]. However, since the 1970s, there has been no change in the survival of patients [3]. As a result, a comprehensive understanding of the biological processes in OS and the factors that contribute to its progression is necessary, and potential therapeutic targets should be identified in clinical research.
Coactivator-associated arginine methyltransferase 1 (CARM1) is a type I arginine methyltransferase involved in autophagy [4], DNA repair [5], mRNA decay [6], and epigenetic regulation [7]. It is a member of the PRMT protein family and acts as an oncogene in lung cancer [8], breast cancer [9], and colorectal cancer [10]. In breast cancer, CARM1 methylates SWI/SNF to activate the c-Myc pathway and enhances breast cancer progression and metastases [9]. Overexpression of CARM1 is correlated with poorly characterized clinicopathologic parameters and molecular subtypes [11]. CARM1 deregulates β-catenin to increase colorectal cell growth and is associated with the development of prostate cancer [10]. It also has been shown that CARM1 can act as a coactivator of transcription factors other than nuclear receptors, such as p53 [12], NF-κB [13], and Cyclin E1 [14]. However, the effects of CARM1 in OS are not yet known.
The β-catenin/cyclinD1 pathway is an important signaling pathway involved in proliferation and differentiation [15]. Initially, β-catenin was discovered as a component of cell-cell adhesive junctions regulated by adenomatous polyposis coli and glycogen synthase kinase-3β (GSK3β) [16]. When Wnt signaling is absent, β-catenin is phosphorylated by GSK-3β and CK1 complexes. Phosphorylated β-catenin tends to be multi-ubiquitinated and then degraded in proteasomes [17]. When Wnt signaling is present, GSK-3β activity is inhibited, and the degradation of β-catenin is blocked [17]. β-catenin accumulates in the cytoplasm and is then translocated into the nucleus [18], where it activates Wnt target genes together with various transcriptional repressors or activators, including the cell cycle regulators MYC and cyclinD1, which are important for cancer development [19].
In this study, we first found that the overexpression of CARM1 is associated with the Enneking stage of OS. The knockdown of CARM1 expression decreased proliferation and affected the cell cycle in OS cell lines. Most importantly, we found that the knockdown of CARM1 could downregulate the activity of GSK3β and decrease the expression of β-catenin and cyclinD1. Therefore, CARM1 could affect Wnt signaling to promote OS proliferation. It may be a novel target for OS treatment.
Results
CARM1 was overexpressed in osteosarcoma cells and was associated with the Enneking stage
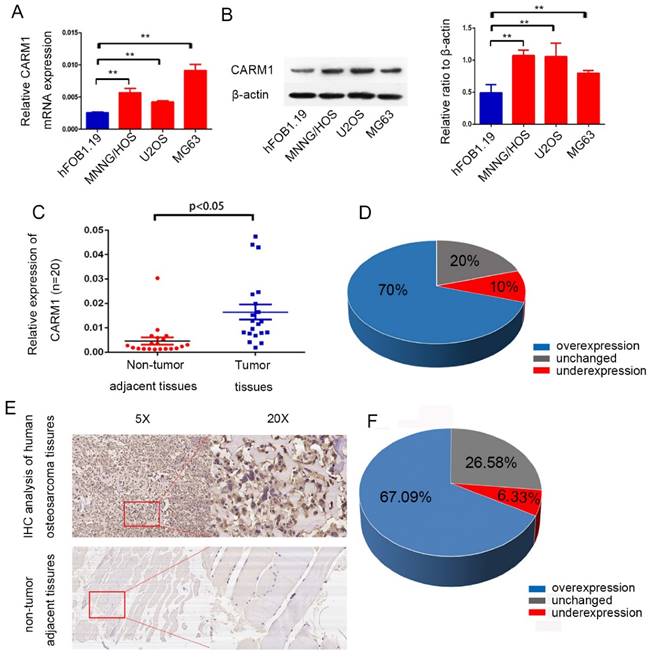
First, we analyzed CARM1 mRNA expression and protein expression profiles in the osteoblast cell line hFob and OS cell lines (MNNG/HOS, U2OS, and MG63). Relative to the hFob cell line, CARM1 was obviously overexpressed in OS cell lines (Figure 1 A,B). To examine the expression of CARM1 mRNA in fresh osteosarcoma patient samples, we collected 20 pairs of primary tumor samples and adjacent non-tumor tissues. Based on a qRT-PCR analysis, CARM1 expression was remarkably higher in tumor tissues (70%) than in control tissues (Figure 1 C, D). To further determine the clinicopathological significance of CARM1 in OS, we performed an immunohistochemical analysis of CARM1 in OS tissues, including 79 paired OS tissues and adjacent non-tumor tissues. A representative image of OS tissues and adjacent non-tumor tissues is shown in Figure 1 E. Microscopic observations indicated that CARM1 is expressed in the cytoplasm and nuclei of OS cells (Figure 1 E). Positive CARM1 expression was more frequent in osteosarcoma tissues (Figure 1 F) with high Enneking stages, and negative CARM1 expression was more common for low Enneking stages (P < 0.05) (Table 1). However, no significant differences in CARM1 expression were observed with respect to patient gender, age, tumor location, or histological classification (Table 1).
Relationship between CARM1 IHC results and clinical information for 79 OS patients.
| Variables | CARM1 expression(N) | |||||
|---|---|---|---|---|---|---|
| Total | Negative | Positive | X^2 | P-values | ||
| Gender | Male | 46 | 15 | 31 | 2.031 | 0.154 |
| Female | 33 | 16 | 17 | |||
| Age | ≤20 | 47 | 18 | 29 | 0.043 | 0.835 |
| >20 | 32 | 13 | 19 | |||
| Tumor location | Femur | 37 | 20 | 17 | 8.862 | 0.065 |
| Tibia | 21 | 7 | 14 | |||
| Humerus | 12 | 1 | 11 | |||
| Fibula | 5 | 2 | 3 | |||
| Others | 4 | 1 | 3 | |||
| Tumor Necrosis Rate | ≥90% | 45 | 17 | 28 | 0.094 | 0.759 |
| <90% | 34 | 14 | 20 | |||
| Histological classification | Osteoblastic | 29 | 12 | 17 | 2.461 | 0.292 |
| Chondroblastic | 28 | 8 | 20 | |||
| Others | 22 | 11 | 11 | |||
| Histological grade | Ⅰ | 12 | 10 | 2 | 11.538 | 0.001 |
| Ⅱ/Ⅲ | 67 | 21 | 46 | |||
| Enneking stage | Ⅰ | 11 | 9 | 2 | 9.818 | 0.007 |
| Ⅱ | 60 | 19 | 41 | |||
| Ⅲ | 8 | 3 | 5 | |||
Depletion of CARM1 reduced in vitro tumor growth
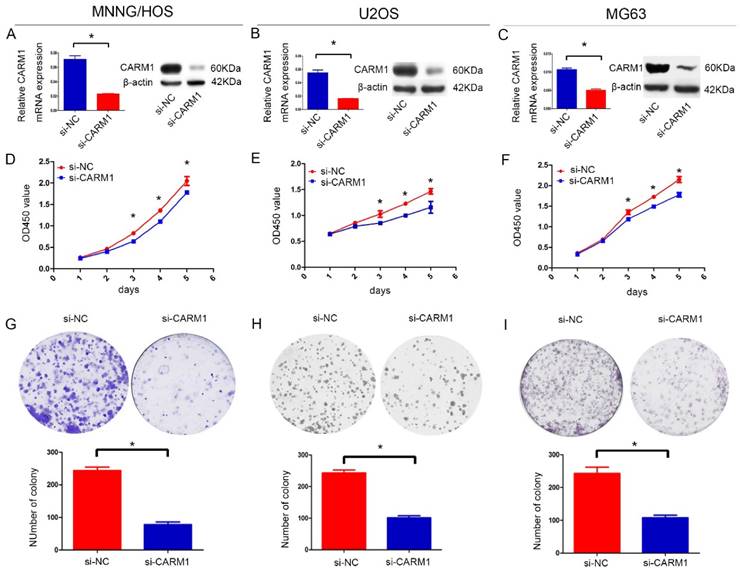
To examine the role of CARM1 in tumor development, we used siRNA targeting CARM1 to downregulate CARM1 expression. qRT-PCR and immunoblotting analyses showed greater than 60% efficiency in the reduction of endogenous CARM1 protein levels when β-actin was used as the internal standard (Figure 2A-C). Cell proliferation was evaluated using a CCK8 assay, which showed that the inhibition of CARM1 suppressed the proliferation of MNNG/HOS, U2OS, and MG63 cells (Figure 2D-F). A colony forming assay was also used as a direct method to detect the proliferation ability of cells. We found that the number of colony forming units was significantly reduced by more than 50% after transfect si-CARM1 in OS cell lines (Figure 2G-I). These data demonstrate that CARM1 is involved in regulating the proliferation of OS cancer cells. However, migration and invasion were not affected in MNNG/HOS cells (Figure S1).
CARM1 was overexpressed in OS cell lines and clinical tissue samples. (A) The expression of CARM1 in the human osteoblastic cell line hFob and OS cell lines (MNNG/HOS, U2OS, and MG63) was examined by qRT-PCR. (B) Western blotting showed the CARM1 was overexpressed in OS cells in comparison to hFob cells. (C) qRT-PCR showed that CARM1 was overexpressed in OS tissues in comparison with corresponding non-tumorous tissues obtained from 20 OS patients. (D) The mRNA expression level of CARM1 was higher in 70% of OS tissues compared to the corresponding non-tumorous tissues. (E) A representative image of immunohistochemical results for paraffin-embedded human OS tissues and the corresponding non-tumor tissues using a CARM1 antibody. (F) Based on an immunohistochemical analysis, in 67.09% of patients, CARM1 expression was higher in OS tissues than in the corresponding non-tumor tissues. The “**” was indicated a significant difference (P<0.01) in the figures.
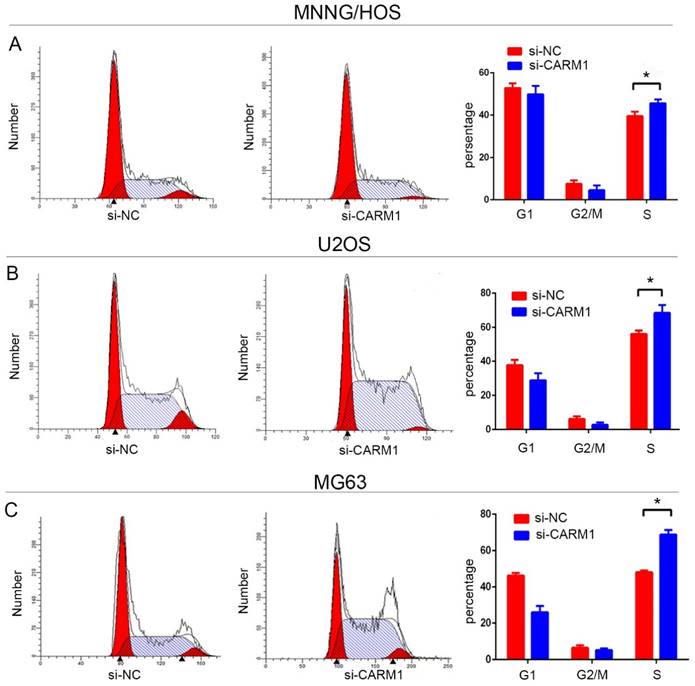
Downregulation of CARM1 expression prolonged the S phase in OS cells
The effects of CARM1 levels on the cell cycle were investigated by flow cytometry. The cell cycle of OS cell lines was compared before and after CARM1 knockdown. The cell populations in S-phase were 39.6 ± 2.0%, 56.1 ± 2.1%, and 48.1 ± 0.9% for negative control siRNA (si-NC)-transfected MNNG/HOS, U2OS, and MG63, and 45.6 ± 1.765%, 68.5 ± 4.6%, and 68.76 ± 2.6% in si-CARM1-transfected MNNG/HOS, U2OS, and MG63 cell lines (Figure 3). These results showed that the S phase was significantly prolonged after transfection with si-CARM1 in OS cell lines (P < 0.05).
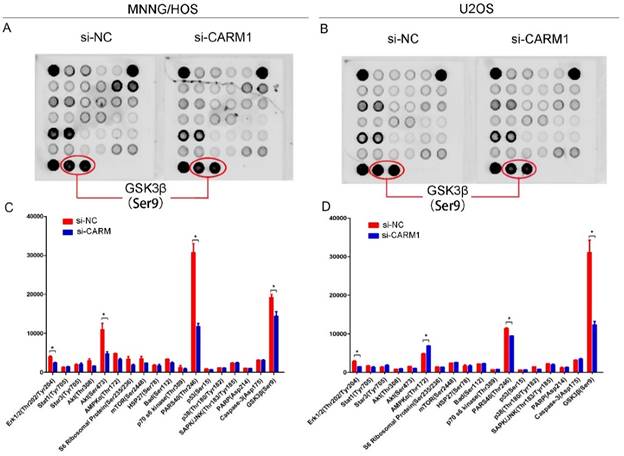
An antibody array indicated that Erk1/2(Thr202/Tyr204), PARS40 (Thr246), GSK3β (Ser9) are affected by CARM1
To further characterize the mechanism by which CARM1 promotes OS cell proliferation, we used a signaling array kit to detect phosphorylation at 18 sites after transfection with si-NC and si-CARM1 in MNNG/HOS and U2OS cells (Figure 4A, B). The array was analyzed using image analysis software. Erk1/2 (Thr202/Tyr204), AKT (Ser473), PARS40 (Thr246), GSK3β (Ser9) exhibited obvious differences between si-NC and si-CARM1-transfected MNNG/HOS cells (Figure 4C), and Erk1/2 (Thr202/Tyr204), AMPKα (Thr172), PARS40 (Thr246), and GSK3β (Ser9) differed in U2OS cells. Accordingly, 3 phosphorylation sites (Erk1/2, PARS40, and GSK3β) differed in both MNNG/HOS and U2OS cells (Figure 4 D).
Downregulation of the expression of CARM1 suppressed the proliferation of OS cells. The mRNA and protein expression levels of CARM1 were inhibited by si-CARM1 in the MNNG/HOS (A), U2OS (B), and MG63 (C) cell lines. Cell growth was detected by a CCK-8 assay. A significant reduction in CARM1 inhibited cell growth in the MNNG/HOS (D), U2OS (E), and MG63 (F) cell lines. The number of colony-forming units decreased when the MNNG/HOS (G), U2OS (H), and MG63 (I) cell lines were transfected with si-CARM1. The “*” was indicated a significant difference (P<0.05) in the figures.
CARM1 promoted OS proliferation via pGSK3β/β-catenin/cyclinD1 signaling
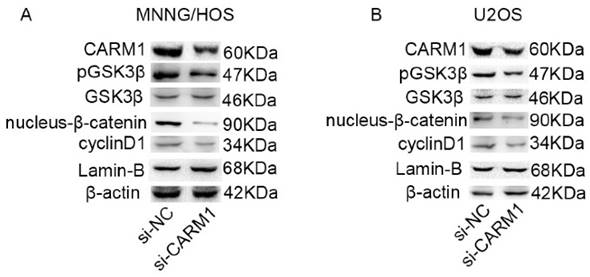
We next examined the exact signaling molecules that contribute to the effects of CARM1 on OS cell proliferation. After siRNA-mediated silencing of CARM1 expression, a western blot assay showed that Erk1/2 (Thr202/Tyr204) and PARS40 (Thr246) expression did not decreased (Figure S2), but the band intensity for pGSK3β (Ser9) decreased by more than 50% compared with expression in control cells. The total GSK3β protein levels were not affected by the silencing of CARM1 expression, but expression levels of β-catenin in nucleus and cyclinD1 decreased substantially in MNNG/HOS (Figure 5A) and U2OS cells (Figure 5B). A previous study showed when GSK-3β is inhibited in Wnt signaling, β-catenin accumulates in nuclei to target cyclinD1, thereby influencing cell cycle [19]. Wnt signaling has been reported to affect neoplasm growth in OS [37]. pcDNA-β-catenin was used to recover the expression of nucleus-β-catenin in si-CARM1 targeted OS cell lines (Figure S3 A, B), and then we found that overexpression of β-catenin could rescue the proliferation of si-CARM1 targeted OS cell lines (Figure S3 C, D). Therefore, these data show that pGSK3β/β-catenin/cyclinD1 signaling is crucial in the process of CARM1-mediated OS cell proliferation.
Discussion
OS is a common malignant bone tumor that often occurs in teenagers. It can grow rapidly and result in lung metastasis in the early stage. Our research was focused on the identification of oncogenes that can be used as therapeutic targets in OS.
The cell cycle was arrested in S phase when CARM1 expression was reduced in MNNG/HOS (A), U2OS (B), and MG63 (C). The results show that the percentage of S phase cells (P < 0.05, n = 3) increased significantly after transfection with si-CARM1 in the MNNG/HOS, U2OS, and MG63 cell lines.
Previous studies have shown that CARM1 methylates substrates involved in transcription processing [20], glutamine metabolism [21], and epigenetic transcriptional regulation [22]. CARM1-knockout mice die during late embryonic development or immediately after birth [24]. Therefore, we believe that CARM1 is a crucial gene in biological processes. At the cellular level, CARM1 plays a complex role in controlling gene expression, with targets that regulate transcription (p300), DNA packaging (histone H3), and mRNA stability and translation (PABP1) [24]. CARM1 is overexpressed in many cancers and is associated with proliferation and metastasis. CARM1 can transactivate many cancer-associated transcription factors, including p53 [12], E2F1 [23], androgen receptor [25], and ERα [26]. Frietze et al. reported that CARM1 can improve estrogen-stimulated breast cancer cell proliferation via the upregulation of E2F1 [23]. A reported that CARM1 binds to the C terminus to affect p53 function [12]. However, the role of CARM1 in OS is not known.
In this study, we examined the physiological function of CARM1 in OS as well as the underlying mechanisms. First, we found that CARM1 is unequivocally overexpressed in OS cell lines and tissues in comparison with corresponding non-tumorous tissues by RT-PCR. Then, we examined CARM1 expression in OS tissues and adjacent normal tissues from 79 patients by immunohistochemistry, and found that CARM1 is overexpressed in tumor tissues and the expression level was related to the histologic grade and Enneking stage of OS tissues. To determine the exact function of CARM1 in OS, we used CARM1-targeted siRNA to interfere with CARM1 expression in OS cell lines. Based on CCK-8 and colony-forming assays, we found that the knockdown of CARM1 significantly reduced the OS cell proliferation. This result is similar to those of studies on hepatocytes [27], lung cancer [28], and colorectal cancer [29], indicating that CARM1 may be an oncogene involved in proliferation and cell processes. The si-CARM1-treated OS cells exhibited a large S phase population. However, decreased expression of CARM1 in MNNG/HOS cells did not influence migration and invasion (Figure S1).
MNNG/HOS and U2OS cells were grown to 80% confluency and were then serum-starved overnight. Cells were either transfected with si-NC or si-CARM1 after 48 h. A PathScan Intracellular Signaling Array Kit (Fluorescent Readout; #7744) was using to analyze the protein extracted from cells. (A) Panel A summarizes the protein expression results for si-NC and si-CARM1 transfection of MNNG/HOS cells. (B) Panel B shows the protein expression results for si-NC- and si-CARM1-transfected U2OS cells. Images were acquired using the LICOR Bio-science® Odyssey® Image System. (C) Expression levels of Erk1/2 (Thr202/Tyr204), Akt (Thr308), PARS40 (Thr246), and GSK3β (ser9) were significantly reduced after si-CARM1 transfection in MNNG/HOS cells. (D) Expression levels of Erk1/2 (Thr202/Tyr204), AMPKα (Thr172), PARS40 (Thr246), GSK3β (ser9) clearly changed after the downregulation of CARM1 expression in U2OS cells.
Western blotting results showed that pGSK3β/β-catenin/cyclinD1 signaling was involved in the process by which CARM1 promotes the proliferation of OS cells. (A) The downregulation of CARM1 expression in MNNG/HOS did not affect GSK3β and pGSK3β, β-catenin, and cyclinD1 were simultaneously downregulated. (B) The gray value of GSK3β was unchanged and the gray values of pGSK3β, β-catenin, and cyclinD1 decreased when the expression of CARM1 was reduced in U2OS cells.
To determine the mechanism by which CARM1 promotes the proliferation of OS cells, a PathScan (®) Intracellular Signaling Array Kit (Fluorescent Readout) was used to detect changes in signaling molecules in MNNG/HOs and U2OS cells before and after CARM1 knockdown. The phosphorylation levels of Erk1/2 (Thr202/Tyr204), PARS40 (Thr246), and GSK3β (Ser9) were downregulated in si-CARM1-treated MNNG/HOS cells and U2OS cells. These results suggested that CARM1 knockdown in OS cells inhibits proliferation via Erk1/2 (Thr202/Tyr204), PARS40 (Thr246), or GSK3β (Ser9) activation. Furthermore, using western blotting, we found that the band intensities of Erk1/2 (Thr202/Tyr204) and PARS40 (Thr246) did not decrease according to the expression of CARM1. However, the band intensities of pGSK3β (Ser9) was significantly downregulated after CARM1 knockdown.
GSK3β is a critically important kinase that phosphorylates and regulates many signaling proteins and transcription factors. Boyle et al. reported that the activation of protein kinase C decreases phosphorylation at sites that are negatively regulated by GSK3β, resulting in increased AP-1 activity [30]. Pulverer et al. reported that myc is negatively regulated by GSK3β-mediated phosphorylation of Thr-58 and Ser-62, resulting in decreased cell growth and transformation [31]. Serine and tyrosine phosphorylation regulate the activity of GSK3β. The phosphorylation of Ser9 of GSK3β results in decreased GSK3β activity [32]. In this study, we found that the knockdown of CARM1 expression could decrease the expression of pGSK3β (Ser9), but the total GSK3β expression did not change. According to a previous study, β-catenin is crucial for cell proliferation [33], cell apoptosis [34] and tumorigenesis [35]. β-catenin could also be activated in OS, playing an important role in tumor proliferation [36] and epithelial-to-mesenchymal transition [37]. GSK3β could be involved in the phosphorylation of β-catenin to active the E3 ubiquitin ligase subunit β-Trcp, which targets beta-catenin for proteasomal degradation [38]. The phosphorylation of GSK3β leads to GSK3β inactivation, resulting in β-catenin accumulation and promoting translocation to the nucleus [39]. Additionally, β-catenin accumulates in the nucleus to promote cyclinD1 expression, thereby affecting cell proliferation and the cell cycle [16]. We extracted the nuclear protein for western blotting. The result confirmed that β-catenin expression was decreased when CARM1 was knocked down in MNNG/HOS and U2OS cells, consistent with previous results obtained using colorectal cancer [29] and prostate cancer cells [10]. Our results suggest that CARM1 could inactivate GSK3β, resulting in the activation of β-catenin/cyclinD1 signaling to promote OS cell proliferation. There are some possible mechanisms between CARM1 and GSK3β phosphorylation. The first hypothesis is as follows. CARM1 has been confirmed to interact with PELP1 [41]. According to another previous study, PELP1 could enhance the phosphorylation of GSK3β and further activate beta-catenin signaling [42]. So we conclude that CARM1 may interact with PELP1 to activate pGSK3β/β-catenin signaling pathway. Secondly, PRMT1 could methylate Axin to enhance the interaction of Axin and GSK3β and then active the Wnt signaling [43]. CARM1 and PRMT1 were both members of PRMT family. So CARM1 may promote the phosphorylation of GSK3β through its interaction with PRMT1. However, the exact mechanism by which CARM1 is involved in the phosphorylation of GSK3β requires further investigation.
Materials and Methods
Cell lines and cell culture
The human osteosarcoma cell lines MNNG/HOS, U2OS, and MG63 were maintained in Dulbecco's Modified Eagle Medium or RPMI-1640 supplemented with 10% (v/v) fetal bovine serum (Gibco BRL, Gaithersburg, MD, USA), 100 U/mL penicillin (Sigma-Aldrich, St. Louis, MO, USA), and 100 mg/mL streptomycin (Sigma-Aldrich). All cell lines were incubated at 37°C in a humidified incubator with 5% CO2.
Clinical data and tumor specimen collection from patients with osteosarcoma
A total of 79 tumor tissues and adjacent non-tumor tissues were collected from OS patients after surgery at Shanghai Jiao Tong University Affiliated Sixth People's Hospital from 2013 to 2015. The tissues were fixed in 10% formalin and embedded in paraffin after resection. All human samples were collected after informed, written consent was obtained in accordance with the Helsinki guidelines. The study was approved by the Ethics Committee of the Shanghai Jiao Tong University Affiliated Sixth People's Hospital (YS-2016-064; February 24, 2016).
RNA isolation and qRT-PCR assay
Total cellular RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA). The NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) was used for RNA quantification. The PrimeScript RT Reagent Kit (TaKaRa Biotechnology, Shiga, Japan) was used to transcribe RNA into cDNA. The primer sequences were as follows: CARM1 Forward: CGCCCTCTACAGCCATGAAG, Reverse: ACTGGATGAGGACGCTGTTG; β-actin: Forward: TTGTTACAGGAAGTCCCTTGCC, Reverse: ATGCTATCACCTCCCCTGTGTG. Quantitative real-time PCR was performed using the Applied Biosystems 7500 Real-Time PCR System (Applera Deutschland, Darmstadt, Germany).
Transfection
Cell culture was performed on 6-well plates until MNNG/HOS grew to 60% confluency. Cells were then transfected for 6 h with si-RNA or pcDNA-β-catenin specific to either MG63 or U2OS (both at 50 nM) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The CARM1 siRNA and the negative Control siRNA was provided by Ribobio (Guangzhou, China). The pcDNA-β-catenin was provided by GENEWIZ (Suzhou, China).
Colony formation assay and cell cycle assay
The si-NC- and si-CARM1-transfected cells were counted using a cell counter. They were then plated at a density of 1 × 103 cells/well in 6-well plates. After 12 days, the cells were washed with phosphate-buffered saline 2 times, fixed with methanol for 30 min, and stained with 0.1% crystal violet solution for 10 min. Visible colonies were counted. The cell cycle was analyzed using ModFit software (BD Biosciences) 48 h after transfection with si-NC and si-CARM1.
Cell proliferation analysis using the Cell Counting Kit-8
Cells treated with siRNA targeting CARM1 (si-CARM1) and negative control (si-NC) were plated at a density of 3 × 103 cells/well in 96-well plates. At the indicated time points after transfection or treatment, 10 μL of CCK-8 solution (Dojindo Molecular Technologies, Kumamoto, Japan) was added to 90 μL of culture medium. The cells were subsequently incubated for 1 h at 37°C and the optical density was measured at 450 nm. Three independent experiments were performed.
Western blotting to detect antibody expression
Protein was extracted, separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes, followed by incubation with the appropriate concentration of various antibodies. The antibodies were as follows: anti-CARM1 (ab128851, NOVUS, NB200-342); anti-GSK3β (#9315; CST, Danvers, MA, USA), anti-GSK3β (ab75814; Abcam), anti-β-catenin (#9562; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-cyclinD1 (ab134175; Abcam), anti-PRAS40 (ab214100; Abcam), anti-ERK1/2(ab214362; Abcam), anti-Lamin B1 (#12586; CST), anti-rabbit IgG (Sigma-Aldrich), or anti-mouse IgG (Sigma-Aldrich).
Immunohistochemistry (IHC)
Paraffin-embedded OS tissue sections were warmed in a 90°C dryer for 30 min. They were dewaxed in xylene for 15 min and washed with absolute ethyl alcohol and double distilled water with 0.3% hydrogen peroxide for 30 min to rinse the immunoperoxidase. Protein Block was used to incubate the tissue sections for 30 min to minimize nonspecific staining. CARM1 antibody was added to the tissues, followed by incubation at room temperature for 1 h. Counterstaining (ab128990; Abcam) was performed and samples were incubated for 30 min. A microscope assay was used to estimate the expression level of CARM1 according to the following scale: 0 (0-5%), 1 (6-25%), 2 (26-50%), 3 (51-75%), or 4 (>75%). Scores of 0 (0-5%) and 1 (6-25%) were considered negative, and scores of 2 (26-50%), 3 (51-75%), and 4 (>75%) were considered positive. For IHC staining score, 10 different optical fields were selected randomly. Each sample was evaluated by 3 different pathologists and a double-blind method was used.
Statistical analysis
All numerical data are presented as means ± standard error and figures were obtained using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). Data analyses were performed using SPSS 16.0 (IBM Corporation, New York, NY, USA). The relationship between CARM1 and Enneking stage was determined by Pearson's chi-squared test with continuity correction. P < 0.05 was considered statistically significant.
Conclusion
In this study, we first determined that CARM1 is overexpressed in OS cells and clinical tissue samples. CARM1 can promote proliferation and affect the cell cycle of OS cells. The novelty of this report is the analysis of the proximal mechanisms underlying the role of CARM1 in OS. Specifically, we found that CARM1 affects the phosphorylation of GSK3β to active β-catenin/cyclinD1 signaling. Many current gene therapy strategies are focused on beta-catenin/cyclinD1 signaling [40]. Accordingly, CARM1 is a potential new target for OS treatment.
Abbreviations
OS: osteosarcoma;
CARM1: Coactivator-associated arginine methyltransferase 1;
GSK3β: Glycogen synthase kinase-3 beta
qRT-PCR: Real-time Quantitative polymerase chain reaction
CCK8: Cell Counting Kit-8
IHC: Immunohistochemistry
Supplementary Material
Supplementary figures.
Ethics Committee Approval
This research was approved by the Ethics Committee of the Shanghai Jiao Tong University Affiliated Sixth People's Hospital (YS-2016-064, 24 February 2016).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gebhardt MC, Ready JE, Mankin HJ. Tumors about the knee in children. Clin Orthop Relat Res. 1990(255):86-110
2. Longhi A, Errani C, De Paolis M, Mercuri M, Bacci G. Primary bone osteosarcoma in the pediatric age: state of the art. Cancer Treat Rev. 2006;32:423-36
3. Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523-32
4. Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ. et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553-7
5. Franek M, Kovaříková A, Bártová E, Kozubek S. Nucleolar Reorganization Upon Site-Specific Double-Strand Break Induction: DNA Repair and Epigenetics of Ribosomal Genes. J Histochem Cytochem. 2016 Sep 28
6. Sanchez G, Bondy-Chorney E, Laframboise J, Paris G, Didillon A, Jasmin BJ. et al. A novel role for CARM1 in promoting nonsense-mediated mRNA decay: potential implications for spinal muscular atrophy. Nucleic Acids Res. 2016;44:2661-76
7. Wang L, Zhao Z, Meyer MB, Saha S, Yu M, Guo A. et al. CARM1 Methylates Chromatin Remodeling Factor BAF155 to Enhance Tumor Progression and Metastasis. Cancer Cell. 2014;25:21-36
8. Jangani M, Poolman TM, Matthews L, Yang N, Farrow SN, Berry A. et al. The Methyltransferase WBSCR22/Merm1 Enhances Glucocorticoid Receptor Function and Is Regulated in Lung Inflammation and Cancer. J Biol Chem. 2014;289:8931-8946
9. Wang X, Roberts CW. CARMA: CARM1 methylation of SWI/SNF in breast cancer. Cancer Cell. 2014;25:3-4
10. Kim YR, Lee BK, Park RY, Nguyen NT, Bae JA, Kwon DD. et al. Differential CARM1 expression in prostate and colorectal cancers. BMC Cancer. 2010;10:197
11. Cheng H1, Qin Y, Fan H, Su P, Zhang X, Zhang H. et al. Overexpression of CARM1 in breast cancer is correlated with poorly characterized clinicopathologic parameters and molecular subtypes. Diagn Pathol. 2013;8:129
12. An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735-48
13. Miao F, Li S, Chavez V, Lanting L, Natarajan R. Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated genetranscription through methylation of histone H3 at arginine 17. Mol Endocrinol. 2006;20:1562-73
14. EI Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D. et al. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci U S A. 2006;103:13351-6
15. Zhang X1, Chen L, Wang Y, Ding Y, Peng Z, Duan L. et al. Macrophage migration inhibitory factor promotes proliferation and neuronal differentiation of neural stem/precursor cells through Wnt/β-catenin signal pathway. Int J Biol Sci. 2013;9:1108-20
16. Nusse R. Wnt signaling. Cold Spring Harb Perspect Biol. 2012:4
17. Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898
18. Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781-810
19. Benham-Pyle BW, Sim JY, Hart KC, Pruitt BL, Nelson WJ. Increasing β-catenin/Wnt3A activity levels drive mechanical strain-induced cell cycle progression through mitosis. Elife. 2016;5:e19799
20. Cheng D, Côté J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71-83
21. Wang YP, Zhou W, Wang J, Huang X, Zuo Y, Wang TS. et al. Arginine Methylation of MDH1 by CARM1 Inhibits Glutamine Metabolism and Suppresses Pancreatic Cancer. Mol Cell. 2016;64:673-687
22. Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem. 2006;281:8476-8485
23. Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301-6
24. Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM. et al. Specific protein methylation defects and gene expression perturbations in coactivator-associatedarginine methyltransferase 1-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6464-8
25. Hong H1, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA. et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101:83-9
26. Sanders DA, Ross-Innes CS, Beraldi D, Carroll JS, Balasubramanian S. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol. 2013;14:R6
27. Yeom CG, Kim DI, Park MJ, Choi JH, Jeong J, Wi A. et al. Insulin-induced CARM1 upregulation facilitates hepatocyte proliferation. Biochem Biophys Res Commun. 2015;461:568-74
28. Elakoum R, Gauchotte G, Oussalah A, Wissler MP, Clément-Duchêne C, Vignaud JM. et al. CARM1 and PRMT1 are dysregulated in lung cancer without hierarchical features. Biochimie. 2014;97:210-8
29. Ou CY, LaBonte MJ, Manegold PC, So AY, Ianculescu I, Gerke DS. et al. A coactivator role of CARM1 in the dysregulation of β-catenin activity in colorectal cancer cell growth and gene expression. Mol Cancer Res. 2011;9:660-70
30. Boyle WJ, Smeal T, Defize LH, Angel P, Woodgett JR, Karin M. et al. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell. 1991;64:573-84
31. Pulverer BJ, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett JR. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59-70
32. Maqbool M, Mobashir M, Hoda N. Pivotal role of glycogen synthase kinase-3: A therapeutic target for Alzheimer's disease. Eur J Med Chem. 2016;107:63-81
33. Zhang X, Chen L, Wang Y, Ding Y, Peng Z, Duan L. et al. Macrophage migration inhibitory factor promotes proliferation and neuronal differentiation of neural stem/precursor cells through Wnt/β-catenin signal pathway. Int J Biol Sci. 2013;9:1108-20
34. Guo X, Zhang L, Zhang Y, Zhang D, Qin L, Dong S. et al. Oxysterol binding protein-related protein 8 inhibits gastric cancer growth through induction of ER stress, inhibition of Wnt signaling and activation of apoptosis. Oncol Res. 2016 Nov 8. [Epub ahead of print]
35. Isoda T, Nakatsu Y, Yamauchi K, Piao J, Yao T, Honda H. et al. Abnormality in Wnt signaling is causatively associated with oxidative stress-induced intestinal tumorigenesis in MUTYH-null mice. Int J Biol Sci. 2014;10:940-7
36. Goldstein SD, Trucco M, Bautista Guzman W, Hayashi M, Loeb DM. A monoclonal antibody against the Wnt signaling inhibitor dickkopf-1 inhibits osteosarcoma metastasis in a preclinical model. Oncotarget. 2016;7:21114-23
37. Cai Y, Cai T, Chen Y. Wnt pathway in osteosarcoma, from oncogenic to therapeutic. J Cell Biochem. 2014;115:625-31
38. Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199-221
39. MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms,and diseases. Dev Cell. 2009;17:9-26
40. Tai D, Wells K, Arcaroli J, Vanderbilt C, Aisner DL, Messersmith WA. et al. Targeting the WNT Signaling Pathway in Cancer Therapeutics. Oncologist. 2015;20:1189-98
41. Monica Mann, Valerie Cortez, Ratna Vadlamudi. PELP1 oncogenic functions involve CARM1 regulation. Carcinogenesis. 2013;34:1468-75
42. Sareddy GR, Zhang Q, Wang R, Scott E, Zou Y, O'Connor JC, Chen Y. et al. Proline-, glutamic acid-, and leucine- rich protein 1 mediates estrogen rapid signaling and neuroprotection in the brain. Proc Natl Acad Sci U S A. 2015;112:E6673-82
43. Cha B1, Kim W, Kim YK, Hwang BN, Park SY, Yoon JW. et al. Methylation by protein arginine methyltransferase 1 increases stability of Axin, a negative regulator of Wnt signaling. Oncogene. 2011;30:2379-89
Author contact
![]() Corresponding author: tjyqccom; Tel.: +86-21-64369181; Fax: +86-21-64701361
Corresponding author: tjyqccom; Tel.: +86-21-64369181; Fax: +86-21-64701361

 Global reach, higher impact
Global reach, higher impact