10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(12):1497-1506. doi:10.7150/ijbs.22531 This issue Cite
Review
Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity
Institute for Translational Medicine, Medical College of Qingdao University, Dengzhou Road 38, Qingdao 266021, China
Received 2017-8-25; Accepted 2017-10-8; Published 2017-11-2
Abstract
Circular RNAs (circRNAs), a novel type of non-coding RNAs (ncRNAs), are ubiquitously expressed in eukaryotic cells during post-transcriptional processes. Unlike linear RNAs, circRNAs form covalent-closed continuous loops without 5' to 3' polarities and poly (A) tails. With advances in high-throughput sequencing technology, numerous circRNAs have been identified in plants, animals and humans. Notably, circRNAs display cell-type, tissue-type and developmental-stage specific expression patterns in eukaryotic transcriptome, which reveals their significant regulatory functions in gene expression. More importantly, circRNAs serve as microRNA (miRNA) sponges and crucial regulators of gene expression. Additionally, circRNAs modulate pre-mRNA alternative splicing and possess protein-coding capacity. CircRNAs exhibit altered expression under pathological conditions and are strongly associated with the development of various human diseases. Interestingly, circRNAs can also induce antiviral immune responses. A recent study found that the delivery of circRNAs generated in vitro activates RIG-I-mediated innate immune responses and provides protection against viral infection. The antiviral dsRNA-binding proteins, NF90/NF110, act as key regulators in circRNA biogenesis. NF90/NF110 are also functional in inhibiting viral replication through binding to viral mRNAs. In this review, we provide a comprehensive overview on the classification, biogenesis and functions of circRNAs. We also discuss the critical role of circRNAs in eliciting antiviral immunity, providing evidence for the potential implications of circRNAs in antiviral therapies.
Keywords: circular RNA, microRNA sponge, antiviral immunity
Introduction
Studies of human genome demonstrate that protein-coding genes only occupy less than 2% of the entire genome [1]. However, it has been proven that more than 62% of genomic DNA serves as a template for transcription, which indicates that there are abundant non-coding RNAs (ncRNAs) in human transcriptome [2, 3]. ncRNAs play important roles in controlling gene expression at both transcriptional and post-transcriptional levels. ncRNAs can be classified into long ncRNAs (lncRNAs) and small ncRNAs (sncRNAs) [4]. LncRNAs with lengths greater than 200 nucleotides (nt) represent the largest part of mammalian non-coding transcriptome [5]. SncRNAs are less than 200 nt in length, consisting of microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), endogenous small interfering RNAs (endo-siRNAs or esiRNAs) and transcription initiation RNAs (tiRNAs) [6].
Circular RNAs (circRNAs), a novel member of the lncRNA kingdom, are generated by non-sequential backsplicing of exons, introns or a combination of both [7-9]. They are characterized by covalently closed loop feature without 5' end caps or 3' Poly (A) tails [10]. CircRNAs are highly stable forms of ncRNAs due to their nuclease resistance properties [11]. CircRNAs were first identified in RNA viruses in the 1970s [12-14]. However, circRNAs were initially regarded as viral genomes or byproducts of pre-mRNA alternative splicing, and thus they did not gain much attention for a long time [15]. Recent developments in high-throughput sequencing technologies and microarray techniques enable scientists to conduct comprehensive analyses on the abundance, composition, expression profile and molecular function of circRNAs [16]. So far, large amounts of circRNAs have been successfully identified in a variety of organisms including plants, animals and human beings [17]. Moreover, circRNAs are evolutionarily conserved across these species [18]. The widespread abundance of circRNAs in eukaryotic cells suggests that circRNAs are not simply accidental byproducts of mis-splicing events, but represent an indispensable part of lncRNA families [19].
The biological function of circRNAs has become a hotspot of scientific research in recent years. A growing body of evidence shows that circRNAs act as miRNA sponges and fulfill regulatory function in gene expression [20]. Remarkably, a variety of circRNAs are aberrantly expressed in specific disease contexts, suggesting their association with the occurrence and development of human diseases [21, 22]. For instance, Sand et al [23] investigated the expression of circRNAs in cutaneous squamous cell carcinoma (cSCC). They identified 322 differentially expressed circRNAs (143 up- and 179 down-regulated) in cSCC. Strikingly, there are 1,603 MREs (miRNA response elements) present in these differentially expressed circRNAs, which highlights that the involvement of circRNAs in cancer progression is strongly related to their interference with miRNA functions.
Another study in chronic thromboembolic pulmonary hypertension (CTEPH) indicates that 351 circRNAs (122 up- and 229 down-regulated) are differentially expressed between the control and CTEPH groups [24]. Among these circRNAs, hsa_circ_0022342 and hsa_circ_0002062 can regulate the activity of miRNAs that are crucial in CTEPH development. Therefore, hsa_circ_0022342 and hsa_circ_0002062 are considered as potential therapeutic targets for CTEPH treatment. Similarly, 183 circRNAs are upregulated and 64 are downregulated in type 2 diabetes mellitus (T2DM) patients with depressive symptoms compared with that in T2DM patients [25]. Several differentially expressed circRNAs are predicted to participate in the pathogenesis of depression in patients with T2DM. Due to their abundance, stability and evolutionary conservation, circRNAs have great potential as novel diagnostic biomarkers as well as therapeutic targets for various diseases [26-28].
Apart from their roles as miRNA sponges, circRNAs perform multiple functions in cellular processes including regulation of alternative splicing and gene expression, scaffolds for the assembly of protein complexes, templates for translation, and modulators of rRNA and tRNA biogenesis. Interestingly, two research groups report the most recent progress on circRNAs and reveal a novel function of circRNAs, linking circRNAs to host-virus interaction. The recognition of circRNAs by the nucleic acid sensor RIG-I is essential for the innate immune signaling [29]. This recognition activates the innate immune responses and provides protection against viral infection. The immune factors NF90/NF110 modulate circRNA biosynthesis and suppress viral infection by interacting with viral mRNAs [30]. Taken together, these findings confirm the intervention of circRNAs in immune regulation and viral infection. CircRNAs can be used to boost immune activation for antiviral therapeutic purposes. However, several intriguing questions require to be clarified: How do circRNAs escape immune surveillance? How do the cellular immune factors interact with circRNAs? How do circRNAs contribute to antiviral immune responses? Therefore, in this review, we summarize the classification, biogenesis and function of circRNAs, and emphasize their implications in antiviral immunity.
Classification and Biosynthesis of circRNAs
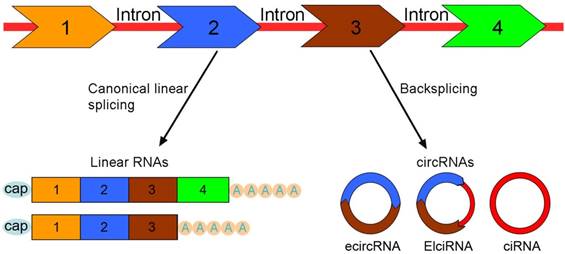
CircRNAs arise from exons, introns, intergenic or untranslational regions by a non-canonical splicing process termed 'backsplicing' [31, 32]. The canonical splicing machinery can control pre-mRNA backsplicing process [33]. The mutations at splice sites that bracket circRNA-forming exons prevent exon circularization, which indicates that the efficiency of exon circularization relies on the presence of canonical splice sites flanking the exons [22]. CircRNAs are mainly divided into three groups based on their components: 1) exonic circRNAs (ecircRNAs) [34]; 2) ElciRNAs or retained-intron circRNAs [35]; 3) circular intronic RNAs (ciRNAs) [36] (Figure 1).
EcircRNAs, consisting of one or more exons, make up the vast majority (over 80%) of circRNAs. EcircRNAs are synthesized by non-sequential backsplicing in which a downstream splice donor of pre-mRNA covalently joins with an upstream splice acceptor [18, 37]. Exons of single-exon circRNAs are much longer than exons in multiple-exon circRNAs [38]. Exons with the length greater than 300 nt are more likely to form circRNAs than short exons, which demonstrates that exon length affects the efficiency of backsplicing [39].
Two potential models of ecircRNA generation, “lariat-driven circularization” and “intron-pairing-driven circularization”, were previously proposed [40]. The former is related to exon skipping, in which the (3') splice acceptor site of an upstream exon is ligated to the (5') splice donor site of a downstream exon and an RNA lariat containing one or more skipped exons is formed. The intronic sequences in the lariat are then removed, resulting in the generation of a mature ecircRNA. The latter is mainly dependent on reverse complementary motifs in introns. Two introns that flank alternatively spliced exons within the nascent RNA transcript form a circular structure by complementary base-pairing. These intronic sequences are subsequently trimmed to generate an ecircRNA.
The categories of circRNAs. Pre-mRNA can be processed through either canonical splicing to produce linear RNAs (left) or backsplicing to generate circRNAs (right). CircRNAs are mainly divided into three categories based on their components. Exonic circRNAs (ecircRNAs) are exclusively composed of exons and represent the largest group of circRNAs. ElciRNAs retain intronic sequences between the back-spliced exons (at least two) and predominantly exist in the nucleus. Circular intronic RNAs (ciRNAs) are generated only from intron lariats and constitute a small class of circRNAs.
The introns bordering circularized exons usually contain reverse complementary Alu repeats, which is essential for intron-pairing-driven circularization [41, 42]. The RNA-editing enzyme, ADAR1, can restrain circRNA biogenesis. ADAR1 binds to dsRNA and catalyzes the conversion of adenosine (A) to inosine (I) [43]. ADAR1 destabilizes the Alu repeat sequences within the introns via mediating RNA editing in Alu regions, which leads to the inhibition of circRNA biogenesis [44]. A genome-wide analysis indicates that introns bracketing circularized exons are much longer than those flanking exons processed by linear splicing [45]. However, in some circumstances, introns between the encircled exons are not spliced out, which results in the formation of retained-intron circRNAs or ElciRNAs [46].
CiRNAs are generated from intron lariats processed by spliceosome-mediated splicing [47]. The intron lariats that escape debranching and degradation after splicing are circularized at the branchpoint 2'-5' linkage [48]. The 3' tail downstream from the branchpoint is removed to produce stable ciRNAs [49]. Recent studies indicate that circRNAs can be synthesized by RNA-binding proteins (RBPs)-mediated circularization [50, 51]. RBPs bring the introns flanking circRNA-forming exons into close proximity to favor exon circularization.
Biological functions of circRNAs
CircRNAs act as novel players in human diseases by sponging miRNAs
Emerging evidence suggests that the biological functions of circRNAs include molecular sponges for miRNAs or RBPs, regulation of alternative splicing and gene expression, templates for translation and probably other unknown roles [46, 51-53]. Notably, the property of circRNAs as miRNA sponges has been extensively studied. miRNAs are a class of small RNAs that negatively regulate the expression of target genes [54]. miRNAs destabilize target mRNAs by binding to the 3' UTRs of mRNAs and interfere with their translation. miRNAs are involved in human diseases and are potential predictive biomarkers and therapeutic targets for many diseases [55-57]. CircRNAs can function as molecular sponges to absorb and sequester miRNAs, and thus they can counteract miRNA-mediated degradation of mRNAs [33]. Based on these clues, circRNAs may serve significant roles in the initiation and progression of human diseases such as cancers, neurological disorders and cardiovascular diseases by acting as miRNA sponges.
The first identified miRNA sponge is ciRS-7, which is generated from the cerebellum degeneration-related protein 1 antisense (CDR1) [58, 59]. CiRS-7 contains over 70 miR-7 binding sites, which enables it acting as an endogenous miR-7 inhibitor and weakening the regulatory effects of miR-7 on target mRNAs. The ciRS-7/miR-7 interaction is highly associated with the development of various cancers including neuroblastoma, astrocytoma, lung carcinoma and cervical cancer [60]. The epidermal growth factor receptor (EGFR), a target of miR-7, is a key factor in the development of multiple cancers [61-63]. Kruppel-like factor 4 (KLF4) is another target of miR-7, which exhibits an inhibitory effect on the growth of cancer cells [64]. CiRS-7 can quench miRNA-7 function and upregulates the expression of EGFR and KLF4, thus promoting cancer development [65]. miR-7 is involved in the pathogenesis of Parkinson's disease by regulating the expression of α-synuclein [66]. In dendrites, ciRS-7 co-localizes with miR-7, which indicates that ciRS-7 could be an important regulator in the development of neurological disorders [22]. A decreased expression of ciRS-7 in the hippocampus of Alzheimer's patients indicates that the deficiency of ciRS-7/miRNA-7 leads to the progression of Alzheimer's disease [67]. Memczak et al. [8] found that upregulation of ciRS-7 or knockdown of miR-7 impairs the midbrain development in zebrafish.
The characterization of ciRS-7 sheds light on the miRNA sponge function of circRNAs. Testis-specific circRNA, Sry (sex-determining region Y), is abundantly present in the testis of adult mouse [52]. Sry circRNA harbors 16 binding sites for miR-138 and influences the expression of miRNA-138 target genes. miR-138 can control the expression of genes related to the initiation, invasion and metastasis of cancers [20]. Therefore, the Sry circRNA/miRNA-138 pathway can be delved for the purpose of cancer diagnosis and therapy.
CircRNA-MYLK is identified as a competitive endogenous RNA molecule (ceRNA) for miR-29a [68]. CircRNA-MYLK can promote the development of bladder cancer via relieving miR-29a-mediated suppression of VEGFA/VEGFR2 signaling pathway. Cir-ITCH (E3 ubiquitin protein ligase) inhibits the regulatory effect of miR-7, miR-17 and miR-214 on target genes involved in Wnt/β-catenin signaling pathway and blocks the growth of esophageal squamous cell carcinoma (ESCC) [69]. Huang et al. [70] showed that the level of cir-ITCH is extremely lower in colorectal cancer (CRC) than that of in the peritumoral tissue. In fact, cir-ITCH has the capability to inhibit the Wnt/β-catenin pathway by elevating the level of ITCH, which suppresses the proliferation of CRC cells. Hsa_circ_0013958 promotes the progression of lung adenocarcinoma by antagonizing the inhibitory effect of miR-134 on the expression of oncogenic cyclin D1 [71]. Similarly, circRNA_100269 regulates the proliferation of gastric cancer cells through sponging its target miRNA (miR-630) [72]. CircTCF25 sequesters miR-103a-3p and miR-107, and enhances the expression of CDK6 [73]. Consequently, circTCF25 can promote the proliferation, migration and metastasis of bladder cancer cells. A recent study found that hsa_circ_001569 is strongly associated with the development of colorectal cancer [74]. Hsa_circ_001569 upregulates the expression of miR-145 target proteins (E2F5, BAG4 and FMNL2), which contributes to the proliferation and invasion of colorectal cancer cells.
Several circRNAs can specifically capture the miRNAs associated with cardiovascular diseases. Heart-related circRNA (HRCR) increases the expression level of ARC by acting as a miR-223 sponge [75]. The upregulation of HRCR protects the heart from pathological hypertrophy. A novel circRNA, circRNA_010567, negatively regulates the activity of miR-141 [76]. The circRNA_010567/miR-141/TGF-β1 axis plays a regulatory role in the pathogenesis of myocardial fibrosis in the diabetic mice. The mitochondrial fission and apoptosis-related circRNA (MFACR) modulates mitochondrial fission and cardiomyocyte apoptosis by suppressing miR-652-3p and upregulating its target MTP18 [77]. CircRNA_000203, derived from mouse Myo9a transcript, is highly active in diabetic mouse myocardium and Ang-II-induced cardiac fibroblasts [78]. CircRNA_000203 selectively sponges miR-26b-5p and blocks its anti-fibrotic effect in cardiac fibroblasts, leading to increased proliferation of cardiac fibroblasts.
The clinical studies reveal the critical role of circRNAs in the development of human diseases. The levels of circ-FBXW7 and its translated protein FBXW7-185aa are decreased in glioblastoma tissues compared with adjacent non-tumorous tissues [79]. Circ-FBXW7 expression is positively linked with the overall survival of glioblastoma patients. Therefore, circ-FBXW7 might be a clinical indicator for glioblastoma prognosis. In gastric cancer (GC) tissues, circRNA_0026 exhibits a significantly differential expression pattern when compared to adjacent normal tissues [80]. CircRNA_0026 is a potentially valuable biomarker for GC diagnosis and therapy.
Another study indicates that circRNA_100338 is highly expressed in patients with hepatocellular carcinoma (HCC) [81]. CircRNA_100338 regulates cell metastasis in liver cancer by acting as an endogenous sponge for miR-141-3p. Accordingly, circRNA_100338 is closely associated with decreased survival time and tumor metastasis in HCC patients. These findings suggest that the circRNA_100338/miR-141-3p pathway is involved in the progression of HCC. In rheumatoid arthritis (RA) patients, the expression levels of circRNA_003524, circRNA_101873, circRNA_103047 and circRNA_104871 are significantly increased, and these circRNAs could serve as potential biomarkers for RA diagnosis [82]. Likewise, a list of circRNAs is significantly upregulated in patients with type 2 diabetes mellitus with proliferative diabetic retinopathy (T2DR) [83]. These circRNAs can be developed into novel targets for T2DR therapy.
Collectively, these studies indicate that circRNAs can sequester miRNAs and block them from acting on the protein-coding mRNAs. The circRNA/miRNA/mRNA axis plays a crucial role in the occurrence and progression of human diseases. CircRNAs possess great potentials as diagnostic biomarkers and therapeutic targets for many diseases. However, there is still a long way to go for the clinical application of circRNAs. For example, the molecular mechanisms underlying the specific interactions among circRNA, miRNA and its target mRNA during the progression of diseases remains to be comprehensively elucidated. In addition, a detailed investigation is essential to delineate the regulation of circRNA biosynthesis and degradation during the initiation and development of pathological disorders. Furthermore, it is important to clearly understand the biological functions of circRNAs under physiological as well as pathological conditions.
CircRNAs regulate the assembly and transport of cellular proteins
CircRNAs act as scaffolds for the assembly of protein complexes [84]. For instance, Foxo3 circular RNA (circ-Foxo3) serves as a scaffold to enable the interaction between cyclin-dependent kinase 2 (CDK2) and CDK inhibitor p21 [85]. The circ-Foxo3/CDK2/p21 complex blocks the cell cycle progression in response to cell overgrowth. CircRNAs also have the capability to sequester cellular proteins from their native subcellular localization [84]. Circ-Foxo3 mainly presents in the cytoplasm [86]. The cytoplasmic circ-Foxo3 binds to inhibitor of differentiation-1 (ID-1), the transcription factor E2F1, hypoxia inducible factor-1α (HIF-1α) and focal adhesion kinase (FAK), and inhibits their translocation, leading to their retention in the cytoplasm. In addition, it has been speculated that circRNAs can function as RBP reservoirs and transporters to deliver RBPs to the specific subcellular locations [26].
CircRNAs regulate alternative splicing and gene expression
The biogenesis of circRNAs negatively affects the efficiency of pre-mRNA alternative splicing and exerts regulatory effects on gene expression [87]. CircRNAs are known to be produced by scrambling of exons during pre-mRNA splicing. After backsplicing, the internal exons are excised to form a circRNA, leaving the remaining pre-mRNA spliced in an alternative pattern [48]. CircRNAs are generally synthesized at the expense of their linear equivalents [88]. Recent studies show that there are competitions between backsplicing and linear splicing due to overlapping dependence on the spliceosomal machinery [89, 90]. As a result, the biogenesis of circRNAs causes a decrease in the level of protein-coding mRNAs. The regulatory mechanisms underlying the choice of pre-mRNA splicing remain unclear and are worthy to be intensively studied.
CircRNAs inhibit parental gene expression through competing with their linear counterparts [51]. A circRNA (circMbl) produced from the second exon of the splicing factor muscleblind (MBL) harbors conserved MBL binding sites in its flanking introns [91]. The binding of MBL to the flanking introns is essential for the circularization of its own second exon, thus promoting the production of circMbl [92]. If MBL protein level is in excess, its mRNA is downregulated by increasing the production of circMbl. Therefore, circMbl functions as MBL sponges to modulate the translation of MBL. CircRNAs also suppress parental gene transcription by occupying RNA binding sites in target genes. SEPALLATA3 (SEP3) exon 6 circRNA strongly binds to its cognate DNA locus and blocks the binding of its linear isoform to the cognate DNA [93]. The formation of RNA: DNA hybrid or R-loop results in the termination of SEP3 gene transcription.
Interestingly, circRNAs can promote the expression of their parental genes in some circumstances. For example, knockdown of circRNA EIF3J and circRNA PAIP2 remarkably reduces the expression level of their parental genes [46]. Two abundantly expressed ciRNAs, ci-ankrd52 and ci-sirt7, are able to enhance the transcription of their parental genes through binding to RNA Pol II complex [94]. Furthermore, EIciRNAs, a class of regulatory circRNAs, binds to RNA Pol II via interacting with U1 small nuclear ribonucleoprotein (snRNP). The interaction between EIciRNAs and RNA Pol II leads to an increase in the mRNA level of their parental genes [95].
It has been found that RBPs are capable of regulating post-transcriptional processes of gene expression [96]. CircRNAs can prevent RBPs binding to the corresponding mRNAs and inhibit the translation of mRNAs. Human antigen R (HuR), an mRNA binding protein, has been reported to regulate the gene expression by interacting with coding and non-coding linear RNAs [97]. The binding of circPABPN1 to HuR blocks its interaction with PABPN1 transcript and thus suppresses PABPN1 translation [98].
CircRNAs possess protein-coding activity
It is well known that circRNAs are mainly produced from exons, which suggests that circRNAs can be potentially translated into proteins. Many research groups found evidence to support this notion. A circRNA, circ-ZNF609, contains a 753-nt open reading frame (ORF) [99]. This circRNA can be translated into functional proteins in a cap-independent and splicing-dependent manner. The translation of a circRNA derived from the muscleblind locus provides strong evidence for the protein-coding capability of circRNAs [100].
In eukaryotes, transcripts can be translated through a cap-independent mechanism [101]. The presence of internal ribosomal entry sites (IRES) is important for the initiation of cap-independent translation [102]. Has_circ_0041407 contains an IRES element as well as protein-coding fragments of MAX network transcriptional repressor (MNT) [103]. Moreover, has_circ_0041407 can be translated into a 31-kDa chimeric protein, which suggests that the IRES sequences drive circRNA translation. Some natural circRNAs can interact with cap-independent translation factors such as eukaryotic initiation factor 3 (eIF3) and N6-Methyladenosine (m6A), suggesting the translational potentials of these circRNAs [104].
CircRNAs modulate generation of rRNAs and tRNAs
The circular form of antisense non-coding RNA in the INK4 locus (circANRIL) can bind to crucial 60S-preribosomal assembly factor, pescadillo ribosomal biogenesis factor 1 (PES1), and blocks ribosomal RNA (rRNA) maturation [105]. In some archaea, circRNAs act as midbodies during rRNA processing. rRNA precursors initially form circular shaped intermediates by cleavage and linkage [106]. The circRNA intermediates are then cleaved into 16S and 23S rRNAs [107]. CircRNAs are also important intermediates in the biogenesis of transfer RNAs (tRNAs). CircRNA intermediates are generated to inverse the order of the 5'- and 3'- regions of permuted tRNAs [108, 109].
CircRNAs are involved in antiviral immunity
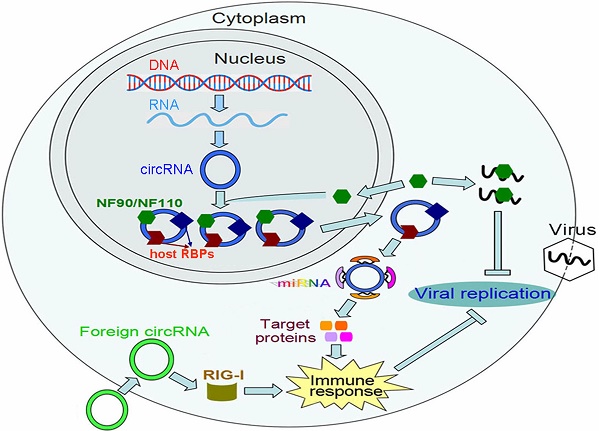
The dsRNA-binding proteins, such as retinoic acid-inducible gene-I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), laboratory of genetics and physiology 2 (LGP2) and Toll-like receptor 3 (TLR3), are key factors in the host antiviral defense system [110, 111]. These proteins sequester viral RNAs to antiviral effector molecules, which consequently leads to the elicitation of antiviral immune responses. Recently, Chen et al. [29] uncovered a novel function of circRNAs in evoking antiviral immune responses. They found that the delivery of purified circRNAs activates RIG-I and confers effective immune protection against viral infection (Figure 2). Further exploration indicated that RIG-I is activated by circRNAs made with self-splicing introns instead of the same circRNAs created with endogenous introns and spliced by cellular splicing machinery. Thus, the activation of immune responses depends on the splicing mechanism of circRNAs. In host cells, a set of diverse RBPs bind to the cellular endogenous circRNAs. In contrast, few proteins interact with 'non-self' circRNAs. The RBPs serve to mark 'self' circRNAs and allow the host cells to distinguish 'self' circRNAs from foreign circRNAs.
RIG-I is known as a dsRNA-binding protein [112]. The endogenous circRNAs are able to evade RIG-I-mediated immune recognition, suggesting that circRNAs possess unknown RIG-I-regulatory elements. This finding also raises several interesting questions. Further researches are needed to find out whether the interaction between specific RBPs and 'self' circRNAs is essential to escape RIG-I recognition. The molecular mechanisms by which RBPs protect 'self' circRNAs from RIG-I sequestration remain to be clarified. The linearization of circRNAs greatly reduces their immune stimulatory activities, which indicates that the circular property of RNAs is vital for the host immune responses. It is intriguing how circRNAs interact with RIG-I despite without 5' or 3' ends.
In a previous study, circRNA sequencing was performed to analyze circRNA expression in avian leukosis virus subgroup (ALV-J) resistant and susceptible chickens [113]. As a result, 12 differentially expressed circRNAs are identified to be upregulated in ALV-J resistant chickens. The miRNA target genes of these highly expressed circRNAs are mainly implicated in immune pathways such as modulation of B cell activation and antigen-presentation process. Grass carp reovirus (GCRV) is a major pathogen that contributes to severe hemorrhagic diseases in grass carp [114]. The role of circRNAs in host-GCRV interactions was explored by applying deep circRNA sequencing [115]. A total of 41 differentially expressed circRNAs with their binding miRNAs are identified in grass carp following GCRV infection. Moreover, the target genes of these miRNAs serve important functions in immune responses, hemostasis, and coagulation and complement cascades. Collectively, the circRNA/miRNA/mRNA network can regulate host immune function and thus provide efficient protection against viral infection.
Another investigation reveals that circRasGEF1B is conserved between human and mouse, and acts as a positive controller of lipopolysaccharide (LPS) response [116]. Notably, circRasGEF1B can regulate the abundance of the intercellular inflammatory adhesion molecule ICAM-1 by modulating the stability of ICAM-1 mRNA. ICAM-1 is essential for the recruitment of leukocytes to inflamed sites and thus plays a critical role in viral pathogenesis [117, 118]. CircRasGEF1B may be instrumental in spurring innate immune responses and protecting host cells against pathogen invasion. Although there is currently limited information about the regulatory role of circRNAs in the host immune responses, we speculate that circRNAs exert multiple functions in immune regulation. Additional investigations are required to throw light on the abundance and functions of circRNAs in the host immune system.
The functional role of circRNAs in antiviral immunity. The immune factors NF90/NF110 promote circRNA biogenesis via interacting with the introns flanking circRNA-forming exons. Upon viral infection, NF90/NF110 are released from the circRNPs and exported from the nucleus to the cytoplasm, which leads to a decreased level of circRNAs. The released NF90/NF110 suppress viral replication through binding to viral mRNAs. In host cells, endogenous circRNAs generally bind to RNA-binding proteins (RBPs). These RBPs serve to mark 'self' circRNAs, which allows host cells to distinguish 'self' circRNAs from foreign circRNAs. Foreign circRNAs can activate RIG-I-mediated innate immune responses, thereby protecting host cells from viral infection. CircRNAs can also act as miRNA sponges and upregulate the expression of target proteins that are involved in immune-related pathways.
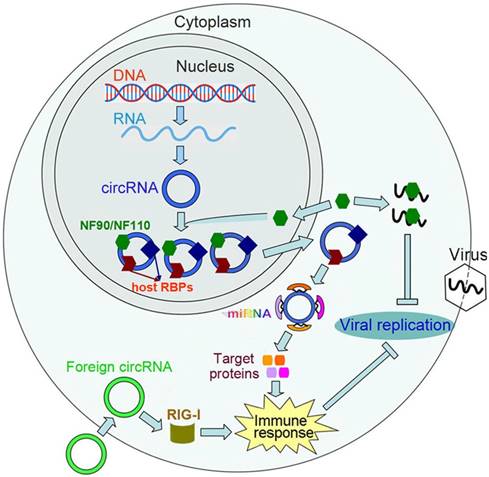
The circRNA biogenesis can be influenced by the host immune system as well as viral infection [30]. The immune factors, NF90 and NF110 (NF90/NF110), play important roles in host immune defense against viral infection [119]. NF90/NF110 bind to inverted repeat regions in the introns flanking the encircled exons and promote pre-mRNA backsplicing by stabilizing intronic RNA hairpin structures. Upon viral infection, NF90/NF110 are rapidly released from circRNPs and exported from the nucleus to the cytoplasm. As a result, the abundance of NF90/NF110 in the nucleus is significantly reduced. The cytosolic retention of NF90/NF110 may attribute to PRK-mediated inhibition of nuclear import or capture of NF90/NF110 by viral mRNAs in the cytoplasm. The released NF90/NF110 can inhibit viral proliferation by binding to viral mRNAs. The nuclear export of NF90/NF110 also leads to a decreased level of circRNAs in the nucleus. These findings provide important clues for the interplay between circRNAs and the cellular immune factors.
In fact, NF90/NF110 exhibit higher affinities for circRNAs than their linear equivalents. However, the specific mechanism by which these immune factors regulate circRNA biogenesis remains to be illustrated. In addition, more detailed studies are required to identify the key structural elements within circRNAs that recruit cellular antiviral immune factors. The impacts of circRNAs on the function of the cellular immune factors also deserve deep exploration. It is certain that more in-depth investigations on the interaction between circRNAs and the host immune system will enrich our knowledge of circRNA biology.
Given the fact that circRNAs were first found in RNA viruses [120], they can be used as templates for viroid or virus replication. In viruses, circRNAs are highly homologous. Therefore, study on circRNAs could provide insights into the origin and evolution of viruses [121]. Viruses are known to exclusively depend on the host cellular machinery to ensure their survival [122]. Viruses have evolved to regulate the cellular environment such as miRNA-mediated gene silence [123]. In this background, circRNAs may function in a pro-viral pattern. Viruses could take advantages of host circRNAs to enhance expression of the cellular or viral proteins that are essential for viral entry, replication and pathogenesis.
The cellular miRNAs play an important part in host-virus interactions by targeting crucial genes involved in immune regulation and viral pathogenesis [124-126]. It has been reported that viruses are capable of exploiting cellular miRNAs to fit their requirements [127]. In virus-infected cells, circRNAs may exert inhibitory functions in viral infection by suppressing the activities of cellular miRNAs. Likewise, the viral miRNAs take part in the development of viral diseases by modulating the cellular processes including apoptosis-related and anti-tumor pathways [128, 129]. In some cases, virus-encoded miRNAs inhibit the expression of viral genes and thus protect virus-infected cells from the host immune surveillance [130]. CircRNAs may effectively induce antiviral immune responses via offsetting the effects of viral miRNAs on target gene expression. Thus, suppressing the function of the cellular or viral miRNAs by circRNAs might represent a promising therapeutic strategy against viral infection.
There are several useful computational resources available for the study of circRNA interactions and functions. A comprehensive database named 'HumanViCe' provides a repository of potential circRNAs that can sponge human or viral miRNAs in virus-infected cells [131]. In HumanViCe, a large number of circRNAs are predicted to contain multiple binding sites for viral miRNAs. For example, a human circRNA, hsa_circ_002048, derived from ANKRD11 transcript harbors 36 binding sites for EBV-encoded miR-BART20-5p. The function analyses of these circRNAs demonstrate that they mainly participate in the cellular pathways that are associated with viral entry, replication as well as virulence. Therefore, HumanViCe serves as a vital resource for further investigation into the function of host circRNAs in viral infection. A web tool, CircInteractome (circRNA interactome), enables scientists to identify putative circRNAs that can interact with miRNAs or RBPs [103]. CircInteractome also provides detailed information about circRNAs and their potential roles in sponging miRNAs or RBPs. It has been proven that some RBPs such as Muscleblind [51] and Quaking [50] are key regulators in circRNA biogenesis. More importantly, RBPs can function as antiviral effectors by triggering host immune responses [132]. An in-depth understanding of the interplay between circRNAs and RBPs would accelerate our efforts to uncover the functional roles of circRNAs in antiviral immunity.
Conclusions and perspectives
CircRNAs, a major class of ncRNAs, were previously regarded as transcriptional byproducts or errors. The recent advancement in high-throughput sequencing reveals that circRNAs are universally present in many species. The biological functions of circRNAs have gained great attention in recent years. Intriguingly, circRNAs can act as miRNA sponges, gene expression modulators, platforms for the assembly of protein complexes, templates for translation, and regulators of rRNA and tRNA biogenesis. Recently, the involvement of circRNAs in the induction of antiviral immunity has been disclosed. CircRNAs can regulate the expression of genes involved in innate immunity and confer protection against viral infection. In addition, cellular immune factors act as key regulators in circRNA biogenesis and are capable of inhibiting viral infection. These findings offer the latest addition to the ever-growing list of circRNA functions.
However, the current understandings and achievements in the study of circRNAs are still limited. There are a large number of circRNAs with unknown functions present in eukaryotic transcriptome. In circRNA biogenesis, the backsplicing process of pre-mRNA is generally coupled with alternative splicing. It is yet unclear how does the splicing machinery select either alternative splicing or backsplicing to generate a circular RNA. The biogenesis and degradation machineries of circRNAs determine their abundance in specific subcellular localization and thus they are responsible for the biological functions of circRNAs in certain cases. Therefore, the clear understanding of underlying mechanisms of circRNA biogenesis and degradation is important. CircRNAs serve as RBP sponges and reservoirs to regulate the abundance of target proteins. However, the complicated interactions between circRNAs and RBPs also need more systematic survey. At present, the functional role of circRNAs in antiviral immunity is poorly understood. Further investigations are warranted to fully elucidate the complex interplays between circRNAs and the host immune system in antiviral defense responses. Continued studies on the implications of circRNAs in antiviral immunity will not only increase our knowledge about the physiological function of circRNAs, but it could also provide novel therapeutic strategies against viral diseases.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81701991, 31430041 and 81230005), Applied Basic Research Programs of Qingdao, China (17-1-1-59-jch) and the China Postdoctoral Science Foundation Funded Project (2016M600520).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860-921
2. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N. et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559-63
3. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74
4. Awan HM, Shah A, Rashid F, Shan G. Primate-specific Long Non-coding RNAs and MicroRNAs. Genomics Proteomics Bioinformatics. 2017;15:187-95
5. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
6. Kota SK, Kota SB. Noncoding RNA and epigenetic gene regulation in renal diseases. Drug Discov Today. 2017;22:1112-22
7. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH. et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103-11
8. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
9. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453-61
10. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233-47
11. Jin X, Feng CY, Xiang Z, Chen YP, Li YM. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7:66455-67
12. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409
13. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
14. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852-6
15. Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 2016:1-7
16. Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242:1136-41
17. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777
18. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838-47
19. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733
20. Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514-21
21. Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R. et al. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci. 2017;4:38
22. Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53:359-65
23. Sand M, Bechara FG, Gambichler T, Sand D, Bromba M, Hahn SA. et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83:210-8
24. Miao R, Wang Y, Wan J, Leng D, Gong J, Li J. et al. Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine (Baltimore). 2017;96:e7354
25. Jiang G, Ma Y, An T, Pan Y, Mo F, Zhao D. et al. Relationships of circular RNA with diabetes and depression. Sci Rep. 2017;7:7285
26. Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6:1167-76
27. Lu D, Xu AD. Mini Review: Circular RNAs as Potential Clinical Biomarkers for Disorders in the Central Nervous System. Front Genet. 2016;7:53
28. Hou X, Wen J, Ren Z, Zhang G. Non-coding RNAs: new biomarkers and therapeutic targets for esophageal cancer. Oncotarget. 2017;8:43571-8
29. Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE. et al. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol Cell. 2017;67:228-38 e5
30. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF. et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67:214-27 e7
31. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
32. Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J. et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2016:1-8
33. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42-51
34. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563-79
35. Shen T, Han M, Wei G, Ni T. An intriguing RNA species-perspectives of circularized RNA. Protein Cell. 2015;6:871-80
36. Yang L. Splicing noncoding RNAs from the inside out. Wiley Interdiscip Rev RNA. 2015;6:651-60
37. Dong Y, He D, Peng Z, Peng W, Shi W, Wang J. et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10:2
38. Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C. et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126
39. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540
40. Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in Cardiovascular Disease: An Overview. Biomed Res Int. 2017;2017:5135781
41. Daniel C, Behm M, Ohman M. The role of Alu elements in the cis-regulation of RNA processing. Cell Mol Life Sci. 2015;72:4063-76
42. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170-7
43. Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321-49
44. Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S. et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870-85
45. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
46. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
47. Wu H, Yang L, Chen LL. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017;33:540-52
48. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829-42
49. Li TR, Jia YJ, Wang Q, Shao XQ, Lv RJ. Circular RNA: a new star in neurological diseases. Int J Neurosci. 2017;127:726-34
50. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-34
51. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
52. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
53. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172-9
54. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
55. Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515-24
56. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460-9
57. Olson EN. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci Transl Med. 2014;6:239ps3
58. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-12
59. Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep. 2015;33:2669-74
60. Zheng XB, Zhang M, Xu MQ. Detection and characterization of ciRS-7: a potential promoter of the development of cancer. Neoplasma. 2017;64:321-8
61. Ribeiro FA, Noguti J, Oshima CT, Ribeiro DA. Effective targeting of the epidermal growth factor receptor (EGFR) for treating oral cancer: a promising approach. Anticancer Res. 2014;34:1547-52
62. Stock AM, Hahn SA, Troost G, Niggemann B, Zanker KS, Entschladen F. Induction of pancreatic cancer cell migration by an autocrine epidermal growth factor receptor activation. Exp Cell Res. 2014;326:307-14
63. Tomao F, Di Tucci C, Imperiale L, Boccia SM, Marchetti C, Palaia I. et al. Cervical cancer: are there potential new targets? An update on preclinical and clinical results. Curr Drug Targets. 2014;15:1107-20
64. Yang WT, Zheng PS. Promoter hypermethylation of KLF4 inactivates its tumor suppressor function in cervical carcinogenesis. PLoS One. 2014;9:e88827
65. Hou LD, Zhang J. Circular RNAs: An emerging type of RNA in cancer. Int J Immunopathol Pharmacol. 2017;30:1-6
66. Recasens A, Perier C, Sue CM. Role of microRNAs in the Regulation of alpha-Synuclein Expression: A Systematic Review. Front Mol Neurosci. 2016;9:128
67. Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet. 2013;4:307
68. Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L. et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305-17
69. Li F, Zhang L, Li W, Deng J, Zheng J, An M. et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001-13
70. Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225
71. Zhu X, Wang X, Wei S, Chen Y, Fan X, Han S. et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284:2170-82
72. Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z. et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9:1585-94
73. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919
74. Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y. et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680-91
75. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B. et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602-11
76. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun. 2017;487:769-75
77. Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN. et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111-20
78. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z. et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. 2017;7:40342
79. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F. et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2018:110
80. Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16:2469-76
81. Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428
82. Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A. et al. Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Rheumatoid Arthritis Patients. Cell Physiol Biochem. 2017;42:651-9
83. Gu Y, Ke G, Wang L, Zhou E, Zhu K, Wei Y. Altered Expression Profile of Circular RNAs in the Serum of Patients with Diabetic Retinopathy Revealed by Microarray. Ophthalmic Res. 2017;58:176-84
84. Fischer JW, Leung AK. CircRNAs: a regulator of cellular stress. Crit Rev Biochem Mol Biol. 2017;52:220-33
85. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846-58
86. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402-12
87. Taborda MI, Ramirez S, Bernal G. Circular RNAs in colorectal cancer: Possible roles in regulation of cancer cells. World J Gastrointest Oncol. 2017;9:62-9
88. Kelly S, Greenman C, Cook PR, Papantonis A. Exon Skipping Is Correlated with Exon Circularization. J Mol Biol. 2015;427:2414-7
89. Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907
90. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381-8
91. Vicens Q, Westhof E. Biogenesis of Circular RNAs. Cell. 2014;159:13-4
92. Chen L, Huang C, Wang X, Shan G. Circular RNAs in Eukaryotic Cells. Curr Genomics. 2015;16:312-8
93. Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G. et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053
94. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-11
95. Eidem TM, Kugel JF, Goodrich JA. Noncoding RNAs: Regulators of the Mammalian Transcription Machinery. J Mol Biol. 2016;428:2652-9
96. Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5'- and 3'-UTR-binding factors. Trends Biochem Sci. 2003;28:182-8
97. Poria DK, Guha A, Nandi I, Ray PS. RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene. 2016;35:1703-15
98. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361-9
99. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37 e9
100. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L. et al. Translation of CircRNAs. Mol Cell. 2017;66:9-21 e7
101. Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859:1245-51
102. Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229-40
103. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34-42
104. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-41
105. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429
106. Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin ML, Huber H, Charpentier B. et al. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 2002;30:921-30
107. Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131-42
108. Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M. et al. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007;318:450-3
109. Soma A. Circularly permuted tRNA genes: their expression and implications for their physiological relevance and development. Front Genet. 2014;5:63
110. Cadena C, Hur S. Antiviral Immunity and Circular RNA: No End in Sight. Mol Cell. 2017;67:163-4
111. Sun L, Liu S, Chen ZJ. SnapShot: pathways of antiviral innate immunity. Cell. 2010;140:436-e2
112. Kell AM, Gale M Jr. RIG-I in RNA virus recognition. Virology. 2015;479-480:110-21
113. Zhang X, Yan Y, Lei X, Li A, Zhang H, Dai Z. et al. Circular RNA alterations are involved in resistance to avian leukosis virus subgroup-J-induced tumor formation in chickens. Oncotarget. 2017;8:34961-70
114. Zhang A, He L, Wang Y. Prediction of GCRV virus-host protein interactome based on structural motif-domain interactions. BMC Bioinformatics. 2017;18:145
115. He L, Zhang A, Xiong L, Li Y, Huang R, Liao L. et al. Deep Circular RNA Sequencing Provides Insights into the Mechanism Underlying Grass Carp Reovirus Infection. Int J Mol Sci. 2017:18
116. Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861-71
117. Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849-53
118. Hubbard AK, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic Biol Med. 2000;28:1379-86
119. Nakadai T, Fukuda A, Shimada M, Nishimura K, Hisatake K. The RNA binding complexes NF45-NF90 and NF45-NF110 associate dynamically with the c-fos gene and function as transcriptional coactivators. J Biol Chem. 2015;290:26832-45
120. Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547-55
121. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558-60
122. Gale M Jr, Tan SL, Katze MG. Translational control of viral gene expression in eukaryotes. Microbiol Mol Biol Rev. 2000;64:239-80
123. tenOever BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. 2013;11:169-80
124. Scaria V, Hariharan M, Maiti S, Pillai B, Brahmachari SK. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3:68
125. Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009;37:1035-48
126. Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25-30
127. Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123-41
128. Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R. et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23
129. Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2008;451:600
130. Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682-6
131. Ghosal S, Das S, Sen R, Chakrabarti J. HumanViCe: host ceRNA network in virus infected cells in human. Front Genet. 2014;5:249
132. Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol. 2014;15:484-91
Author contact
![]() Corresponding authors: Kun Wang; Mailing address: Institute for Translational Medicine, Medical College of Qingdao University, Dengzhou Road 38, Qingdao 266021, China. Tel.: +86-532-82991791; E-mail: wangk696com and Peifeng Li; Mailing address: Institute for Translational Medicine, Medical College of Qingdao University, Dengzhou Road 38, Qingdao 266021, China. Tel.: +86-532-82991791; E-mail: peifliedu.cn
Corresponding authors: Kun Wang; Mailing address: Institute for Translational Medicine, Medical College of Qingdao University, Dengzhou Road 38, Qingdao 266021, China. Tel.: +86-532-82991791; E-mail: wangk696com and Peifeng Li; Mailing address: Institute for Translational Medicine, Medical College of Qingdao University, Dengzhou Road 38, Qingdao 266021, China. Tel.: +86-532-82991791; E-mail: peifliedu.cn

 Global reach, higher impact
Global reach, higher impact