10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(3):237-252. doi:10.7150/ijbs.22868 This issue Cite
Research Paper
Molecular Networks of Postia placenta Involved in Degradation of Lignocellulosic Biomass Revealed from Metadata Analysis of Open Access Gene Expression Data
Department of Biology, Lakehead University, 955 Oliver Road, Thunder Bay, Ontario, P7B 5E1, Canada.
Received 2017-9-18; Accepted 2018-1-9; Published 2018-2-9
Abstract
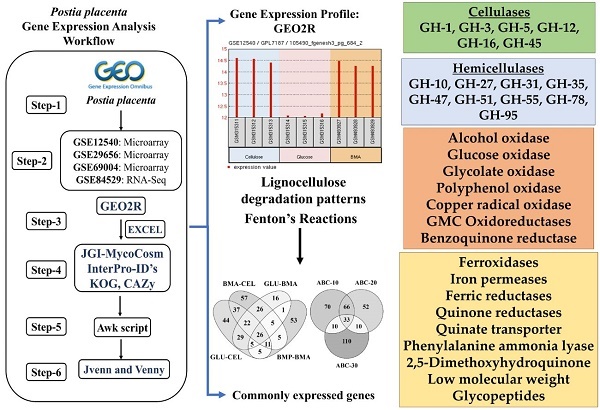
To understand the common gene expression patterns employed by P. placenta during lignocellulose degradation, we have retrieved genome wide transcriptome datasets from NCBI GEO database and analyzed using customized analysis pipeline. We have retrieved the top differentially expressed genes and compared the common significant genes among two different growth conditions. Genes encoding for cellulolytic (GH1, GH3, GH5, GH12, GH16, GH45) and hemicellulolytic (GH10, GH27, GH31, GH35, GH47, GH51, GH55, GH78, GH95) glycoside hydrolase classes were commonly up regulated among all the datasets. Fenton's reaction enzymes (iron homeostasis, reduction, hydrogen peroxide generation) were significantly expressed among all the datasets under lignocellulolytic conditions. Due to the evolutionary loss of genes coding for various lignocellulolytic enzymes (including several cellulases), P. placenta employs hemicellulolytic glycoside hydrolases and Fenton's reactions for the rapid depolymerization of plant cell wall components. Different classes of enzymes involved in aromatic compound degradation, stress responsive and detoxification mechanisms (cytochrome P450 monoxygenases) were found highly expressed in complex plant biomass substrates. We have reported the genome wide expression patterns of genes coding for information, storage and processing (KOG), tentative and predicted molecular networks involved in cellulose, hemicellulose degradation and list of significant protein-ID's commonly expressed among different lignocellulolytic growth conditions.
Keywords: Postia placenta, Brown-rot decay, Gene expression, NCBI-Gene Expression Omnibus (GEO), Lignocellulose, Fenton's reaction
Introduction
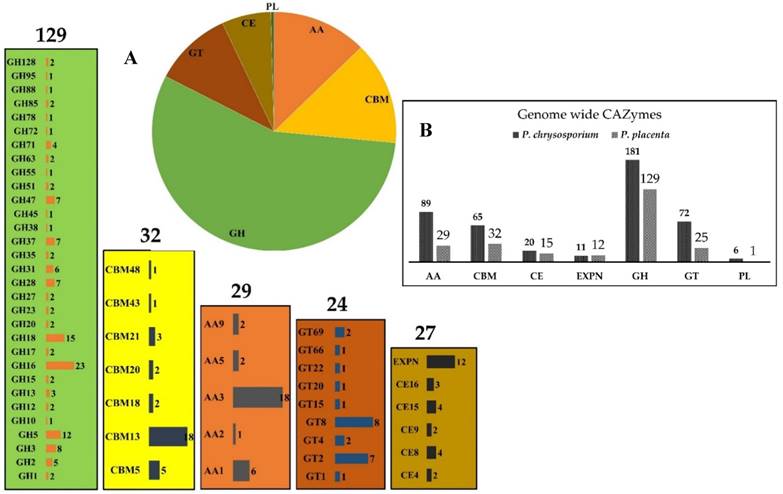
Postia placenta a filamentous basidiomycete, which causes brown rot decay of the wood, and is one of the major destructors of wood based constructions. P. placenta also exhibits the unique decaying property of brown rot fungi by rapidly breaking down cellulosic units of plant biomass with-out efficiently depolymerizing lignin units [1]. Whereas white rot fungi such as Phanerochaete chrysosporium attack plant biomass by efficiently degrading lignin followed by carbohydrate units of the plant biomass. Collectively these fungi are major inhabitants of forest biomass thus, playing a major role in carbon cycling and recycling of nutrients [1, 2]. Apart from their contrasting decay patterns, both P. chrysosporium and P. placenta belong to order Polyporales and are classified under Phlebia clade [3]. Phylogenetic studies conducted earlier have reported that brown rot fungi have progressively evolved from white rot fungi, the morphological properties and their choice of substrates also supports the above fact [4] (Fig. S1). Martinez et al (2009), have completed the total genome sequence of P. placenta and performed genome wide transcriptome and secretome analysis, through which he has revealed the functional properties of various genes regulated during the process of wood decay [1]. This study has revealed a surprising fact that P. placenta genome codes for fewer number of cellulases (glycoside hydrolases) and other CAZymes when compared to P. chrysosporium. Present day JGI MycoCosm database harbours the genomes of P. placenta MAD 698-R v1.0 and P. placenta MAD-698-R-SB12 v1.0, coding for about 242 CAZymes and 324 CAZymes respectively [1] (Fig. 1). Though brown-rot fungi are well known for extensive cellulose depolymerization, P. placenta genome reveals that number of genes coding for cellulases are less when compared to P. chrysosporium. Thus, it is highly significant to understand the molecular mechanisms responsible for lignocellulose degradation by P. placenta.
Studies were also being conducted to understand and reveal the genes encoding enzymes involved in Fenton's reactions employed during lignocellulose degradation [5]. Brown rot fungi generally involve several non-enzymatic pathways to commence the Fenton's reactions which are required for the generation of ROS, OH*, H2O2 etc. The probable mechanisms involved are a) location and solubilization of iron (Fe (oxyhydroxide) complexes in plant wood cell lumen, which is reduced to ferrous iron) [6, 7], b) hydrogen peroxide generation (reduction of molecular oxygen and oxidation of methanol)[8-10], c) iron reducing agents (reduction of Fe3+ to Fe 2+ by reductants like 2,5-dimethoxy hydroquinone) [5, 6, 11, 12]. When compared to commercial cellulase producers like Trichoderma reesei, brown rot fungi such as P. placenta lacks genes coding for cellulases/carbohydrate binding domains, exocellobiohydrolases [1]. According to Ratto et al (1997), free radicals generated from Fenton's reactions will initially attack the cell walls, followed by breakdown of native and holocellulose happens through action of hydrolytic enzymes (hemicellulases and endoglucanases) [5, 13]. It is well-known that brown-rot fungi only modify lignin without significantly altering or degrading the structure of lignin, recent studies show that brown rot fungi cause extensive demethylation, oxidation of side chains, depolymerization and also can potentially repolymerize lignin, however brown-rotted lignin is observed in polymeric form supporting earlier statement [5, 14, 15]. However, exact mechanisms involved in iron reduction and homeostasis, extracellular H2O2 and free radical generation and enzymes involved in lignocellulose degradation were unclear.
Pictorial representation of CAZymes distribution (GH-glycoside hydrolases, GT-glycosyl transferases, AA-auxiliary activity, CBM-carbohydrate binding domains, CE-carbohydrate esterases, EXPN-expansin like protiens, PL-polysaccharide lyases) in P. placenta MAD-698- Rv1.0 (A) and the top number denotes for the number of genes coding for particular class of enzymes and each bar internally shows different sub-classes of enzymes and the number of genes encoding for the corresponding enzymes, (B) Comparison of genome wide CAZymes between P. placenta and P. chrysosporium
Martinez et al (2009) and Wymelenberg et al (2010) have conducted a genome level transcriptome and proteome studies on P. placenta by culturing it on wood-derived microcrystalline cellulose, glucose (sole carbon source) and ball milled aspen, supplemented with Highley's basal salt medium (GSE12540). These studies have reported following facts A) P. placenta genome lacks genes encoding for exocellobiohydrolases and carbohydrate binding domains (commonly observed in cellulolytic microorganisms), B) several genes encoding for hemicellulases β-1-4 endoglucanases, iron and quinone reductases, extracellular iron Fe(II) and H2O2 generating oxidases were found to be highly expressed when P. placenta was cultured in cellulose medium. Wymelenberg et al (2011) has performed a microarray study to reveal the gene expression of P. chrysosporium and P. placenta colonized on ball milled aspen and pine. This study has reported that gene expression patterns of these fungi were significantly influenced by type of wood species (BMA or BMP) it colonizes and differences in gene expression patterns of these fungi reveal their preferences for carbon sources and their central decaying properties [16]. Daniel et al (2011) has developed a method for solubilization and multidimensional 1H-13C NMR spectroscopy study to analyze the degradation of aspen wood by P. placenta. Obtained results showed that P. placenta majorly degraded the content of principal aryl glycerol-β-aryl ether interunit linkages of the lignin, by employing reactive oxygen species (ROS) free hydroxy radicals (OH*) obtained through extracellular Fenton system [17]. According to Micales (1991), oxalic acid which is highly produced and found accumulated in huge quantities by brown rot fungi, also might involve in iron chelation, pH gradient, free radical formation and acid hydrolysis of cellulose and hemicellulosic units etc [18]. In order to estimate the quantity of reactive oxygen species (ROS) produced by P. placenta laccase during the process of wood degradation, Wei et al (2010) has performed proteomic and gene expression studies of laccase and reported that oxidation of every hydroquinone molecule results in production of single perhydroxyl radical [19]. Oleksandr et al (2016) has performed, a genome wide transcriptome study to understand the effect of chemical composition of wood (Populus trichocarpa) on the gene expression of P. placenta and P. chrysosporium. Results obtained by them has proved that gene expression of these fungi is significantly influenced by the wood substrate composition and the incubation periods [20]. Recently Jiwei et al (2016) has performed a genome wide transcriptome study to examine differential expression of lignin oxidizing components involved in ROS generation earlier than differential expression of genes encoding for glycoside hydrolases [21]. Higher expression of ROS generating lignin oxidizing components in hyphal front, followed by higher expression of genes encoding for glycoside hydrolases (GH) in lower hyphal front regions confirms the fact [21]. Zhang et al (2016), has developed a spatial mapping method to resolve temporal sequence in P. placenta by considering thin sections of directionally colonized wood wafers [21].
Understanding the fundamental molecular mechanisms employed by P. placenta during wood decay will significantly help the growing biofuel industries to develop novel and efficient methods for the breakdown of carbohydrate units (cellulose and hemicellulose) by selective modification of lignin (by demethoxylation and limited ring cleavage). In this study, we have performed gene expression metadata analysis on publicly available microarray and RNA-Seq datasets performed on P. placenta to understand the common gene expression patterns required during the progression of Fenton's reactions, oxalate metabolism, H2O2 and free radical generations, lignocellulose degradation (CAZymes and oxidoreductases).
Data retrieval
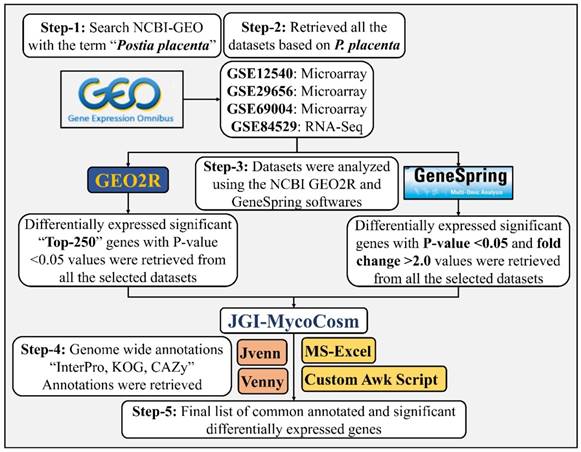
Gene expression datasets used in our present study were retrieved from NCBI GEO (Gene Expression Omnibus is a public repository for gene expression datasets) by using the term “Postia placenta”. Present day GEO repository resides four functional genomics datasets of P. placenta out of which three were microarray datasets (GSE12540, GSE29656, GSE69004) and one was RNA-Seq (GSE84529) dataset. All the microarray datasets were based on NimbleGen P. placenta MAD-698 whole genome microarray platform and cultured on different growth substrates. Dataset GSE12540 was based on P. placenta cultured on microcrystalline cellulose (Avicel), glucose, 0.5% (w/v) ball milled aspen (BMA) as sole carbon source and supplemented with Highley's basal medium [1, 22]. Similarly, GSE29656 dataset, P. placenta was cultured on 0.5% (w/v) ball milled white pine (Pinus strobus), 0.5% (w/v) ball milled bigtooth aspen (Populus grandidentata) as the sole carbon source and supplemented with Highley's basal medium for macro and micronutrients [16]. In dataset GSE69004, P. placenta was cultured on chemically distinct A (high lignin-low glucose), B (high glucose-low lignin) and C (average lignin-average glucose) Populus trichocarpa wood substrates [20]. The dataset GSE84529 P. placenta was cultured on wood wafers cut in dimensions (60 x 25 x 2.5mm) where the largest face of the wood wafers is cross sectioned and the tangential plane was arranged to be in contact with P. placenta mycelium, hyphal growth was allowed till 50 mm up on the wafers and later sectioned to A) 0-5 mm B) 15-20 mm C) 30-35 mm RNA-Seq analysis [21]. All the experimental conditions in these datasets were designed with three replicates each and complete details about the gene expression platforms and sample information were listed in Table-1.
Data analysis methodology
Microarray GSE12540, GSE29656 and GSE69004 datasets were analyzed using GEO2R and customized programming scripts. The literature and supplementary information for the corresponding datasets were studied to understand the experimental design for performing the statistical analysis. Following options were applied when using GEO2R website A) autodetect option (for log transformation of the data), B) box-whisker plot (samples and value distributions), C) submitter provided annotations (for gene level annotations) and D) Benjamini & Hochberg (False discovery rate correction) for multiple testing correction of p-values was used. Once the samples are grouped according to their experimental conditions, Top 250 option was used for obtaining statistically significant genes expressed in each dataset. We have retrieved Top 1000 differentially expressed genes to understand P. placenta gene expression by using the function “Save all results” and the obtained results were analyzed further. The ranking of differentially expressed genes (Top 250) and statistical analysis in GEO2R internally happens through limma package and the statistically significant genes were ranked based on their corrected p-value (0.05). Thus, obtained statistically significant genes were analyzed by the supplier provided annotations Protein-ID and Best hit BlastP. Gene and protein level annotations of P. placenta MAD-698R were retrieved from MycoCosm (fungal genome repository) [23, 24]. Custom linux based scripts were written to retrieve the annotations for differentially expressed gene list using KOG, GO and InterPro annotations of P. placenta MAD-698R v1. We have also used other analysis options available in JGI-MycoCosm such as Gene Ontology (GO)[25, 26], EuKaryotic Orthologous Groups (KOG) [27] and CAZy [28, 29] for analyzing the results obtained. The differentially expressed gene lists of the respective experimental conditions were compared using Venny 2.1 [30] and Jvenn [31] softwares. We have retrieved sample level FPKM (Fragments Per Kilobase of transcripts per Million) values, P-values and log fold change values from supplementary files provided for GSE84529 dataset, gene list was sorted based on their P-values and top 1000 genes were retrieved and compared among the conditions. The data analysis was similar as explained earlier in our previous studies [32-34].
Simultaneously, the datasets were also analyzed using GeneSpring® v.14.8 software. Gene expression datasets were retrieved using the option “Import NCBI GEO experiment” by saving the GEO sample files in the local folder. The experiments were created as generic single color by applying the following preprocessing conditions “Threshold value set at 1.0”, “Normalization using shift 75th percentile”, sample values were log base 2 transformed and baselined to the median of all samples. The experimental conditions were retrieved from the corresponding GEO experiment and literature, was used for the grouping the samples. The samples were filtered using “Probesets by expression” with the parameters set to data filter on normalized data and filter by percentile (upper percentile set to 100.0 and lower percentile 20.0) respectively. Based on the experimental conditions one-way Anova and Moderated T-test was performed respectively. However, for the dataset GSE84529, the samples were retrieved, and the experiment was created without any preprocessing steps. We have performed fold change analysis on the grouped samples using the FPKM values and the transcripts differentially expressed >2.0 were retrieved for the analysis.
The detailed step by step workflow used for the metadata analysis of P. placenta gene expression datasets were showed in Fig. 2.
Platform details of Postia placenta gene expression data:
| Accession ID | Substrate | Platform | Samples | Ref |
|---|---|---|---|---|
| GSE84529 | Wood wafers | Illumina HiSeq (Postia placenta) | 9 | [21] |
| GSE69004 | Poplar wood stems | NimbleGen_UW/FPL Postia placenta MAD-698 whole genome 37K expression array version 1 | 23 | [20] |
| GSE29656 | Ball milled aspen Ball milled pine | NimbleGen_UW/FPL Postia placenta MAD-698 whole genome 37K expression array version 1 | 6 | [16] |
| GSE12540 | Glucose Cellulose Ball milled aspen | NimbleGen_UW/FPL Postia placenta MAD-698 whole genome 37K expression array version 1 | 9 | [1, 22] |
Customized step by step workflow used for the metadata analysis of Postia placenta gene expression datasets.
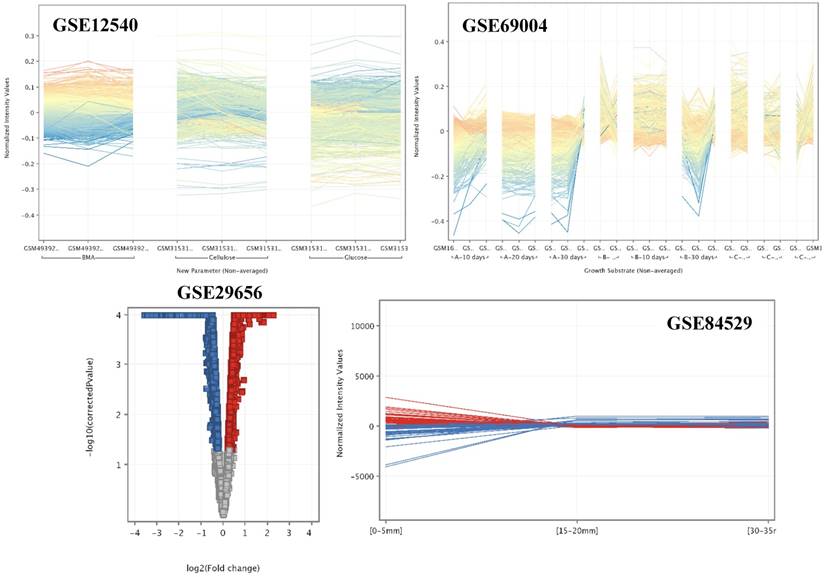
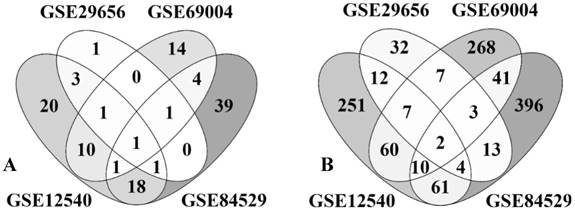
The rapid cellulolytic capacity of the brown rot fungi P. placenta surely will be credited to the potential cellulolytic enzymes and uncompromising Fentons reactions. Apart from these enzymes P. placenta also secretes a large list of aromatic compound degrading and detoxifying enzymes. Statistical analysis of the P. placenta gene expression datasets based on the provided experimental conditions has resulted in 5174, 7519, 6390 and 10,754 differentially expressed transcripts among GSE12540, GSE29656, GSE69004 and GSE84529 datasets respectively (Fig. 3).
Genes encoding for Carbohydrate Active Enzymes (CAZymes) and Metabolism
The genome of the P. placenta MAD-698 Rv1 harbors around 245 CAZymes, when compared to the genome of P. chrysosporium RP78 (450 CAZymes), several genes encoding for glycoside hydrolase and glycosyl transferase classes were reduced to 1 or absent. Genes encoding for various classes of glycoside hydrolases were found to be absent in the genome of P. placenta such as GH6, GH7, GH9, GH11, GH25, GH30, GH43, GH53, GH74, GH79, GH89, GH-92, GH115, GH125, GH131, GH133 and GH135. Several other GH-classes were reduced to one gene such as GH-10, GH38, GH45, GH55, GH72, GH78, GH88 and GH95 [1]. Similarly, P. placenta genome codes only for 24 glycosyl transferase encoding genes which were classified under GT (1), GT15 (1), GT20 (1), GT22 (1), GT66 (1), GT4 (2), GT69 (2), GT2 (7) and GT8 (8) classes and several GT-classes were found to be absent such as GT3, GT5, GT17, GT21, GT24, GT31, GT32, GT33, GT35, GT39, GT41, GT48, GT49, GT50, GT57, GT58, GT59, GT76 and GT90 [1]. Also, genes coding for polysaccharide lyase class 8, carbohydrate esterase class 1 and carbohydrate binding module class 1, 35 and 50 were found to be absent in P. placenta genome [1]. The statistically significant differentially expressed genes obtained under different growth conditions were compared to find the genes which were expressed commonly. When P. placenta was cultured on simple culture medium containing cellulose, glucose and ball-milled aspen growth conditions in GSE12540 dataset. Several genes encoding CAZymes such as glycoside hydrolases (GH-1, 2, 3, 5, 8,10, 16, 18, 27, 28, 31, 35, 37, 51, 55, 71, 88, 95 and 128), glycosyl transferases (GT-8), carbohydrate esterase (CE-4 and CE-8), auxiliary activity (GMC oxidoreductases, ferroxidases) EXPN related proteins were commonly expressed. Similarly, when P. placenta was cultured on ball milled aspen (BMA) and ball milled pine (BMP), CAZymes such as glycoside hydrolase classes 16,18, 28, 55, 71, glycosyl transferases class-1, and laccase encoding genes were highly and differentially expressed. In gene expression datasets GSE69004 and GSE84529, P. placenta was cultured on natural plant biomass growth substrates. In GSE69004, genes encoding for CAZymes GH-5, GH-16, GH-18, GH-71 and GH-128, carbohydrate binding module CBM-18 and CBM-21 and carbohydrate esterase class CE-4 were found be commonly expressed among all the conditions of A-B-C-10 days, A-B-C-20 days and A-B-C-30 days. Similarly, in GSE84529 dataset, genes encoding for CAZymes GH-1, 2, 3, 5, 8, 12 13, 16, 17, 18, 28, 51 and 78 GT-20, CE-8, 9 and 15, CBM-13 and GMC oxidoreductases, AA-3 were found to be commonly expressed among the growth conditions (Fig. 3). On comparison of Top-1000 differentially expressed genes from the datasets GSE12540, GSE29656, GSE69004 and GSE84529, CAZymes encoding genes for GH-2, GH-3, GH-5, GH-16, GH-18, GH-27, GH-28, GH-55, GH-71 and GH-95, GT-1 were found to be commonly expressed among the datasets.
Profile plot (GSE12540, GSE69004 and GSE84529) and volcano plots (GSE29656) of the significant and differentially expressed genes among the conditions.
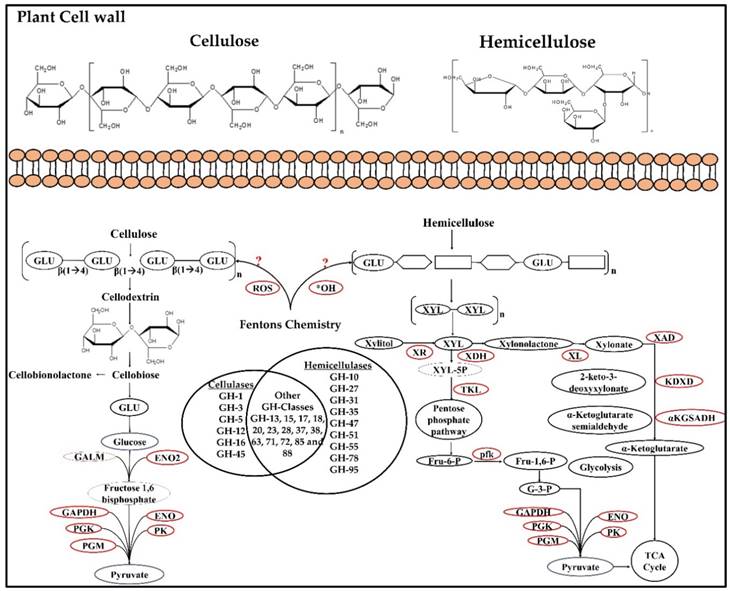
In dataset GSE12540, when P. placenta was cultured on glucose, cellulose and ball milled aspen several glycoside hydrolases involved in cellulose and hemicellulose degradation were found to be highly expressed. Genes encoding for cellulases GH-1, GH-3, GH-5, GH-12, GH-16 were found to be highly up-regulated in ball milled aspen followed by cellulose growth mediums. Similarly, genes encoding for hemicellulases GH-10, GH-27, GH-31, GH-35, GH-47, GH-51, GH-55 and GH-95 were also found to be up-regulated mostly in ball milled aspen followed by cellulose growth mediums. Based on the previous reports it is known that microorganisms release several carbohydrate esterases for the deacetylation of carbohydrates, genes encoding for acetyl xylan esterase CE-4 and pectin methyl esterase CE-8 and carbohydrate esterase type-B were highly expressed in ball milled aspen followed by cellulose growth medium. We have observed that in dataset GSE29656, where P. placenta was cultured on complex plant cell wall materials of ball milled aspen and ball milled pine, out of top-1000 differentially expressed genes only few CAZymes encoding genes were found. Genes encoding for glycosyl transferase class-1, glycoside hydrolases GH-18 (Ppl118230), GH-28 were highly up regulated in ball milled pine, GH-18 (Ppl1107968), and GH-28, in ball milled aspen GH-16 GH55 and GH71, carboxyl esterase type B and laccase (AA1) were found to be highly expressed.
When cultured on composite natural plant biomass growth substrates (GSE69004), containing chemically modified Populus trichocarpa cell wall components. We have observed that the gene expression patterns of P. placenta fluctuated upon different incubation periods. The top-1000 genes obtained for the 10-days incubation period of all the three conditions showed genes encoding for cellulases GH-3, GH-5, GH-16, CBM-18 and CBM21, hemicellulases GH-2, GH-27, GH-47, GH-55, GH-95 and CE-4 were highly expressed in low lignin-high glucose and average lignin-average glucose conditions. While with 20- days incubation period samples genes encoding for cellulases GH-5, GH-16, CBM-18, CBM-21 and hemicellulases GH-35, GH-47, CE-4. Similarly, 30-days incubation samples genes coding for hemicellulases GH-37, GH-55 along with GH-16, GH-128 and GT-20 encoding genes.
Enzymes coding Fentons Reaction
Absence of genes encoding for lignin (LiP) and manganese peroxidase (MnP) is one of the major reason for minimal the lignin degrading abilities of P. placenta. Interestingly, P. placenta is completely dependent on the hydroxy radical (OH*) and ferric ion (Fe3+) generated through Fentons reaction for the process of lignin modification (or) depolymerization [6, 35]. Previous studies have reported that Fentons reactions in wood rotting basidiomycetes is deployed by three different mechanisms a) cellobiose dehydrogenase based reactions b) small glycopeptide catalyzed reactions generating hydroxy radicals c) cyclic redox based reactions by low molecular weight redox compounds (such as quinones and oxalates) [35]. Fentons reaction is majorly dependent on a) extracellular hydrogen peroxide generation and b) metabolite and enzyme based reduction of iron from ferric state to ferrous state (cellobiose dehydrogenase) [1]. However, absence of genes encoding for cellobiose dehydrogenase makes P. placenta solely dependent on the extracellular fungal metabolites such as hydroquinones and low molecular weight glycopeptides to catalyze the reduction of iron [1]. Hydrogen peroxide generating enzymes mainly glucose oxidase, copper radical oxidase, alcohol oxidase, glycolate oxidase and polyphenol oxidase were observed among the P. placenta gene expression datasets. Similarly, enzymes involved in iron reduction and homeostasis mainly quinone reductases, quinone transporters, phenylalanine ammonia lyase, ferric reductases, iron permeases and ferroxidases were significantly expressed among the datasets. In GSE12540 dataset, genes encoding for copper radical oxidase, 1,4-benzoquinone reductase, polyphenol oxidase, phenylalanine ammonia lyase, iron permease, ferroxidase and multicopper oxidase were found to be highly expressed in cellulose, ball milled aspen growth substrates. Similarly, in GSE29656 dataset genes encoding copper radical oxidase, polyphenol oxidase, phenylalanine ammonia lyase, iron permease, multicopper oxidase and ferroxidase were found to be differentially expressed in ball milled aspen and 1,4-benzoquinone reductase, ferric reductase encoding genes were expressed in ball milled pine growth substrates. Results obtained from GSE69004 dataset showed that genes encoding for GMC oxidoreductases, iron permease were highly expressed in A 10, 20, 30 (low glucose-high lignin) growth substrates, alcohol oxidase, glucose oxidase and glycolate oxidase were highly expressed in low lignin- high glucose and average glucose-average lignin growth substrates.
Lignin Degrading and Detoxifying Enzyme systems
Due to the lack of genes encoding for ligninolytic peroxidases, the process of lignin modification or depolymerization is majorly dependent on other auxiliary activity enzymes such as multicopper oxidases, laccases and oxidoreductases. Genes encoding for auxiliary enzymes such as ferroxidase, multicopper oxidase, laccase, GMC Oxidoreductases, alcohol oxidases, glyoxal oxidase 1,4-benzoquinone reductases, polyphenol oxidase and lytic polysaccharide monoxygenases were found to be highly expressed under different culture conditions as mentioned above. Apart from the auxiliary enzymes several other enzymes involved in aromatic compound degradation and metabolism were also found to be highly expressed among the datasets. In GSE12540 dataset, genes encoding for aromatic ring hydroxylase, catechol dioxygenase, intradiol ring dioxygenase, Tannase and feruloyl esterase, taurine catabolism dioxygenase, 2-nitropropane dioxygenase, 4-coumarate coenzyme A ligase, O-methyl transferase, FAD-linked oxidoreductase, alpha aminoadipate reductase, aldo/keto reductases, alcohol dehydrogenases, zinc alcohol dehydrogenases were found to be highly expressed in cellulose and ball milled aspen growth substrates. In GSE29656 dataset, genes encoding for aldehyde dehydrogenase, acyl-coA thioesterase, homocitrate synthase, flavodoxin, epoxide hydrolase, HMG-CoA lyase, isoflavone reductase, FAD monoxygenase, NADH flavin dependent oxidoreductase, short chain dehydrogenase, transketolase, terpene synthase, zinc alcohol dehydrogenase, 4-coumarate coA ligase, O-methyl transferase and various genes encoding for cytochrome P450 class monoxygenases were highly expressed in ball milled pine growth substrate. However, genes encoding for esterases, Tannase and feruloyl esterase, phenylalanine ammonia lyase, polyphenol oxidase, laccase and copper radical oxidase were found to be highly expressed in ball milled aspen growth substrates. In GSE69004 dataset, where P. placenta was cultured on Populus trichocarpa with chemically distinct growth substrates genes encoding for aldo keto reductases, cytochrome c, several genes coding for cytochrome P450, dienelactone hydrolases, E-class P450 group IV, induced cAMP protein, iron permease, NADH flavin oxidoreductase, class-I auxiliary activity enzymes were found to be expressed in high lignin-low glucose conditions (A10, A20 and A30). Several genes encoding for acyl-CoA dehydrogenase, carboxyl esterase type B, several genes encoding for cytochrome c, cytochrome b5, dienelactone hydrolase, esterase, haloacid dehalogenase/epoxide hydrolase, mandelate racemase, Metallophosphoesterase, FAD monoxygenase, oxidoreductase, Thiolase, UbiA prenyltransferase, alcohol oxidase, 2-nitropropane dioxygenase, flavin monoxygenase and peroxidase were found to be highly expressed among low lignin-high glucose and average lignin-average glucose growth conditions (Supplementary information-S2).
In GSE84529 dataset among the top 1000 differentially expressed genes, we have commonly observed genes encoding for aromatic ring hydroxylase, 2-nitropropane dioxygenase, flavoprotein monoxygenase, carboxylesterase type B, esterase, flavoprotein monoxygenase, aldo/keto reductase, zinc alcohol dehydrogenase, short chain dehydrogenase/reductase, 2OG-Fe(II) oxygenase superfamily, isoflavone reductase, GMC oxidoreductase, generic and O-methyltransferase, UbiA prenyltransferase and D-isomer specific 2-hydroxyacid dehydrogenase in the early (0mm to 5mm) and late (15mm-20mm) conditions. Along with above mentioned enzymes several genes encoding for cytochrome P450 monoxygenases were found to be highly expressed in early (0mm to 5mm) and few in late (15mm to 20 mm) conditions (Supplementary information-S2). According to Zhang et al (2016), lignin oxidizing and Fenton chemistry related enzymes required for the production of H2O2 and iron reduction and homeostasis were found to be highly expressed during early decay (0mm to 5mm) process [21]. This study also reported that during early decay phase (0mm to 5mm) genes coding for lignin oxidation, Fenton chemistry, cytochrome P450 enzymes were highly expressed when compared to late decay phases (15mm to 20mm and 30mm to 35mm) where genes coding for various CAZymes, sugar metabolism, Fenton chemistry and aldo keto reductases were found to be differentially expressed [21] (Fig. 4).
The comparison of significant differentially expressed gene lists obtained from all the datasets have revealed that overall 40 genes encoding for CAZymes were found to be common with each dataset sharing few common CAZymes (Supplementary information-S2). Common auxiliary activity enzymes include laccase (Pspl1_111314), ferroxidase (Pspl1_109824), GMC oxidoreductase (Pspl1_92024 and Pspl1_27847). Similarly, common cellulolytic enzymes include GH-3 (Pspl1_128500 and Pspl1_46915), GH-5 (Pspl1_121831, Pspl1_116199, Pspl1_121713 and Pspl1_57386), GH-16 (Pspl1_112941, Pspl1_62300, Pspl1_94601 and Pspl1_51311). Finally, common hemicellulolytic enzyme classes include GH-27 (Pspl1_120395), GH-31 (Pspl1_117029), GH-35 (Pspl1_127993), GH-51 (Pspl1_100251 and Pspl1_94557), GH-55 (Pspl1_105490) and GH-95 (Pspl1_105952) (Supplementary information-S2). Several genes encoding for aromatic compound degrading and metabolizing enzymes were found to be commonly expressed among the datasets. Importantly, genes encoding for cytochrome P450 monoxygenases (Pspl1_9739, Pspl1_98329, Pspl1_89741, Pspl1_92219 and Pspl1_21733), aromatic ring hydroxylase (Pspl1_90902, Pspl1_22746 and Pspl1_23052), dioxygenases including 2-nitropropane dioxygenase (Pspl1_24756, Pspl1_28683), intradiol dioxygenase (Pspl1_34850) and taurine catabolism dioxygenase (Pspl1_89958) (Supplementary information-S2) (Fig. 5).
Four-way and three-way Venn diagrams showing the commonly expressed statistically significant CAZymes among the gene expression datasets A) glucose-cellulose, glucose-BMA, BMA-cellulose and GSE29656 B) high lignin and low glucose (A) high glucose and low lignin (B) average lignin and average glucose at incubation periods of 10-days, 20-days and 30-days, C) 15mm-20mm vs 0mm-5mm, 30mm-35mm vs 0mm-5mm and 30mm-35mm vs 15mm-20mm D) GSE29656, GSE69004, GSE12540 and GSE84529 datasets.
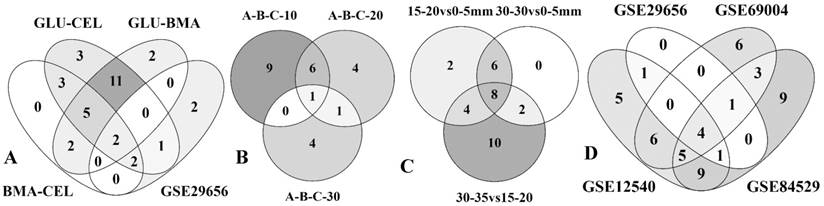
Four-way and three-way Venn diagrams showing the commonly expressed statistically significant enzymes among the gene expression datasets A) glucose-cellulose, glucose-BMA, BMA-cellulose and GSE29656 B) high lignin and low glucose (A) high glucose and low lignin (B) average lignin and average glucose at incubation periods of 10-days, 20-days and 30-days, C) 15mm-20mm vs 0mm-5mm, 30mm-35mm vs 0mm-5mm and 30mm-35mm vs 15mm-20mm D) GSE29656, GSE69004, GSE12540 and GSE84529 datasets.
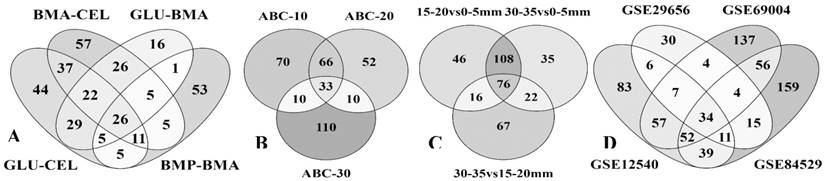
Four-way Venn diagrams showing the commonly expressed statistically significant protein-Ids among the gene expression datasets A) CAZymes among all the datasets B) All InterPro-IDs among all the datasets.
Genes encoding for Information, Storage and Processing processes
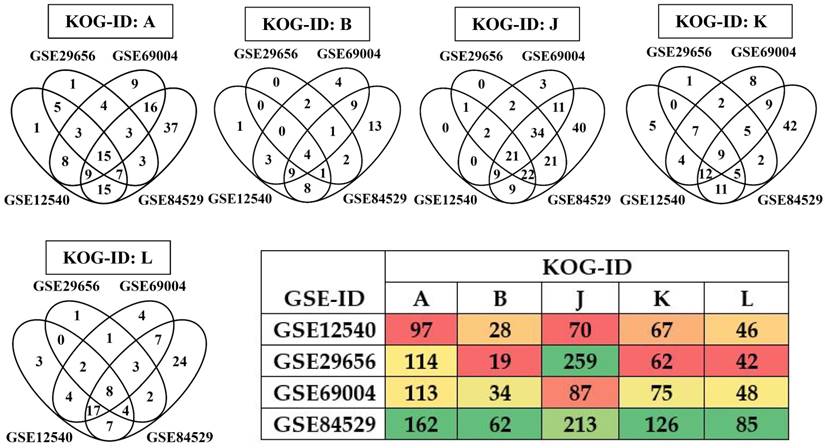
Expression of the lignocellulolytic enzymes are controlled and regulated by genes encoding for the information, storage and processing processes. The eukaryotic orthologous group have classified these genes into RNA processing and modification (KOG: A), chromatin structure and dynamics (KOG: B), Translation, ribosomal structure and biogenesis (KOG: J), Transcription (KOG: K) and Replication, recombination and repair (KOG: L) groups. We have separated the differentially expressed gene list in to the KOG groups A, B, J, K and L using the KOG classification list retrieved from MycoCosm database for P. placenta. We have observed that a total of 308, 496, 357 and 648 transcripts were significantly expressed among GSE12540, GSE29656, GSE69004 and GSE84529 datasets respectively (Fig. 7). Total of 37 (A), 15 (B), 89 (J), 38 (K) and 36 (L) were found to be commonly expressed among the gene expression datasets (Supplementary Information-S3) (Fig. 7).
Results obtained from these gene expression studies has revealed that total of 15 proteins encoding for RNA processing and modification were found to be common and significantly expressed RNA helicase nonsense mRNA reducing factor, splicing coactivator SRm160/300, RNA directed polymerase QDE1, polyadenylation factor, dsRNA-specific nuclease Dicer and ribonuclease, mRNA capping enzyme, mRNA decay protein, RNA helicase BRR2, RNA binding protein p54nrb and polyadenylate binding protein. Similarly, four genes encoding for chromatin structure and dynamics structural maintenance of chromosome protein 4, Zn-finger-MYND type, chromosome condensation complex and ubiquitin component Cue were common and significantly expressed. 21 genes encoding for translation, ribosome biogenesis and structure were common and significantly expressed 40S ribosomal protein S6, SA(P40), 60S ribosomal protein L10, L3, L28, Ribosomal proteins L7A, L18e, L23, L5, S25, L6E, S14, S10, L32, translation initiation factors eIF-5B, eIF-3a, eIF-4G and mRNA export factor. 9 genes encoding for chromodomain helicase, transcription initiation factor, HMG1/2, nuclear receptor coregulator SMRT, transcription regulator XNP, Ssu72, bZIP, TGF-beta and SMAD protein, HLH-transcription factor was commonly expressed in transcription class. Finally, in replication, recombination, repair class 8 genes encoding for 3-methyladenine DNA glycosidase, exonuclease, replication factor C, apurinic/ apyrimidinic endonuclease, DNA replication-MCM7 factor, A/G-specific DNA glycosylase, excision repair protein RAD14/XPA and Nudix family hydrolase were commonly expressed among all the datasets (Supplementary Information-S3). Majority of the genes encoding for information storage and processing were highly expressed in glucose, cellulose, ball milled aspens, A (low glucose-high lignin) and 0mm05mm and 30mm-35mm growth substrates (Supplementary Information-S3).
Four-way Venn diagrams showing the commonly expressed statistically significant genes encoding for KOG groups A (RNA Processing and modification), B (Chromatin structure and dynamics), J (Translation, ribosomal structure and biogenesis), K (Transcription) L (Replication, recombination and repair) and heatmap showing total list of significant information storage and processing groups among the datasets.
Discussion
Genome wide evolutionary studies conducted earlier have revealed that during transition from white-rot to brown-rot, basidiomycete fungi have experienced an extensive gene loss. The whole genome studies have revealed that P. placenta MAD-698-R v1 has suffered complete reduction of various genes coding for glycoside hydrolases such as GH-6, GH-7, GH-10, GH-11 and GH-61 [1]. Though the genome of P. placenta lacks genes encoding for various essential lignocellulolytic CAZymes the decay patterns of P. placenta suggest the rapid depolymerization of cellulose and hemicellulose units through modifying the lignin units. During initial degradation process P. placenta significantly reduces the degree of polymerization (DP) of cellulose from 1800-2000 glycosyl units to 150-200 units [36]. However, due to its large structure cellulases cannot enter through the pores of wood, thus brown rot fungi employ other oxidoreductases and Fenton's system for the depolymerization of plant cell wall structures. Unlike other cellulose degrading microorganisms, P. placenta lack genes coding for conserved CBH1(GH7) and CBH2 (GH6) exocellobiohydrolases, carbohydrate binding module class-1 and cellulase binding endoglucanases. Hence, P. placenta must accomplish the process of cellulose and hemicellulose degradation through a set of endoglucanases, exoglucanases, β-glucosidases and several other hemicellulases. Previous gene expression studies of P. placenta conducted on cellulose reported various genes coding for hemicellulases, laminarinases, chitinases, but it is still not clear whether these enzymes can attack crystalline cellulose [1]. It is well known that P. placenta genome lacks the sequences encoding for major ligninolytic such as lignin (LiP), manganese (MnP) and versatile (VP) peroxidases. However, the P. placenta genome was found to possess gene sequences coding for a low redox potential peroxidase (Protein-ID: 50226) which lacks the Mn(II) oxidation sites but possess a tryptophan residue which is involved in substrate oxidation similar to LiP and VP [1]. These genomic evidences support the stronger involvement of Fentons chemistry during depolymerization or modification of the lignocellulosic biomass.
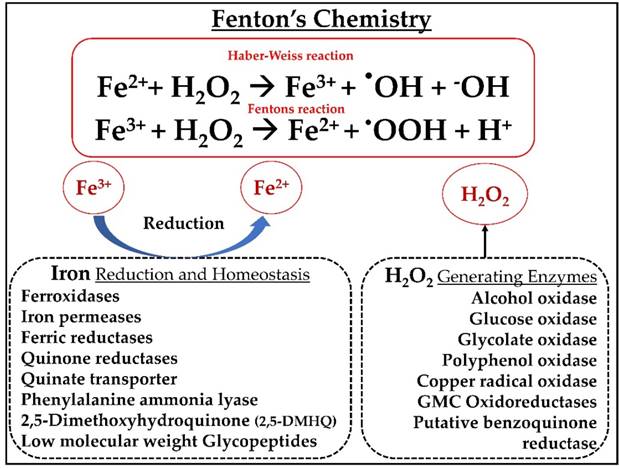
The process of Fentons chemistry in P. placenta is employed by a set of genes encoding for enzymes involved in hydrogen peroxide generation and iron reduction and homeostasis. Generation of non-specific highly powerful oxidative hydroxy radicals with a half-life period around 9 to 10 seconds by Fentons chemistry will play a crucial role in the depolymerization and modification of lignocellulosic components of plant cell wall [5, 6]. Studies have reported that Fentons chemistry is highly dependent on three factors they are iron (Fe3+), hydrogen peroxide (H2O2) and pH [5]. Earlier studies have reported that brown rot fungi might also depend on the insoluble iron oxyhydroxide complexes for the uptake of iron and reduction [6, 7]. We have commonly observed the genes encoding for hydrogen peroxide generating and iron homeostasis and reduction among various datasets. Expression of genes involved in the process of iron reduction and homeostasis such as ferroxidase, phenylalanine ammonia lyase, catalase, iron permease, flavin monoxygenases, dioxygenases quinone transporters and reductases proves the importance of Fentons system. According to Berry et al (1997), brown rot fungi secretes oxalic acid extracellularly which might be involved in solubilizing the iron from its naturally occurring iron oxyhydroxide complexes of the plant wood [6]. As reported by Suong oh et al (2006), dissolution of iron oxides (protonating OH groups resulting in weak Fe-O bond) is significantly influenced by the cellular pH conditions [37]. Several theories have been proposed on the process of iron chelation with oxalic acid under lower pH conditions by forming soluble and stable oxalate-iron complexes which further diffuse into the plant cell wall through wood lumen [5-7]. Genes encoding for alcohol and methanol oxidases which are also involved in the extracellular production of H2O2 were found to be highly expressed among the datasets. Studies have also reported that low molecular weight compound such as 2,5-dimethoxyhydroquinone are employed during the process of lignin depolymerization [38] (Fig. 8).
However, till date the exact reaction mechanisms involved during the depolymerization or modification of lignocellulosic biomass is not well understood. In our present analysis, we have clearly observed significant expression of genes encoding for enzymes involved in Fentons reaction. Apart from the above mentioned enzymatic fungal Fe3+ reductants, non-enzymatic low molecular weight iron reductants such as 2,5-dimethyoxyhydroquinone (2,5-DMHQ) were also reported to be significantly involved in depolymerization of lignocellulose components. Previous studies have observed the presence of 2,5-DMHQ in the cultures of Gleophyllum trabeum[38], Postia placenta [19], Serpula lacrymans [11] and also in various cultures of Gleophyllum species [11, 12, 39]. The low molecular weight 2,5DMHQ reduces fungal Fe3+ to Fe2+ by simultaneously generating a radical of semiquinone, which can reduce oxygen to *OOH later upon dismutation reaction it can generate H2O2 or it can reduce an extra Fe3+ to Fe2+ resulting in quinone formed due to original Fe3+ reductant [9, 39]. Along with these mechanisms there are other two iron reduction mechanisms a) cellobiose dehydrogenase dependent and b) low molecular weight glycopeptides. However, the genome of P. placenta lacks the genes encoding for cellobiose dehydrogenase (CDH) which rules out the CDH dependent iron reduction [1]. According to Martinez et al (2009), Low molecular weight glycopeptides were found to be expressed in the cultures of P. placenta [1]. Based on the results obtained in our present study, we have listed the common significant enzymes obtained among all the datasets which were listed in the Table 2.
Common differentially expressed significant class of enzymes among different growth conditions of gene expression datasets.
| Enzyme class | Commonly Expressed P. chrysosporium Genes among the datasets |
|---|---|
| Cellulose Degradation | GH-1, GH-3, GH-5, GH-12, GH-16 and GH-45 |
| Hemicellulose Degradation | GH-10, GH-27, GH-31, GH-35, GH-47, GH-51, GH-55, GH-78 and GH-95 |
| Carbohydrate Metabolism | GH-2, GH-13, GH-15, GH-17, GH-18, GH-20, GH-23, GH-28, GH-37, GH-38, GH-63, GH-71, GH-71, GH-72, GH-85 and GH-88 |
| Lignin Degradation | Auxiliary Activity Enzymes: Laccase, Ferroxidase, Multicopper oxidase, Low redox potential lignin peroxidase, Glucose-Methanol-Choline (GMC) oxidoreductase, Alcohol oxidase, Glyoxal oxidase, Lytic polysaccharide monoxygenase, Chloroperoxidase Aromatic compound degrading: Intradiol dioxygenases, Aromatic ring hydroxylase, Epoxide hydrolase, Cytochrome P450 Monoxygenase, Alcohol dehydrogenase, Dioxygenase, 2-nitropropane dioxygenase, Flavin containing monoxygenase, Iron reductases, Catalase, Alcohol/methanol oxidases, Haloacid dehalogenase, Oxidoreductase, Tannase and feruloyl esterase, Esterase/ lipase/thioesterase, Short-chain dehydrogenase/ reductase, D-isomer specific 2-hydroxyacid dehydrogenase, Beta-ketoacyl synthase, 2-oxo acid dehydrogenase, Aldo/keto reductase, Aldehyde dehydrogenase, Alkyl hydroperoxide reductase, FAD- linked oxidase, Thiolase, Carbohydrate esterases, Glycosyl transferases |
| Fenton's Chemistry | Ferroxidase, Ferric reductase, Iron permease, Quinone reductase, Quinone transporters, phenylalanine ammonia lyase, low molecular weight glycopeptides, alcohol oxidase, glucose oxidase, glycolate oxidase, polyphenol oxidase, copper radical oxidase, 1, 4-Benzoquinone reductase |
| Detoxification &Stress Responsive enzymes | 2OG-Fe(II) oxygenase superfamily, Argonaut and dicer complex, Cytochrome P450, E-class P450, Thioesterase, flavin monoxygenase, Glutathione-S-transferase, Thiolase, Ubiquitin, Universal stress protein, Flavodoxin, Epoxide hydrolase, Isoflavone reductase, FAD monoxygenase, NADH flavin oxidoreductase, Dienelactone hydrolase, Thioredoxin, Thaumatin, Ferredoxin, Beta lactamase, Catalase, UbiA prenyltransferase, Universal stress protein, Ubiquitin system component, Cytochrome b5, Cytochrome c, amidohydrolase |
Tentative representation of Fenton's reaction system observed in P. placenta mainly includes H2O2 generating and Iron reduction-homeostasis enzyme systems found to be commonly observed among the gene expression datasets.
Wood decaying fungi majorly depend on the holocellulose for the carbon and energy requirements, however genome wide studies have revealed that cellulolytic systems of P. placenta and other brown rot fungi vary significantly when compared with popular fungi such as Trichoderma reesei and Phanerochaete chrysosporium (which employ an efficient set of hydrolytic enzymes involving endo and exo glucanases, beta-glucosidases and cellobiose dehydrogenases [5]. Despite the loss of various genes encoding for glycoside hydrolases in the genome P. placenta has found to cause rapid and significant depolymerization of holocellulose by significantly decreasing the degree of polymerization to 150-200 through reducing the amorphous regions of cellulose. Brown rot fungi likely opens the cell wall structure by extensive removal of hemicelluloses resulting in a 20% of weight loss which increases the accessibility of the cellulose [5, 12, 40, 41]. According to Cohen et al (2005), some fungi the absence of cellobiose dehydrogenase is compensated by the expression of functionally similar endoglucanase [42]. These earlier proposed reports suggest that hydrolytic enzymes including endoglucanases and hemicellulases contribute to the complete holocellulose depolymerization released after primary attack of the cell wall [5]. As reported by Ratto et al (1997), higher hydrolysis rates were observed when P. placenta and T. reesei were cultured on spruce sawdust containing growth substrates, after the initial Fenton reaction chemistry [13]. Ratto et al (1997) have also reported that initial oxidation will significantly increase the hydrolysable nature of cellulose by endoglucanases secreted by brown rot fungi [13]. Jung et al (2015), have conducted a study by mimicking Fenton reaction conditions on rice straw degradation, results obtained in this study have showed that composition of lignin and xylan present in the rice straw was significantly reduced by the Fenton reaction systems [43]. Results obtained in our present analysis is in accordance with previous studies and supports the highly dependent nature of brown rot fungi on Fentons reaction system for the process of lignocellulose degradation. We have also observed that genes encoding for hemicelluloses were highly up-regulated during the cellulolytic conditions, when cultured on both simple synthetic and complex natural growth substrates (Fig. 9).
Studies conducted in the past have reported that reactive hydroxy radicals are involved in the polymerization and depolymerization of lignin [44, 45]. According to Goodell et al (1997), brown rot fungi exhibits similar mechanism for the process of lignin depolymerization or modification, using chelator mediated Fenton reaction [5, 6]. Various studies have strongly reported that brown rot fungi affected wood experience extensive oxidative demethylation [15, 46-48] and side chain oxidation [15, 48, 49]. During the process of lignin modification various important processes were reported earlier which includes a) partial aromatic ring cleavages [48] b) aromatic ring hydroxylation [48] c) limited side chain hydroxylation and Cβ-ether cleavage [49] d) formation of aryl derivative side chains (aryl-o-aryl, aryl-aryl etc.) [14]. It is well-known that unlike its counterparts P. placenta (brown rot fungi) do not secrete lignin degrading auxiliary enzymes such as LiP, MnP [1]. However it was reported that S2 layer of the secondary cell wall was found to contain brown rotted lignin whereas the above mentioned ligninolytic auxiliary enzymes were expected to fail in penetration and causing attack [50]. Although brown rot fungi (P. placenta) affected lignin undergoes the above-mentioned modifications, it was reported that lignin does not lose its polymeric nature, which proves the fact that brown rot fungi are weak lignin degraders. Studies have also reported that reactive hydroxy radicals released due to Fenton reaction system might also lead to the repolymerization of lignin [14, 15, 51-54]. These reporting's from the previous studies were observed in our study, genes encoding for various enzymes involved in aromatic compound degrading enzymes and iron reduction-homeostasis reactions of Fenton system were found to be highly expressed in P. placenta during the ligninolytic conditions. Along with the genes encoding for lignocellulolytic enzymes several detoxification and stress responsive related enzymes were found to be highly expressed during the ligninolytic conditions suggesting the release of highly toxic intermediates and conditions due to lignin modification.
Tentative network of genes involved in coding for enzymes involved during cellulose and hemicellulose degradation.
Genes classified under information storage and processing are involved in controlling the expression of lignocellulolytic and detoxification enzymes. Present day genome of P. placenta encodes total of 1035 genes encoding for information storage and processing (KOG group) which are further classified into 296 (KOG: A), 103 (KOG: B), 253 (KOG: J), 244 (KOG: K) and 139 (KOG: L) [1]. The eukaryotic KOG process RNA processing and modification decides the cellular fate of transcripts. Several studies conducted in the past have clearly explained that mRNA is considerably pretreated by 5' capping, splicing and 3' end processing before the mRNA gets exported. The above reported commonly expressed genes RNA helicase, RNA polymerase QDE1 and RNA binding protein are involved in the initiation process. Genes encoding for mRNA guanylyl and methyl transferases are involved in 5' end capping, splicing coactivator, dsRNA specific nuclease Dicer and related ribonuclease, non-sense mediated mRNA decay and polyadenylation factor complex protein are involved in elongation and termination of the transcripts [55-57]. Together these genes directly control the expression, localization, stability and export of the mature transcripts. Organization of large eukaryotic genomes into stable chromosomes requires a wide range of enzymes classified under chromatin structure and dynamics class. Maintaining the structural dynamics of eukaryotic chromatin and organizing it in stable chromosomes is of primary importance for the cell otherwise, it might bring various restrictions in various DNA related processes. Expression of histone methyl and acetyl transferases, chromatin condensation and SWI-SNF remodeling complex commonly among the gene expression datasets, expression of these genes is directly involved in regulation and control of various plant cell wall degrading enzymes. Several genes encoding for 40S, 60S ribosomal proteins, eukaryotic initiation factors, translation initiation factors mentioned in Supplementary information-S3. Similarly, genes involved in base excision repair, nucleotide excision repair, mismatch repair and homologous recombination were found to be commonly expressed among all the datasets (Supplementary information-S3). Results obtained from the gene expression studies that when P. placenta was cultured on natural plant biomass growth substrates expression of plant cell wall degrading and detoxifying enzymes are strongly influenced and regulated at various cellular and nuclear levels. However, the research must be conducted to understand the exact cellular mechanisms required for the expression of plant cell wall degrading enzymes. Previous studies have revealed that ascomycetes code for a wide range of transcription factors out of which zinc binuclear cluster proteins (XYR1, ACE2, XlnR and XLR1), multimeric proteins (HAP2, HAP3, HAP5), two zinc binuclear cluster (CLR-1/2), three Cys2-His2 Zn-finger (PacC) are well known as positive regulators of cellulolytic and hemicellulolytic enzymes. Whereas, ACE1, CRE1, CRE-A, CRE-B and CRE-C are considered as negative regulators of cellulolytic and hemicellulolytic enzymes [58]. Genes coding for the aforementioned transcription factors were not significantly expressed among the P. placenta gene expression datasets, however genes encoding for other transcription factors have been observed among the datasets. Studies must be conducted to understand the functional involvement of the common significantly expressed information storage and processing genes (Supplementary information-S3) [34].
Conclusion
Genome wide transcriptome and proteome studies of P. placenta conducted in the last decade have revealed about the functional properties of lignocellulolytic enzymes and various CAZymes. We have performed an extensive metadata analysis on the P. placenta gene expression datasets (GSE12540, GSE29656, GSE84529 and GSE69004) to understand the common gene expression patterns involved during the lignocellulose degradation. For the first time, we have reported the genome wide common expression patterns of P. placenta when cultured on different plant biomass growth substrates. We have clearly observed the strong dependency of P. placenta on the Fenton's reaction system for the modification and degradation of lignocellulosic components. The higher expression of genes encoding for hemicellulolytic glycoside hydrolases even under cellulolytic conditions proves the synchronized action of both cellulolytic and hemicellulolytic hydrolytic enzymes. Based on the reports from previous plant biomass degradation studies and the results obtained in our present study clearly show that, P. placenta exhibits stronger ability to degrade cellulose and hemicellulose by selectively modifying lignin. Even though P. placenta has experienced an evolutionary loss of genes encoding for CAZymes, it is well known for its rapid cellulolytic and lignin modifying abilities which can be credited to its outstanding oxidizing capacities. The significant differentially expressed genes from all the datasets were compared and the protein Id's of the common highly expressed genes were also reported. However, level of gene expression does not completely explain the lignocellulose degradation mechanisms employed by the P. placenta, as the gene expression studies are the snapshot of the cell at a particular moment. Simultaneously, several functionally uncharacterized proteins were also found to be highly expressed among the P. placenta datasets. Further studies must be conducted to exactly understand and reveal the functional roles of these commonly observed significant genes and various other uncharacterized proteins. Understanding the rapid degradation of cellulosic and hemicellulosic units by brown rot fungi will significantly benefit the growing biofuel and biorefining industries, through exploring its unprecedented oxidizing abilities.
Supplementary Material
Supplementary Information-S1.
Supplementary Information-S2.
Supplementary Information-S3.
Acknowledgements
This work was supported by Natural Sciences and Engineering Research Council of Canada Funding (RGPIN-2017-05366) to Wensheng Qin and Ontario Trillium Scholarship (OTS) to Ayyappa Kumar Sista Kameshwar.
Availability of data and materials
All the datasets considered in our present study is publicly available from NCBI GEO repository.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP. et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proceedings of the National Academy of Sciences. 2009;106:1954-9
2. Schilling JS, Kaffenberger JT, Liew FJ, Song Z. Signature wood modifications reveal decomposer community history. PloS one. 2015;10:e0120679
3. Binder M, Hibbett DS, Larsson KH, Larsson E, Langer E, Langer G. The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Systematics and Biodiversity. 2005;3:113-57
4. Hibbett DS, Donoghue MJ. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Systematic biology. 2001:215-42
5. Arantes V, Goodell B. Current understanding of brown-rot fungal biodegradation mechanisms: a review. Deterioration and protection of sustainable biomaterials. 2014;1158:3-21
6. Goodell B, Jellison J, Liu J, Daniel G, Paszczynski A, Fekete F. et al. Low molecular weight chelators and phenolic compounds isolated from wood decay fungi and their role in the fungal biodegradation of wood. Journal of Biotechnology. 1997;53:133-62
7. Xu G, Goodell B. Mechanisms of wood degradation by brown-rot fungi: chelator-mediated cellulose degradation and binding of iron by cellulose. Journal of biotechnology. 2001;87:43-57
8. Hirano T, Tanaka H, Enoki A. Extracellular substance from the brown-rot basidiomycete Tyromyces palustris that reduces molecular oxygen to hydroxyl radicals and ferric iron to ferrous iron. Journal of the Japan Wood Research Society (Japan). 1995
9. Kerem Z, Jensen KA, Hammel KE. Biodegradative mechanism of the brown rot basidiomycete Gloeophyllum trabeum: evidence for an extracellular hydroquinone-driven fenton reaction. FEBS letters. 1999;446:49-54
10. Daniel G, Volc J, Filonova L, Plíhal O, Kubátová E, Halada P. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Applied and environmental microbiology. 2007;73:6241-53
11. Shimokawa T, Nakamura M, Hayashi N, Ishihara M. Production of 2, 5-dimethoxyhydroquinone by the brown-rot fungus Serpula lacrymans to drive extracellular Fenton reaction. Holzforschung. 2004;58:305-10
12. Suzuki MR, Hunt CG, Houtman CJ, Dalebroux ZD, Hammel KE. Fungal hydroquinones contribute to brown rot of wood. Environmental microbiology. 2006;8:2214-23
13. Rättö M, Ritschkoff A-C, Viikari L. The effect of oxidative pretreatment on cellulose degradation by Poria placenta and Trichoderma reesei cellulases. Applied microbiology and biotechnology. 1997;48:53-7
14. Yelle DJ, Ralph J, Lu F, Hammel KE. Evidence for cleavage of lignin by a brown rot basidiomycete. Environmental microbiology. 2008;10:1844-9
15. Martínez AT, Rencoret J, Nieto L, Jiménez-Barbero J, Gutiérrez A, del Río JC. Selective lignin and polysaccharide removal in natural fungal decay of wood as evidenced by in situ structural analyses. Environmental microbiology. 2011;13:96-107
16. Wymelenberg AV, Gaskell J, Mozuch M, BonDurant SS, Sabat G, Ralph J. et al. Gene expression of wood decay fungi Postia placenta and Phanerochaete chrysosporium is significantly altered by plant species. Applied and Environmental Microbiology. 2011 AEM. 00508-11
17. Yelle DJ, Wei D, Ralph J, Hammel KE. Multidimensional NMR analysis reveals truncated lignin structures in wood decayed by the brown rot basidiomycete Postia placenta. Environmental Microbiology. 2011;13:1091-100
18. Micales JA. Localization and induction of oxalate decarboxylase in the brown-rot wood decay fungus Postia placenta. International biodeterioration & biodegradation. 1997;39:125-32
19. Wei D, Houtman CJ, Kapich AN, Hunt CG, Cullen D, Hammel KE. Laccase and its role in production of extracellular reactive oxygen species during wood decay by the brown rot basidiomycete Postia placenta. Applied and environmental microbiology. 2010;76:2091-7
20. Skyba O, Cullen D, Douglas CJ, Mansfield SD. Gene expression patterns of wood decay fungi Postia placenta and Phanerochaete chrysosporium are influenced by wood substrate composition during degradation. Applied and environmental microbiology. 2016;82(14):4387-400 doi: 10.1128/AEM.00134-16
21. Zhang J, Presley GN, Hammel KE, Ryu J-S, Menke JR, Figueroa M. et al. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proceedings of the National Academy of Sciences. 2016. 2016 08454
22. Wymelenberg AV, Gaskell J, Mozuch M, Sabat G, Ralph J, Skyba O. et al. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Applied and environmental microbiology. 2010;76:3599-610
23. Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R. et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research. Epub. 2013:42 (Database issue)(D):699-704. doi: 10.1093/nar/gkt1183
24. Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP. et al. Fueling the future with fungal genomics. Mycology. 2011;2:192-209
25. Botstein D, Cherry JM, Ashburner M, Ball C, Blake J, Butler H. et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25-9
26. Consortium GO. Gene ontology consortium: going forward. Nucleic acids research. 2015;43:D1049-D56
27. Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV. et al. The COG database: an updated version includes eukaryotes. BMC bioinformatics. 2003;4:41
28. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic acids research. 2009;37:D233-D8
29. Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic acids research. 2014;42:D490-D5
30. Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007
31. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC bioinformatics. 2014;15:1
32. Kameshwar AKS, Qin W. Metadata Analysis of Phanerochaete chrysosporium Gene Expression Data Identified Common CAZymes Encoding Gene Expression Profiles Involved in Cellulose and Hemicellulose Degradation. International Journal of Biological Sciences. 2017;13:85-99
33. Kameshwar AKS, Qin W. Gene expression metadata analysis reveals molecular mechanisms employed by Phanerochaete chrysosporium during lignin degradation and detoxification of plant extractives. Current Genetics. 2017:1-18
34. Kameshwar AKS, Qin W. Analyzing Phanerochaete chrysosporium gene expression patterns controlling the molecular fate of lignocellulose degrading enzymes. Process Biochemistry. 2017
35. Grinhut T, Salame TM, Chen Y, Hadar Y. Involvement of ligninolytic enzymes and Fenton-like reaction in humic acid degradation by Trametes sp. Applied microbiology and biotechnology. 2011;91:1131-40
36. Highley TL, Illman BL. Progress in understanding how brown-rot fungi degrade cellulose. Biodeterioration Abstracts. 1991;5:231-44
37. Lee SO, Tran T, Park YY, Kim SJ, Kim MJ. Study on the kinetics of iron oxide leaching by oxalic acid. International Journal of Mineral Processing. 2006;80:144-52
38. Korripally P, Timokhin VI, Houtman CJ, Mozuch MD, Hammel KE. Evidence from Serpula lacrymans that 2, 5-dimethoxyhydroquinone is a lignocellulolytic agent of divergent brown rot basidiomycetes. Applied and environmental microbiology. 2013;79:2377-83
39. Newcombe D, Paszczynski A, Gajewska W, Kröger M, Feis G, Crawford R. Production of small molecular weight catalysts and the mechanism of trinitrotoluene degradation by several Gloeophyllum species. Enzyme and Microbial Technology. 2002;30:506-17
40. Fackler K, Stevanic JS, Ters T, Hinterstoisser B, Schwanninger M, Salmén L. Localisation and characterisation of incipient brown-rot decay within spruce wood cell walls using FT-IR imaging microscopy. Enzyme and microbial technology. 2010;47:257-67
41. Schilling JS, Tewalt JP, Duncan SM. Synergy between pretreatment lignocellulose modifications and saccharification efficiency in two brown rot fungal systems. Applied microbiology and biotechnology. 2009;84:465
42. Cohen R, Suzuki MR, Hammel KE. Processive endoglucanase active in crystalline cellulose hydrolysis by the brown rot basidiomycete Gloeophyllum trabeum. Applied and environmental microbiology. 2005;71:2412-7
43. Jung YH, Kim HK, Park HM, Park Y-C, Park K, Seo J-H. et al. Mimicking the Fenton reaction-induced wood decay by fungi for pretreatment of lignocellulose. Bioresource technology. 2015;179:467-72
44. Barr DP, Aust SD. Mechanisms white rot fungi use to degrade pollutants. Environmental Science & Technology. 1994;28:78A-87A
45. Agosin E, Jarpa S, Rojas E, Espejo E. Solid-state fermentation of pine sawdust by selected brown-rot fungi. Enzyme and microbial technology. 1989;11:511-7
46. Filley T, Cody G, Goodell B, Jellison J, Noser C, Ostrofsky A. Lignin demethylation and polysaccharide decomposition in spruce sapwood degraded by brown rot fungi. Organic Geochemistry. 2002;33:111-24
47. Kirk TK. Effects of a brown-rot fungus, Lenzites trabea, on lignin in spruce wood. Holzforschung-International Journal of the Biology, Chemistry, Physics and Technology of Wood. 1975;29:99-107
48. Kirk TK, Adler E. Methoxyl-deficient structural elements in lignin of sweetgum decayed by a brown-rot fungus. Acta Chem Scand. 1970;24:90
49. Koenig AB, Sleighter RL, Salmon E, Hatcher PG. NMR structural characterization of Quercus alba (white oak) degraded by the brown rot fungus, Laetiporus sulphureus. Journal of wood chemistry and technology. 2010;30:61-85
50. Eriksson K-EL, Blanchette R, Ander P. Microbial and enzymatic degradation of wood and wood components. Springer Science & Business Media. 2012
51. Arantes V, Qian Y, Kelley SS, Milagres AM, Filley TR, Jellison J. et al. Biomimetic oxidative treatment of spruce wood studied by pyrolysis-molecular beam mass spectrometry coupled with multivariate analysis and 13C-labeled tetramethylammonium hydroxide thermochemolysis: implications for fungal degradation of wood. JBIC Journal of Biological Inorganic Chemistry. 2009;14:1253
52. Arantes V, Jellison J, Goodell B. Peculiarities of brown-rot fungi and biochemical Fenton reaction with regard to their potential as a model for bioprocessing biomass. Applied Microbiology and Biotechnology. 2012;94:323-38
53. Hyde SM, Wood PM. A mechanism for production of hydroxyl radicals by the brown-rot fungus Coniophora puteana: Fe (III) reduction by cellobiose dehydrogenase and Fe (II) oxidation at a distance from the hyphae. Microbiology. 1997;143:259-66
54. Arantes V, Milagres AM, Filley TR, Goodell B. Lignocellulosic polysaccharides and lignin degradation by wood decay fungi: the relevance of nonenzymatic Fenton-based reactions. Journal of industrial microbiology & biotechnology. 2011;38:541-55
55. Hocine S, Singer RH, Grünwald D. RNA processing and export. Cold Spring Harbor perspectives in biology. 2010;2:a000752
56. Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Progress in nucleic acid research and molecular biology. 2000;66:1-40
57. Busch H, Reddy R, Rothblum L, Choi Y. SnRNAs, SnRNPs, and RNA processing. Annual review of biochemistry. 1982;51:617-54
58. Amore A, Giacobbe S, Faraco V. Regulation of cellulase and hemicellulase gene expression in fungi. Current genomics. 2013;14:230-49
Author contact
![]() Corresponding author: Wensheng Qin, Email: wqinca, Tel: 807-343-8467
Corresponding author: Wensheng Qin, Email: wqinca, Tel: 807-343-8467

 Global reach, higher impact
Global reach, higher impact