ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2017; 13(12):1470-1478. doi:10.7150/ijbs.21312 This issue Cite
Research Paper
CRISPR-offinder: a CRISPR guide RNA design and off-target searching tool for user-defined protospacer adjacent motif
1. Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education & Key Laboratory of Swine Genetics and Breeding of Ministry of Agriculture, College of Animal Science and Technology, Huazhong Agricultural University, Wuhan 430070, P. R. China;
2. The Cooperative Innovation Center for Sustainable Pig Production, Huazhong Agricultural University, Wuhan 430070, P. R. China;
3. The International Peace Maternity & Child Health Hospital of China Welfare Institute (IPMCH), Shanghai Jiao Tong University School of Medicine, Shanghai 200025, P. R. China;
4. Shanghai Key Laboratory of Reproductive Medicine, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, P. R. China;
5. iGeneTech Bioscience Co., Ltd., Beijing 102206, P. R. China.
* The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint first authors.
Abstract
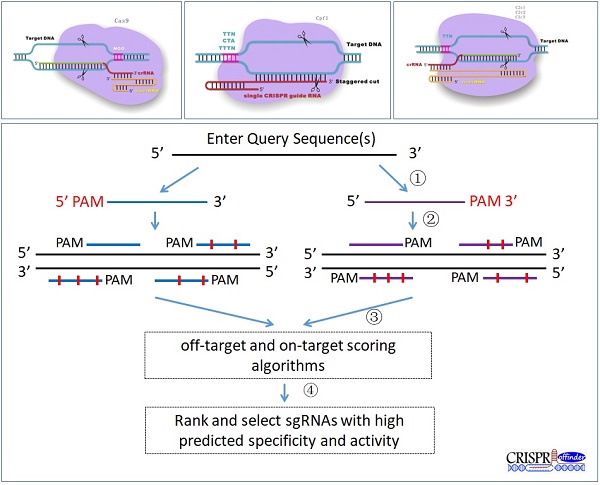
Designing efficient and specific CRISPR single-guide RNAs (sgRNAs) is vital for the successful application of CRISPR technology. Currently, a growing number of new RNA-guided endonucleases with a different protospacer adjacent motif (PAM) have been discovered, suggesting the necessity to develop a versatile tool for designing sgRNA to meet the requirement of different RNA-guided DNA endonucleases. Here, we report the development of a flexible sgRNA design program named “CRISPR-offinder”. Support for user-defined PAM and sgRNA length was provided to increase the targeting range and specificity. Additionally, evaluation of on- and off-target scoring algorithms was integrated into the CRISPR-offinder. The CRISPR-offinder has provided the bench biologist a rapid and efficient tool for identification of high quality target sites, and it is freely available at
Keywords: genome editing, user-defined protospacer adjacent motif, off-target.
