10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(6):633-643. doi:10.7150/ijbs.25018 This issue Cite
Review
Salivary Exosomes: Emerging Roles in Systemic Disease
1. Department of Orthodontics, Peking University School and Hospital of Stomatology, Beijing,100081, China
2. Central Laboratory, Peking University School and Hospital of Stomatology, Beijing, 100081, China
3. Department of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology, Beijing 100081, China
Received 2018-1-18; Accepted 2018-3-9; Published 2018-4-30
Abstract
Saliva, which contains biological information, is considered a valuable diagnostic tool for local and systemic diseases and conditions because, similar to blood, it contains important molecules like DNA, RNA, and proteins. Exosomes are cell-derived vesicles 30-100 nm in diameter with substantial biological functions, including intracellular communication and signalling. These vesicles, which are present in bodily fluids, including saliva, are released upon fusion of multivesicular bodies (MVBs) with the cellular plasma membrane. Salivary diagnosis has notable advantages, which include noninvasiveness, ease of collection, absence of coagulation, and a similar content as plasma, as well as increased patient compliance compared to other diagnostic approaches. However, investigation of the roles of salivary exosomes is still in its early years. In this review, we first describe the characteristics of endocytosis and secretion of salivary exosomes, as well as database and bioinformatics analysis of exosomes. Then, we describe strategies for the isolation of exosomes from human saliva and the emerging role of salivary exosomes as potential biomarkers of oral and other systemic diseases. Given the ever-growing role of salivary exosomes, defining their functions and understanding their specific mechanisms will provide novel insights into possible applications of salivary exosomes in the diagnosis and treatment of systemic diseases.
Keywords: saliva, exosome, biomarker, proteomics, genomics
Introduction
As a biological fluid, saliva is composed of more than 99% water and less than 1% proteins, electrolytes, and other low molecular weight components [1]. In the oral cavity, saliva primarily comes from three pairs of major salivary glands, the parotid, submandibular, and sublingual glands, and 300-400 minor salivary glands [2]. It plays a pivotal role in mastication, lubrication, swallowing, and digestion, protecting the integrity of oral tissues [3]. In addition, saliva also provides clues for local and systemic diseases and conditions because molecules like DNA, RNA, proteins, metabolites, and microbiota are not only present in blood but also in saliva. Therefore, changes in the concentrations of these molecules could be useful as potential biomarkers to predict or detect diseases as well as monitor the therapeutic response [4].
Exosomes are cell-derived vesicles, 30-100 nm in diameter, generated by the endosomal pathway and released through exocytosis of multivesicular bodies (MVBs) to the extracellular space and circulation [5]. Exosomes mediate interactions with the cellular environment, and exist extensively in bodily fluids, such as blood, urine, saliva, bile, cerebrospinal fluid, breast milk, epididymal fluid, semen, ascites, amniotic fluid, and sputum [6]. Exosomes are spherical and, in addition to lipids, the lipid bilayer contains polysaccharides and protein receptors [7, 8]. The content of exosomes is diverse, and includes lipids, enzymes, proteins, RNAs and, in particular, microRNAs (miRNAs) [9], indicating that exosomes play an important role in intracellular communication and in delivering molecular signals from one cell to another at proximal or distal locations [10, 11]. Exosomes are involved in intracellular communication, immune regulation, RNA processing and degradation, tumour promotion, propagation of prion proteins and retroviruses, and can serve as drug delivery vehicles [12-14]. The biological functions of exosomes largely rely on interactions between the exosome and its target cell.
Currently, salivary diagnosis is drawing increasing attention as it is noninvasive, easy to collect, and exhibits greater patient compliance compared with other methods. Additionally, the contents of saliva are similar to those of plasma, owing to the physical interaction between saliva and blood sources [15]. Moreover, saliva has advantages over blood as a bodily fluid as it does not coagulate, which makes it easier to be processed than blood. However, using whole saliva has several disadvantages, including contamination, variability, and the presence of proteins such as amylase that can mask other proteins with low expression. Therefore, salivary exosomes have been investigated to overcome these barriers. A previous study showed that exosomes containing proteins, miRNAs, and mRNAs (exosome shuttle RNA) in various bodily fluids including plasma, malignant ascites, urine, amniotic fluid, and saliva could serve as a novel platform for diagnosis [16]. Notably, extracellular vesicles (EVs) have been found to cross epithelial barriers such as the blood-brain barrier via transcytosis, indicating that they may play a role in transporting RNAs of systemic origin from blood into saliva [17, 18]. Machida et al. [19] identified miR-1246 and miR-4644 in salivary exosomes, which are highly expressed in serum exosomes, as potential biomarkers of pancreatobiliary tract cancer. Moreover, few studies on humans have reported the role of exosomes in other bodily fluids, such as urine, in systemic diseases. For example, the study of urinary exosomes has been limited to kidney-related diseases, such as polycystic kidney disease [20] and aldosteronism [21], and rarely related to other systemic diseases. Therefore, uncovering the role of salivary exosomes may be a superior strategy to exploring the role of exosomes in other bodily fluids. It would be interesting to investigate the role of exosomes in saliva and to understand the mechanisms underlying their regulation in systemic diseases, providing novel insights into the possible applications of salivary exosomes in the noninvasive diagnosis and treatment of systemic diseases. In this review, we first discuss the characteristics and composition of salivary exosomes, as well as the processes of endocytosis and secretion. Then, we describe strategies for the isolation of exosomes from human saliva and the emerging role of salivary exosomes as potential biomarkers in oral and other systemic diseases.
Characteristics of salivary exosomes
Similar to exosomes released in other bodily fluids [22], electron micrographs have revealed that salivary exosomes are rounded cup-shaped vesicles with an average diameter of 30-100 nm and abundant CD63 immunoreactivity on the surface [23]. According to their distinctive size and protein composition, salivary exosomes in whole saliva can be classified into two types, exosomes I and II. The average diameter of exosome I is 83.5 nm, while exosome II is 40.5 nm, as determined by electron microscopy [24]. Both types of exosomes contain exosomal biomarkers, which include Alix, tumour susceptibility gene 101 (TSG101), and heat shock protein (HSP) 7, and both contain immunoglobulin (Ig) A and the polymeric Ig receptor (pIgR) [24]. Overall, 101 and 154 proteins have been identified in exosomes I and II, respectively, and 68 proteins with common markers (CD63, Alix, Tsg101, and Hsp70) overlap between the two groups. Nearly 40% of these proteins are secretory proteins, including serum albumin or extracellular proteins such as Ig chains, suggesting that salivary exosomes originate from circulating lymphocytes and intravascular fluid [25]. Xiao et al. [26] performed proteomic profiling of salivary microvesicles with an average diameter of 100-1000 nm and identified 63 proteins in salivary microvesicles, of which 35 were exclusively identified in microvesicles by comparison with 491 proteins identified in the parotid exosome proteome [27], indicating that salivary exosomes contain their own unique proteins. Exosomes II primarily express dipeptidyl peptidase IV (DPP IV), which is also known as CD26, and is metabolically active in cleaving the chemokines CXCL11 and CXCL12 [24], implying that exosome II is involved in the degradation of polypeptides. DPP IV, carbonic anhydrase 6, cystatin family proteins, IgG, Fc binding protein, and galectin-3 binding protein are highly expressed in type II exosomes, while ezrin, moesin, radixin, Rab GDP dissociation inhibitor β, α-enolase, guanine nucleotide-binding protein Gi/Gs/Gt subunit β-1, and annexins are only expressed in exosomes I. The heterogeneous structure of salivary exosomes may imply that exosomes originate from different parts of the salivary glands. For example, galectin-3 binding protein is only located on the surface of ductal epithelial cells, while pIgR localises mostly in acinar and ductal epithelial cells. Exosomes I and II both contain IgA, indicating that they are possibly both involved in immune responses. Ogawa et al. [24] implanted exosomes I and II into different mice and observed that mice implanted with exosomes II generated antibodies to DPP IV more successfully, indicating a better immunogenicity of exosome II and that exosomes I and II exhibit different proteomic properties. It has been reported that saliva can trigger factor VII-mediated coagulation of human plasma and that salivary exosomes can significantly shorten the clotting time of exosome-depleted plasma. Western blot analysis and transmission electron microscopy with immunogold labelling have been used to further identify the initiator of coagulation activation (tissue factor) in salivary exosomes. Moreover, in the presence of anti-factor VII, salivary exosome-induced shortening of clotting time is abolished, indicating that salivary exosomes serve as the first step in the process of wound healing to facilitate hemostasis [28].
Composition of salivary exosomes
The secretion of ribonucleoproteins, which are immune antigens involved in systematic rheumatic diseases in exosomes of non-neoplastic salivary gland epithelial cells, was first detected in 2005 [29]. The combined use of electron microscopy, immunoblotting, and immunoprecipitation techniques has resulted in the identification of numerous salivary exosomes. Gonzalez et al. [27] used multidimensional protein identification technology (MudPIT) to catalog 491 proteins in the exosome fraction of human parotid saliva. Among these proteins, 265 were observed in ductal saliva from the parotid glands and 72 proteins overlapped with urinary exosome proteins, indicating that exosomes of different origins exhibit some degree of similarity. Ogawa et al. [30] identified DPP IV, galectin-3, and IgA may influence the immune response. Vesicular DPP IV was metabolically active, cleaving substance P and glucose-dependent insulinotropic polypeptide to release N-terminal dipeptides, indicating that salivary exosomes participate in the catabolism of bioactive peptides and play a regulatory role in the local immune response in the oral cavity. The exosomal membrane proteins CD9, CD63, CD81, as well as intracellular proteins like HSP70 and TSG101, are considered markers of exosomes. Salivary exosomes also contain various RNAs, most of which are small RNAs. Because of the small size of noncoding RNAs (ncRNAs), these molecules are not as susceptible as mRNAs to degradation by ribonucleases, and are thus much more stable. It has been reported that salivary miRNAs and P-Element induced wimpy testis (PIWI)-interacting RNAs (piRNAs) are predominantly found in exosomes [31]. Bahn et al. [15], using high-throughput RNA sequencing, found that the most abundant types of small RNAs in human cell-free whole saliva are Piwi-interacting RNAs, miRNAs, and small nucleolar RNAs, accounting for 7.5%, 6.0%, and 0.02%, of all small RNAs, respectively. Therefore, non-coding RNAs have been extensively investigated as regulators of diverse biological functions. Ogawa et al. [32] used next-generation sequencing technology to investigate the compositions of protein-coding RNAs and long non-protein-coding RNAs in exosomes and whole saliva. The results showed that exosomes I and II and WS express miRNAs differently and that most of the miRNAs expressed in WS are not expressed in exosomes. In exosomes, a component of the RNA-induced silencing complex GW182 is important for the stability of exosomal RNAs. In exosomes I and II and WS, the miRNA hsa-mir-378 is associated with cell survival, tumour growth, and angiopoiesis [33]. Gallo et al. [31] reported that miR-22, miR-202, and miR-1237d are predominant in salivary exosomes. However, the specific functions and mechanisms of these exosomal RNAs require further exploration.
Endocytosis and secretion of salivary exosomes
Exosomes exert their biological functions via endocytosis and secretion of their contents into host cells. In salivary exosomes, this process is similar to the endocytosis and secretion of exosomes in other bodily fluids. Endocytosis is the first step (Figure 1). The internalized material delivered to early endosomes is sorted to at least three possible destinations irrespective of the route of entry. One is for degradation through maturation into MVBs and fusion with lysosomes, which are acidic compartments containing hydrolytic enzymes able to digest complex macromolecules [34]. The second is for recycling. According to the time that passes from internalization to exposure at the cell surface, or release to the extracellular medium in the case of luminal soluble factors, recycling processes can be divided into a rapid and slower pathway. The third destination is secretion, which will be described in detail below. Phagocytosis, which occurs primarily in phagocytic cells, or receptor and raft-mediated endocytosis, are part of the endocytic process [35-37]. It has been shown that RAW 264.7 macrophage cells internalize exosomes derived from K562 and MT4 cell lines through actin-mediated and phosphatidylinositol 3-kinase (PI3K) and dynamin 2-dependent pathways [35]. Similarly, the internalized and engulfed exosomes are merged with endosomes of the recipient cell and can be transported to neighboring cells in pancreatic cancer [38]. Receptor-mediated endocytosis occurs via the classical or non-classical pathway. The classical pathway is mediated by caveolin or clathrin membrane proteins (cavoelae are specialized lipid rafts). Exosomes derived from virus-infected cells have been shown to be internalized by target cells via caveolin-dependent endocytosis. Knockdown of the calveolin 1 (CAV1) gene significantly reduced exosome uptake, indicating that caveolin membrane proteins are involved in endocytosis [39]. Bone marrow-derived mesenchymal stromal cells were reported to take up PC12 cell-derived exosomes via clathrin-mediated endocytosis and alter gene expression through the transfer of miR-21 [40]. Additionally, the endocytosis of exosomes induced secretion of pro-inflammatory cytokines by placental cells, demonstrating an exosome-dependent change in placental phenotype. On the other hand, non-classical endocytosis of exosomes occurs independently of membrane proteins. It has been reported that exosome uptake by glioblastoma cells occurs via lipid raft-mediated endocytosis and is dependent on extracellular signal-regulated kinase-1/2 and HSP27 [37]. Another form of exosome-cell interaction is the adhesion of exosomes to a potential docking site on target cells. This interaction is facilitated by the presence of transmembrane proteins on the surface of exosomes. Dendritic cell-derived exosomes express intercellular adhesion molecule-1, major histocompatibility complex, and co-stimulatory molecules that enable exosomes to interact with target cells via their respective signalling receptors [41-43]. By interacting with recipient cells, exosomes can transfer their cargo, which can regulate recipient cell function. This can orchestrate diverse signalling pathways and mediate a broad range of physiological and pathological conditions. Cellular responses to the microenvironment play a decisive role in determining the concentration and composition of exosomes. This has opened up new avenues for biomarker discovery and therapeutic interventions [44-46].
Exosome endocytosis and secretion. Early endosomes are formed after internalization when the receptor binds the ligand. Proteins in the early endosome can either be recycled to the plasma membrane or become part of the luminal vesicle (ILV) of multivesicular bodies (MVBs). ILVs of MVBs are generated by budding from the limiting membrane and enter the lumen of endosomes. Several molecules, such as RAB11 and RNA35, are involved in recycling and early sorting endosomes. MVBs can fuse with lysosomes or the plasma membrane, releasing their contents to the extracellular medium. RAB11 and RAB27 are linked with the late endosomal and secretory compartments. Neighboring cells can internalize the cytoplasmic contents of the exosomes, which can also mature into MVBs.
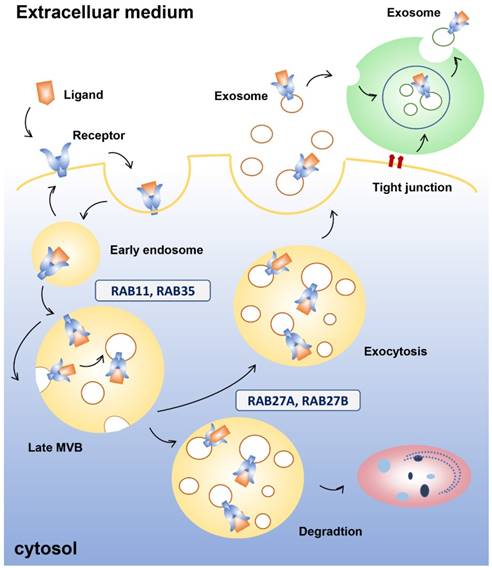
A number of genes are involved in exosomal secretion [47], which ensure that exosomes are transported to the right place at the right time. First, proteins in the early exosome, which has two primary mechanisms of transformation following endocytosis in which the receptors bind the ligands, can either combine with the plasma membrane or become part of the luminal vesicle (ILV) of MVBs. ILVs of MVBs are generated by budding from the limiting membrane and enter the lumen of endosomes. MVBs undergo degradation in lysosomes or fuse with the plasma membrane to undergo exocytosis. The endosomal sorting complexes required for the transport machinery, which is used in transport and ubiquitination, generate the degradative MVBs. Exocytic MVBs fuse with the plasma membrane and exosomes are released into the microenvironment. Neighboring cells can internalize the cytoplasmic content of exosomes, which can also mature into MVBs [48, 49]. During secretion, the RAB family of small GTPase proteins are involved in different steps that lead to membrane fusion, including vesicle budding, vesicle and organelle mobility through cytoskeleton interaction, and docking of vesicles to their target compartment, which is required for eventual fusion of the two membranes [50]. For example, RAB11 has been shown to control transferrin receptor and HSP70-containing exosome release in K562 cells [51]. In HeLa-CIITA cells, depletion of RAB5A, RAB9A, RAB2B, and more remarkably RAB27A and RAB27B, decreased exosome secretion, while depletion of RAB11A or RAB7 did not [52]. Additionally, RAB27A was confirmed to be involved in vesicle secretion in numerous tumour cell lines such as mammary carcinoma [53], murine melanoma [54], and human squamous carcinoma cells [55]. Moreover, different RABs are involved in the docking and fusion machineries, indicating the existence of different multivesicular compartments within the cell. RAB27A and RAB27B have been linked with the late endosomal and secretory compartments, while RAB11 and RAB35 have been linked with the recycling and early sorting endosomes [56]. This evidence suggests that different MVBs could produce different subpopulations of EVs.
Soluble NSF-attachment protein receptor (SNARE) complexes are important in the fusion of lipid bilayers [56]. Though the SNARE proteins SNAP-23, VAMP7, and VAMP8 participate in Ca2+-regulated fusion of secretory lysosomes with the plasma membrane in different cell types [57, 58], they are also essential in the fusion of MVBs with the plasma membrane during exosome secretion. VAMP7 is required for the release of acetylcholinesterase-containing EVs in K562 erythroleukemia cells [59]. When VAMP7 was inhibited in MDCK cells, lysosomal secretion was impaired, although there was no effect on exosome release [60]. More recently, the R-SNARE protein YKT6 was reported to be necessary for the secretion of exosomes containing the WNT3A morphogen in HEK293 cells [61].
Isolation and identification of exosomes in saliva
Multiple methods have been described for the isolation of exosomes, including differential centrifugation, density gradient centrifugation, size exclusion chromatography, filtration, polymer-based precipitation, immunological separation, and commercial exosome isolation kits. [62]. For salivary exosome isolation, there are two primary methods: the classical, physical-based method of ultracentrifugation (UC) [63], and ExoQuick-TC™ (EQ), a chemical-based agent designed to precipitate exosomes [64]. It is technically difficult to isolate exosomes from biological materials because the isolations are easily contaminated with non-exosomal proteins and the cytomembrane can discharge exosomes as well as many other kinds of subcellular bodies. UC is believed to obtain minimally contaminated exosome pellets, but the process is complicated, prolonged, and requires specialized equipment. When EQ is incubated with saliva, intact exosomes can be obtained and this method is considered suitable and efficient for the precipitation of salivary exosomes from small volumes of saliva. However, EQ tends to be associated with considerably more biological impurities (non-exosomal-related proteins/microvesicles) compared with UC [64].
Isolation methods have been developed with the extensive study of salivary exosomes. Kim et al. [65] isolated EVs from the saliva of mouse models with human melanoma using an aqueous two-phase system (ATPS). Most EVs moved into the dextran phase after centrifugation when the ATPS was composed of polyethylene glycol and dextran [66, 67]. With this method, EVs are recovered from cell-free saliva more easily and efficiently than by conventional isolation methods such as UC.
After isolation, transmission electron microscopy is used to identify specific exosomes, which have been described as having a “cup-shaped” morphology. Furthermore, using the ultrasensitive technique of low-force atomic force microscopy, exosomal structure, chemistry, and mechanics can be examined [68]. In addition, on continuous sucrose gradients, exosomes equilibrate at densities between 1.13 and 1.19 g/ml [63]. Exosome-specific markers are important for characterizing exosomes and differentiating them from other EVs. However, the development of novel techniques for the study of exosomes is necessary, and this will aid in the discovery of additional exosomal biomarkers and their use in the diagnosis of various diseases.
Correlation between salivary exosomes and systemic diseases
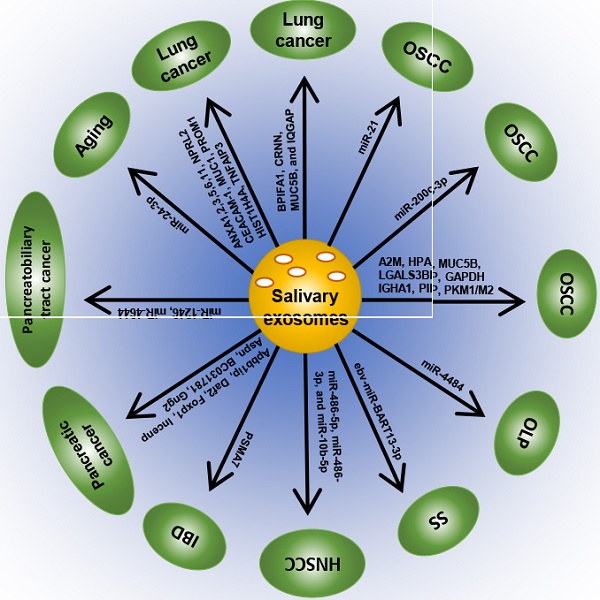
Accumulating evidence has revealed that salivary exosomes are correlated with oral diseases like oral cancer [67], oral lichen planus (OLP) [68], Sjögren's syndrome (SS) [69] as well as other systemic diseases like inflammatory bowel disease (IBD) [70], pancreatic cancer [71], pancreatobiliary tract cancer [19], lung cancer [72] and so on. Several investigations have been carried out to explore the aberrant expression and underlying mechanisms of salivary exosomes, which may be considered as potential diagnostic and therapeutic biomarkers for systemic diseases (Table 1; Figure 2).
Exosome biomarkers for systemic diseases
| diseases | biomarker | type | Expression | Sample | Methods | Reference |
|---|---|---|---|---|---|---|
| OSCC | miRNA-21 | miRNA | Upregulated | OSCC cell line | miRNA sequencing | Li et al. [78] Liu et al. [79] |
| miR-200c-3p | miRNA | Upregulated | OSCC cell line | integrated microarray | Kawakubo-Yasukochi et al. [80] | |
| A2M, HPA, MUC5B, LGALS3BP, IGHA1, PIP, PKM1/M2, GAPDH | proteins | differentially expressed | saliva | Mass spectrometry analysis and proteomics data analysis | Winck et al. [81] | |
| OLP | miR-4484 | miRNA | Upregulated | saliva | miRNA microarray | Byun et al. [69] |
| SS | ebv-miR-BART13-3p | miRNA | transferred from B cells to salivary epithelial cells | salivary glands of SS patients | / | Gallo et al. [84] |
| HNSCC | miR-486-5p, miR-486-3p, and miR-10b-5p | miRNAs | Upregulated | Both HNSCC cell line and saliva | miRNA-sequencing | Langevin et al. [92] |
| IBD | PSMA7 | proteins | Upregulated | saliva | LC-MS/MS | Zheng et al. [71] |
| Pancreatic cancer | Apbb1ip, Daf2, Foxp1, Incenp, Aspn, BC031781, Gng2 | mRNA | Upregulated | rodent model with pancreatic cancer | micorarray | Lau et al. [72] |
| Pancreatobiliary tract cancer | miR‑1246, miR‑4644 | miRNA | Upregulated | saliva | RT‑qPCR | Machida et al. [19] |
| aging process. | miR-24-3p | miRNA | Upregulated | saliva | microarray | Machida et al. [95] |
| Lung cancer | ANXA1, ANXA2, ANXA3, ANXA5, ANXA6, ANXA11, NPRL2, CEACAM-1, MUC1, PROM1, HIST1H4A and TNFAIP3. | proteins | Upregulated | saliva | Sun et al. [73] | |
| BPIFA1, CRNN, MUC5B, and IQGAP | proteins | Upregulated | saliva | Sun et al. [96] |
A2M = alpha-2-macroglobulin; ANX A1,2,3,5,6,11= annexin A1,2,3,5, 6, 11; BPIFA1 = BPI fold-containing family A member 1; CEACAM1 =carcinoembryonic antigen-related cell adhesion molecule 1; CRNN = Cornulin; ebv-miR-BART13-3p = Epstein Barr Virus (EBV) -specific microRNA; GAPDH = glyceraldehyde-3-phosphate dehydrogenase; HIST1H4A = histone H4; HNSCC = head and neck squamous cell carcinoma; HPA = haptoglobin alpha chain; IBD = inflammatory bowel disease; IGHA1 = immunoglobulin alpha-1 chain c region; IQGAP1 = Ras GTPase-activating-like protein; LC-MS/MS = liquid chromatograph-mass spectrometer; LGALS3BP = galectin-3-binding protein; miRNA = microRNA; mRNA = message RNA; MUC1 = mucin 1; mucin-5B = MUC5B; NPRL2 = nitrogen permease regulator 2-like protein; OLP = Oral lichen planus; OSCC = oral squamous cell carcinoma; PIP = prolactin-inducible protein; PKM1/M2 = pyruvate kinase isozymes M1/M2; PROM1 = prominin-1; PSMA7 = proteasome subunit alpha type 7; RT qPCR = quantitative real time PCR; SS = Sjögren's syndrome; TNFAIP3 = tumour necrosis factor alpha-induced protein 3.
Salivary exosomes as potential biomarkers of oral diseases
As mentioned above, saliva contains a variety of proteins, enzymes, and immunoglobulins, which form the first line of defense. Proteomic analysis of whole human saliva has demonstrated that exosomes are rich in IgA and plgR, which play an important anti-inflammatory role [24]. Oral squamous cell carcinoma (OSCC) is the most prevalent oral cancer, accounting for more than 90% of oral cancers and 24% of all head and neck cancers and has a poor prognosis [73]. This cancer is a major challenge as it is often diagnosed at an advanced stage and more than 50% of OSCC patients exhibit lymph node metastasis. [74, 75]. Though advances in surveillance and treatment strategies have been made, the survival rate of OSCC patients has not significantly improved and is still below 50% [76]. Zlotogorski-Hurvitz et al. [67] showed that the morphological and molecular features of exosomes differ between oral cancer patients and healthy individuals, providing an early diagnostic tool for detecting malignant changes in high-risk patients. Li et al. [77] found that exosomes derived from hypoxic OSCC cells increased the migration and invasion of OSCC cells in a HIF-1α- and HIF-2α-dependent manner. miRNA sequencing of normoxic and hypoxic OSCC-derived exosomes indicated that 108 miRNAs were differentially expressed. Among them, miR-21 was the most significantly upregulated miRNA under hypoxic conditions. This study showed that a hypoxic microenvironment could stimulate tumour cells to generate miR-21-rich exosomes that are delivered to normoxic cells to promote prometastatic behavior. In addition, exosomes containing the miR-21 targets phosphatase and tensin homolog (PTEN) and programmed cell death protein 4 (PDCD4) exhibit cisplatin resistance in OSCC [78]. Moreover, Kawakubo-Yasukochi et al. [79] indicated that exosomes containing miR-200c-3p derived from a highly invasive OSCC line could induce a similar phenotype in non-invasive counterparts. In proteomic profiling analyses, Winck et al. [80] identified 381 proteins from salivary EVs in OSCC and healthy groups and found that 8 were differentially expressed, including alpha-2-macroglobulin (A2M), haptoglobin alpha chain (HPA), mucin 5B (MUC5B), galectin-3-binding protein (LGALS3BP), immunoglobulin alpha-1 chain c region (IGHA1), prolactin-inducible protein(PIP), pyruvate kinase isozymes M1/M2 (PKM1/M2), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Oral lichen planus (OLP) is a chronic inflammatory oral mucosal disease with unclear cause. Byun et al. [68] examined salivary exosomal miRNAs of patients with OLP and found that miR-4484 were significantly upregulated and, thus, could be candidates for diagnosing and elucidating the pathogenesis of OLP. Sjögren's syndrome (SS), first described in 1933 by the Swedish physician Henrik Sjögren [81], is a chronic autoimmune disorder with the characteristics of dry mouth (xerostomia) and dry eyes (keratoconjunctivitis sicca) [82]. Hu et al. [69] identified 16 peptides (10 upregulated and 6 downregulated in primary SS) and 27 mRNAs whose expression was significantly altered in whole saliva of primary SS patients compared with healthy individuals. These candidate genomic and proteomic biomarkers may provide insight into the clinical detection of primary SS once the functions of these miRNAs in the pathogenesis of oral inflammatory diseases have been validated. Gallo et al. [83] demonstrated that Epstein Barr virus-specific miRNA (ebv-miR-BART13-3p) could be transferred from B cells to salivary epithelial cells through exosomes in SS patients. However, the correlation between salivary exosomes and SS has never been investigated. Overall, identification of disease-specific biomarkers in saliva is an easy, rapid, and non-invasive approach for molecular diagnosis; hence, the study of exosomes in saliva can prompt further investigation into the therapeutic value of exosome inhibition in oral diseases.
MicroRNA (miR)-21 in hypoxic oral squamous cell carcinoma (OSCC)-derived exosomes is significantly upregulated under hypoxic conditions, and the hypoxic microenvironment stimulates tumour cells to generate miR-21-rich exosomes to be delivered to normoxic cells to promote prometastatic behaviors. On the other hand, exosomes containing miR-21 target phosphatase and tensin homolog (PTEN), and programmed cell death protein 4 (PDCD4), exhibit cisplatin resistance in OSCC. Moreover, miR-200c-3p in exosomes derived from a highly invasive OSCC line can induce a similar phenotype in non-invasive counterparts to promote OSCC invasion. The expression of proteasome subunit α type 7 (PSMA7) in salivary exosomes in patients with inflammatory bowel disease (IBD) showed significant differences between patients with IBD and healthy controls. During aging, miR-24-3p was identified as a novel candidate biomarker of aging, which could affect the MAPK signaling pathway to increase susceptibility to age-dependent alterations in the immune and inflammatory status, or it could induce apoptosis of salivary glands during aging, decreasing the production of saliva.
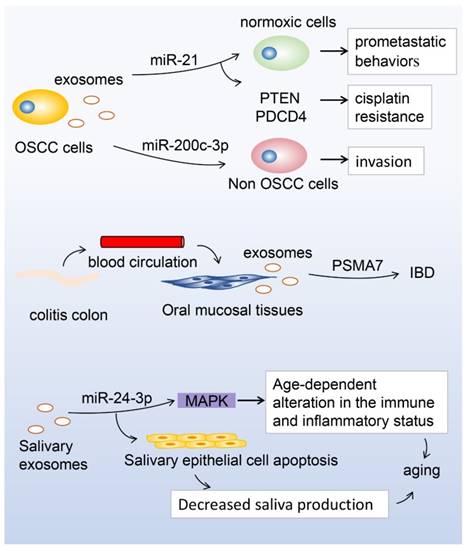
Correlation between salivary exosomes and other systemic diseases
Currently, the study of salivary biomarkers has developed beyond oral diseases [84-86] to systemic diseases [87, 88], which has broadened the potential for systemic disease detection [89, 90]. Recently, Langevin et al. [91], using miRNA sequencing, found that several salivary exosomal miRNAs, including miR-486-5p, miR-486-3p, and miR-10b-5p, were substantially overexpressed among a subset of head and neck squamous cell carcinoma patients compared to cancer-free controls, highlighting the use of salivary exosomal miRNAs as potential noninvasive cancer biomarkers. Zheng et al. [70] used liquid chromatography tandem mass spectrometry to identify the contents of salivary exosomes in patients with inflammatory bowel disease (IBD), which is an intestinal immune-dysfunctional disease with unknown cause. More than 2000 proteins were detected in salivary exosomes from patients with IBD and proteasome subunit α type 7 (PSMA7) was significantly different between patients with IBD and healthy controls, providing a promising biomarker that could allow patients to avoid the pain of colonoscopy. Lau et al. [92] found that salivary gland cells secreted exosome-like microvesicles encapsulating both protein and mRNA, and the interaction with breast cancer-derived exosome-like microvesicles communicated with and activated the transcriptional machinery of salivary gland cells, altering the mRNA and protein content of salivary gland cell-derived exosome-like microvesicles. By developing a pancreatic cancer mouse model, Lau et al. [71] showed that pancreatic tumor-derived exosomes are mechanistically involved in the development of pancreatic cancer-discriminatory salivary transcripttomic biomarkers, providing a mechanistic link between discriminatory salivary biomarkers and distal tumor. In addition, miR-1246 and miR-4644 in salivary exosomes may be potential biomarkers for pancreatobiliary tract cancer [19]. Katsiougiannis et al. [93] previously demonstrated that suppression of exosomes at the distal tumour site of pancreatic ductal adenocarcinoma ablated development of the salivary biomarker profile. Furthermore, they found that salivary exosomes in pancreatic tumor-bearing mice exhibited a suppressive effect on the reduction of the tumor-killing ability of natural killer cells. Moreover, Machida et al. [94] used microarray analysis to identify six miRNAs (miR-24-3p, miR-371a-5p, miR-3175, miR-3162-5p, miR-671-5p, and miR-4667-5p) that were significantly differently expressed during aging. Among them, miR-24-3p was identified as a novel candidate biomarker of aging by comparing total RNA obtained from 15 young and 13 old individuals using reverse transcription quantitative polymerase chain reaction. However, further validation is required to confirm whether miR-24-3p in salivary exosomes is a suitable biomarker of aging. Sun et al. [72] compared the proteomic profiles of saliva samples of lung cancer patients and healthy subjects and found that 63 proteins were unique to lung cancer patients. Further Gene Ontology analyses showed that 12 proteins were lung cancer-related biomarkers, including 6 annexin family members (annexin A1, A2, A3, A5, A6, and A11), nitrogen permease regulator 2-like protein (NPRL2), carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), histone H4 (HIST1H4A), mucin 1 (MUC1), prominin-1 (PROM1), and tumour necrosis factor alpha-induced protein 3 (TNFAIP3). Recently, Sun et al. [94] identified 25 proteins originally from distal organ cells in the salivary exosomes of lung cancer patients, 5 of which were lung-related proteins. Four of these, BPI fold-containing family A member 1 (BPIFA1), cornulin (CRNN), MUC5B, and Ras GTPase-activating-like protein (IQGAP1), were confirmed by Western blot, indicating that salivary exosomes harbour informative proteins that might be used for the detection of lung cancer in a noninvasive manner. Furthermore, Yang et al. [96] implanted human lung cancer H460 cells expressing hCD63-GFP into nude mice to follow the circulation of tumour cell-specific proteins and mRNAs in the form of exosome-like microvesicles (ELMs) in vivo, and found that ELMs carry tumour cell-specific mRNA and protein from blood to saliva, strengthening the link between distal tumour progression and the discovery of saliva biomarkers through ELMs. Moreover, Gonzalez-Begne et al. [27] identified proteins in salivary exosomes involved in Alzheimer's disease, neurodegenerative disorders, Parkinson's disease, cholera infection, Huntington's disease, long-term depression, prion disease, dentatorubropallidoluysian atrophy (DRPLA), glioma, type I and II diabetes, and different types of carcinomas (renal cell carcinoma, pancreatic carcinoma, prostate cancer, small cell lung cancer, bladder cancer, and melanoma), making exosomes an attractive source for exploring biomarkers.
Database and bioinformatics analysis of exosomes
Exosomes contain RNAs, proteins, lipids, and metabolites that are reflective of the cell type of origin. ExoCarta (http://www.exocarta.org) is a manually curated database containing exosomal proteins, RNAs, and lipids from both published and unpublished exosomal studies [97]. The method of exosomal purification and characterization as well as the biophysical and molecular properties are listed in the database with the aim of helping biomedical scientists assess the quality of exosomal preparation and the corresponding data. ExoCarta (version 3.1) contains information on 11,261 protein entries, 2375 mRNA entries, and 764 miRNA entries from 134 exosomal studies. This free web-based community resource is helpful to researchers in the study of molecular signatures (proteins/RNAs/lipids) specific to certain tissues/cell types derived from exosomes. Moreover, to analyze numerous proteins, RNAs, and lipids contained in exosomes, non-commercial tools such as FunRich can be used to identify over-represented groups of molecules [98]. Currently, with the advent of next generation sequencing technologies, investigations into the role of exosomes in cancer and other systemic diseases are increasing. Recently, bioinformatics-based analysis of RNA sequencing data of exosomes extracted from Trypanosoma cruzi has demonstrated a link between vesicles with important gene products and the probability of identifying biomarkers of Chagas disease [99].
Conclusion and Perspectives
In 2008, a breakthrough was made in the field of EVs that glioblastoma microvesicles contain cancer-specific mutant mRNA (EGFRvIII), suggesting that tumour-derived microvesicles may serve as an important diagnostic tool [100]. Since this discovery, much progress has been made in the investigation of exosomes, owing to their unique roles in intracellular communication and signalling. More importantly, exosomes can transport bioactive molecules like proteins, lipids, mRNAs, and miRNAs from one cell to another, which has been linked to the pathogenesis of various diseases. Salivary diagnosis has notable advantages, including noninvasiveness, ease of collection, absence of coagulation, a more positive impact on patient compliance, and a similar content as plasma, as well as their possible use in the diagnosis of systemic diseases. However, the use of salivary diagnosis has a number of limitations, including contamination as well as the presence of proteins such as amylase that may mask other proteins with low expression. Salivary exosomes can overcome these disadvantages and are attracting increasing attention not only in the field of cancer-derived exosomes, but also in other systemic disease-related exosomes such as inflammatory bowel disease and aging. Salivary exosomes have been recognized as exhibiting aberrant expression of various components in a disease-specific manner. However, the majority of exosomes remain to be investigated and specific molecular mechanisms remain to be elucidated. It remains unclear whether salivary exosomes are correlated with various systemic diseases. Discovering potential interactions may lead to novel discoveries of systemic disease signalling pathways affected by altered salivary exosome expression. Furthermore, knowledge of exosome functions would deepen our understanding of the therapeutic use of salivary exosomes in systemic diseases. In the future, given scientists' endeavours and the application of new methods, the underlying mechanisms of salivary exosomes may well be elucidated, providing novel insights that will aid in the treatment of systemic diseases.
Abbreviations
A2M: alpha-2-macroglobulin; AFM: atomic force microscopy; ANX A1,2,3,5,6,11: annexin A1,2,3,5, 6, 11; ATPS: aqueous 2-phase system; BBB: blood-brain barrier; BPIFA1: BPI fold-containing family A member 1; CAV1: calveolin 1; CEACAM1: carcinoembryonic antigen-related cell adhesion molecule 1; CRNN: cornulin; CSF: cerebrospinal fluid; DEX: dextran; DPP IV: dipeptidyl peptidase IV; DRPLA: Dentatorubropallidoluysian atrophy; EBV: epstein Barr Virus; ECM: extracellular matrix; ELMs: exosome-like microvesicles; ESCRT: endosomal sorting complex required for transport; EVs: extracellular vesicles; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; HIST1H4A: histone H4; HPA: haptoglobin alpha chain; HNSCC: head and neck squamous cell carcinoma; HSP: heat shock protein; IBD: inflammatory bowel disease; Ig: immunoglobulin; IGHA1: immunoglobulin alpha-1 chain c region; ILVs: internal luminal vesicle; lncRNAs: long non-protein-coding RNAs; IQGAP1: Ras GTPase-activating-like protein; LC-MS/MS: liquid chromatograph-mass spectrometer; LGALS3BP: galectin-3-binding protein; miRNA: microRNA; miRNAs: microRNAs; mRNAs: messenger RNAs; MUC1: mucin 1; MUC5B: mucin 5B; MudPIT: multidimensional protein identification technology; MVB: multivesicular body; MVBs: multivesicular bodies; ncRNAs: noncoding RNAs; NPRL2: nitrogen permease regulator 2-like protein; OLP: Oral lichen planus; OSCC: Oral squamous cell carcinoma; PIP: prolactin-inducible protein; PKM1/ M2: pyruvate kinase isozymes M1/M2; PROM1: prominin-1; pcRNAs: protein-coding RNAs; PDAC: pancreatic ductal adenocarcinoma; PDCD4: programmed cell death protein 4; PEG: polyethylene glycol; PI3K: phosphati-dylinositol 3-kinase; pIgR: polymeric immunoglobulin receptor; piRNAs: P-Element induced wimpy testis (PIWI)-interacting RNAs; PM: plasma membrane; PTEN: phosphatase and tensin homolog; RNases: ribonucleases; SGECs: salivary gland epithelial cells; SNARE: soluble NSF-attachment protein receptor; SS: Sjögren's syndrome; TF: tissue factor; TSE: transmissible spongiform encephalopathies; TSG101: tumour susceptibility gene 101; TNFAIP3: tumour necrosis factor alpha-induced protein 3; UC: ultracentrifugation; WS: whole saliva; EQ: ExoQuick-TCTM.
Acknowledgements
This study was financially supported by grants from the National Natural Science Foundation of China (No. 81670957; 81402235; 81772876; 81700938), the seed grants from the PKU School of Stomatology for PostDoc (YS0203), the grant of Peking University School and Hospital of Stomatology (PKUSS20140104), and the Beijing Natural Science Foundation (No. 7172239).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Soini HA, Klouckova I, Wiesler D, Oberzaucher E, Grammer K. et al. Analysis of volatile organic compounds in human saliva by a static sorptive extraction method and gas chromatography-mass spectrometry. J Chem Ecol. 2010;36:1035-1042
2. Aps JK, Martens LC. Review: the physiology of saliva and transfer of drugs into saliva. Forensic Sci Int. 2005;150:119-131
3. Slavkin HC. Toward molecularly based diagnostics for the oral cavity. J Am Dent Assoc. 1998;129:1138-1143
4. Lee JM, Garon E, Wong DT. Salivary diagnostics. Orthod Craniofac Res. 2009;12:206-211
5. Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113:3365-3374
6. Yamada T, Noshima Y, Matsuda T, Ishiguro N. Comparison of methods for isolating exosomes from bovine milk. J Vet Med Sci. 2012;74:1523-1525
7. van den Boorn JG, Dassler J, Coch C, Schlee M, Hartmann G. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev. 2013;65:331-335
8. Tan SS, Yin Y, Lee T, Lai RC, Yeo RW, Zhang B. et al. Therapeutic MSC exosomes are derived from lipid raft microdomains in the plasma membrane. J Extracell Vesicles. 2013:2
9. Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184:28-41
10. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-383
11. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569-579
12. Ritchie AJ, Crawford DM, Ferquson DJ, Burthem J, Roberts DJ. Normal prion proteins is expressed on exosomes isolated from human plasma. Br J Haematol. 2013;163:678-680
13. Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery- a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013;31:543-551
14. Zheng X, Chen F, Zhang J, Zhang Q, Lin J. Exosome Analysis: A Promising Biomarker System with Special Attention to Saliva. J Membr Biol. 2014;247:1129-1136
15. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y. et al. The landscape of microRNA, piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61:221-230
16. Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86
17. Schneider A, Simons M. Exosomes: vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 2013;352:33-47
18. Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R. et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. 2015;32:2003-2014
19. Machida T, Tomofuji T, Maruyama T, Yoneda T, Ekuni D, Azuma T. et al. miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep. 2016;36:2375-2381
20. Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, Tammachote R. et al. Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol. 2009;20:278-288
21. van der Lubbe N1, Jansen PM, Salih M, Fenton RA, van den Meiracker AH, Danser AH. et al. The phosphorylated sodium chloride cotransporter in urinary exosomes is superior to prostasin as a marker for aldosteronism. Hypertension. 2012;60:741-748
22. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368-13373
23. Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One. 2010;5:e8577
24. Ogawa Y, Miura Y, Harazono A, Kanai-Azuma M, Akimoto Y, Kawakami H. et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol Pharm Bull. 2011;34:13-23
25. Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol Pharm Bull. 2008;31:1059-1062
26. Xiao H, Wong DT. Proteomic analysis of microvesicles in human saliva by gel electrophoresis with liquid chromatography-mass spectrometry. Anal Chim Acta. 2012;723:61-67
27. Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE. et al. Proteomic Analysis of Human Parotid Gland Exosomes by Multidimensional Protein Identification Technology (MudPIT). J Proteome Res. 2009;8:1304-1314
28. Berckmans RJ, Sturk A, van Tienen LM, Schaap MC, Nieuwland R. Cell-derived vesicles exposing coagulant tissue factor in saliva. Blood. 2011;117:3172-3180
29. Kapsogeorgou EK, Abu-Helu RF, Moutsopoulos HM, Manoussakis MN. Salivary gland epithelial cell exosomes: a source of autoantigenic ribonucleoproteins. Arthritis Rheum. 2005;52:1517-1521
30. Zhao Y, Yin LY, Bi RR, Ma P, Li SB. Current advance in saliva-derived exosome study. Zhonghua Kou Qiang Yi Xue Za Zhi. 2016;51:765-769
31. Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLos One. 2012;7:e30679
32. Ogawa Y, Tsujimoto M, Yanoshita R. Next-Generation Sequencing of Protein-Coding and Long Non-Protein-Coding RNAs in Two Types of Exosomes Derived from Human Whole Saliva. Biol Pharm Bull. 2016;39:1496-507
33. Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol Pharm Bull. 2013;36:66-75
34. Bainton D.F. The discovery of lysosomes. J. Cell Biol. 1981;91:66s-76s
35. Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675-87
36. Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108
37. Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM. et al. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013;288:17713-17724
38. Tian T, Zhu YL, Hu FH, Wang YY, Huang NP, Xiao ZD. Dynamics of exosome internalization and trafficking. J Cell Physiol. 2013;228:1487-95
39. Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H. Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J Virol. 2013;87:10334-10347
40. Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH. et al. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem. 2014;289:22258-22267
41. Segura E, Guérin C, Hogg N, Amigorena S, Théry C. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J Immunol. 2007;179:1489-1496
42. Nolte-'t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113:1977-1981
43. Munich S, Sobo-Vujanovic A, Buchser WJ, Beer-Stolz D, Vujanovic NL. Dendritic cell exosomes directly kill tumour cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology. 2012;1:1074-1083
44. An T, Qin S, Xu Y, Tang Y, Huang Y, Situ B. Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles. 2015;4:27522
45. Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL. et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90
46. Hosseini M, Khatamianfar S, Hassanian SM, Nedaeinia R, Shafiee M, Maftouh M. et al. Exosome-encapsulated microRNAs as potential circulating biomarkers in colon cancer. Curr Pharm Des. 2016;23:1705-1709
47. Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumga¨rtel C, Schekman R. et al. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495-500
48. Simpson RJ, Lim J, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267-283
49. Chlebowski A, Lubas M, Jensen TH, Dziembowski A. RNA decay machines: the exosome. Biochim Biophys Acta. 2013;1829:552-560
50. Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513-525
51. Savina A, Fader CM, Damiani MT, Colombo MI. Rab11 promotes docking and fusion of multivesicular bodies in a calciumdependent manner. Traffic. 2005;6:131-143
52. Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A. et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19-30
53. Bobrie A, Krumeich S, Reyal F. et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumour microenvironment and can promote tumour progression. Cancer Res. 2012:72 4920-4930
54. Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883-891
55. Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N. et al. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013;5:1159-1168
56. Rao SK, Huynh C, Proux-Gillardeaux V, Galli T, Andrews NW. et al. Identification of SNAREs involved in synaptotagmin VIIregulated lysosomal exocytosis. J Biol Chem. 2004;279:20471-20479
57. Tiwari N, Wang CC, Brochetta C, Ke G, Vita F, Qi Z. et al. VAMP-8 segregates mast cell-preformed mediator exocytosis from cytokine trafficking pathways. Blood. 2008;111:3665-3674
58. Puri N, Roche PA. Mast cells possess distinct secretory granule subsets whose exocytosis is regulated by different SNARE isoforms. Proc Natl Acad Sci U S A. 2008;105:2580-2585
59. Fader CM, Sanchez DG, Mestre MB, Colombo MI. TI-VAMP/ VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793:1901-1916
60. Proux-Gillardeaux V, Raposo G, Irinopoulou T, Galli T. Expression of the Longin domain of TI-VAMP impairs lysosomal secretion and epithelial cell migration. Biol Cell. 2007;99:261-271
61. Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14:1036-1045
62. Boriachek K, Islam MN, Möller A, Salomon C, Nguyen NT, Hossain MSA6. et al. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small. 2018;14(6):1-21 doi: 10.1002/smll.201702153
63. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30:3.22.1-3.22.29
64. Zlotogorski-Hurvitz A, Dayan D, Chaushu G, Korvala J, Salo T, Sormunen R. et al. Human saliva-derived exosomes: comparing methods of isolation. J Histochem Cytochem. 2015;63:181-189
65. Kim J, Shin H, Park J. RNA in Salivary Extracellular Vesicles as a Possible Tool for Systemic Disease Diagnosis. J Dent Res. 2017;96:938-944
66. Kim J, Shin H, Kim J, Kim J, Park J. Isolation of high-purity extracellular vesicles by extracting proteins using aqueous two-phase system. PLoS One. 2015;10(Suppl 6e):0129760
67. Zlotogorski-Hurvitz A, Dayan D, Chaushu G, Salo T, Vered M. Morphological and molecular features of oral fluid-derived exosomes: oral cancer patients versus healthy individuals. J Cancer Res Clin Oncol. 2016;142:101-110
68. Byun JS, Hong SH, Choi JK, Jung JK, Lee HJ. Diagnostic profiling of salivary exosomal microRNAs in oral lichen planus patients. Oral Dis. 2015;21:987-993
69. Katsiougiannis S, Wong DT. The Proteomics of Saliva in Sjögren's Syndrome. Rheum Dis Clin North Am. 2016;42:449-56
70. Zheng X, Chen F, Zhang Q, Liu Y, You P, Sun S. et al. Salivary exosomal PSMA7: a promising biomarker of inflammatory bowel disease. Protein Cell. 2017;8:686-695
71. Lau C, Kim Y, Chia D, Spielmann N, Eibl G, Elashoff D. et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288:26888-26897
72. Sun Y, Xia Z, Shang Z, Sun K, Niu X, Qian L. et al. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci Rep. 2016;6:24669-24680 doi: 10.1038/srep24669
73. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90
74. Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26:645-62
75. Kowalski LP, Sanabria A. Elective neck dissection in oral carcinoma: a critical review of the evidence. Acta Otorhinolaryngol Ital. 2007;27:113-7
76. Neville BW, Day TA. Oral cancer and precancerous lesions. CA Cancer J Clin. 2002;52:195-215
77. Li L, Li C, Wang S, Wang Z, Jiang J, Wang W. et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016;76:1770-1780
78. Liu T, Chen G, Sun D, Lei M, Li Y, Zhou C. et al. Exosomes containing miR-21 transfer the characteristic of cisplatin resistance by targeting PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai). 2017;49:808-816
79. Kawakubo-Yasukochi T, Morioka M, Hazekawa M, Yasukochi A, Nishinakagawa T, Ono K. et al. miR-200c-3p spreads invasive capacity in human oral squamous cell carcinoma microenvironment. Mol Carcinog. 2017
80. Winck FV, Prado Ribeiro AC, Ramos Domingues R, Ling LY, Riaño-Pachón DM, Rivera C. et al. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci Rep. 2015;5:16305
81. Sjogren H. Zur Kenntnis der Keratoconjunctivitis sicca. Acta Ophthalmol. 1933;11(Suppl 2):1-151
82. Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE. et al. European Study Group on Classification Criteria for Sjögren's Syndrome: Classification criteria for Sjögren's syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61:554-558
83. Gallo A, Jang SI, Ong HL, Perez P, Tandon M, Ambudkar I. et al. Targeting the Ca(2+) Sensor STIM1 by Exosomal Transfer of Ebv-miR-BART13-3p is Associated with Sjögren's Syndrome. EBioMedicine. 2016;10:216-26
84. Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J. et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246-6252
85. Hu S, Wang J, Meijer J. et al. Salivary proteomic and genomic biomarkers for primary Sjogren's syndrome. Arthritis Rheum. 2007;56:3588-3600
86. Li Y, St John MA, Zhou X, Ieong S, Xie Y, Zhou H. et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442-8450
87. Streckfus CF, Bigler LR, Zwick M. The use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to detect putative breast cancer markers in saliva: a feasibility study. J Oral Pathol Med. 2006;35:292-300
88. Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH. et al. Salivary Transcriptomic Biomarkers for Detection of Resectable Pancreatic Cancer. Gastroenterology. 2009;138:949-957
89. Kaufman E, Lamster IB. The diagnostic applications of saliva-a review. Crit Rev Oral Biol Med. 2002;13:197-212
90. Gao K, Zhou H, Zhang L, Lee JW, Zhou Q, Hu S. et al. Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS One. 2009;4(e):5875
91. Langevin S, Kuhnell D, Parry T, Biesiada J, Huang S, Wise-Draper T. et al. Comprehensive microRNA-sequencing of exosomes derived from head and neck carcinoma cells in vitro reveals common secretion profiles and potential utility as salivary biomarkers. Oncotarget. 2017;8:82459-82474
92. Lau CS, Wong DT. Breast cancer exosome-like microvesicles and salivary gland cells interplay alters salivary gland cell-derived exosome-like microvesicles in vitro. PLoS One. 2012;7(3e):33037
93. Katsiougiannis S, Chia D, Kim Y, Singh RP, Wong DT. Saliva exosomes from pancreatic tumor-bearing mice modulate NK cell phenotype and antitumour cytotoxicity. FASEB J. 2017;31:998-1010
94. Machida T, Tomofuji T, Ekuni D, Maruyama T, Yoneda T, Kawabata Y. MicroRNAs in Salivary Exosome as Potential Biomarkers of Aging. Int J Mol Sci. 2015;16:21294-21309
95. Sun Y, Huo C, Qiao Z, Shang Z, Uzzaman A, Liu S. et al. Comparative Proteomic Analysis of Exosomes and Microvesicles in Human Saliva for Lung Cancer. J Proteome Res. 2018
96. Yang J, Wei F, Schafer C, Wong DT. Detection of tumour cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLoS One. 2014;9(e):110641
97. Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K. et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J Mol Biol. 2016;428:688-692
98. Pathan M, Keerthikumar S, Ang CS, Gangoda L. Technical brief funrich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597-2601
99. Gaur P, Chaturvedi A. Mining SNPs in extracellular vesicular transcriptome of Trypanosoma cruzi: a step closer to early diagnosis of neglected Chagas disease. PeerJ. 2016;4(e):2693
100. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470-1476
Author contact
![]() Corresponding authors: Weiran Li, Department of Orthodontics, Peking University School and Hospital of Stomatology, 22 South Zhongguancun Avenue, Haidian District, Beijing, 100081, China; Tel/Fax: +86-10-82195332; E-mail: weiranliedu.cn and Yunfei Zheng, Department of Orthodontics, Peking University School and Hospital of Stomatology, 22 South Zhongguancun Avenue, Haidian District, Beijing, 100081, China; Tel/Fax: +86-10-82195332; E-mail: yunfei_zhengedu.cn
Corresponding authors: Weiran Li, Department of Orthodontics, Peking University School and Hospital of Stomatology, 22 South Zhongguancun Avenue, Haidian District, Beijing, 100081, China; Tel/Fax: +86-10-82195332; E-mail: weiranliedu.cn and Yunfei Zheng, Department of Orthodontics, Peking University School and Hospital of Stomatology, 22 South Zhongguancun Avenue, Haidian District, Beijing, 100081, China; Tel/Fax: +86-10-82195332; E-mail: yunfei_zhengedu.cn

 Global reach, higher impact
Global reach, higher impact