10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(10):1196-1210. doi:10.7150/ijbs.25023 This issue Cite
Research Paper
Scalable Generation of Mesenchymal Stem Cells from Human Embryonic Stem Cells in 3D
1. Faculty of Health Sciences, University of Macau, Taipa, Macau, China.
2. Department of Orthopedics, First Affiliated Hospital, Guangzhou Medical University, Guangzhou, Guangdong, China.
3. Department of Hematology, Guangdong General Hospital, Guangzhou, Guangdong, China.
4. Department of Biology, University of Hartford, West Hartford, Connecticut, USA.
* These authors contributed equally
Received 2018-1-18; Accepted 2018-5-18; Published 2018-7-1
Abstract
Human embryonic stem cell (hESC) derived mesenchymal stem cells (EMSC) are efficacious in treating a series of autoimmune, inflammatory, and degenerative diseases in animal models. However, all the EMSC derivation methods reported so far rely on two-dimensional (2D) culture systems, which are inefficient, costive and difficult for large-scale production. HESC, as an unlimited source, can be successively propagated in spheroids. Here, we demonstrate that hESC spheroids can directly differentiate into MSC spheroids (EMSCSp) within 20 days in one vessel without passaging and the system is scalable to any desired size. EMSCSp can further differentiate into osteocytes and chondrocytes in spheres or demineralized bone matrix. EMSCSp also retains immune-modulatory effects in vitro and therapeutic effects on two mouse models of colitis after dissociation. Compared to EMSC differentiated in monolayer, EMSCSp-derived cells have faster proliferation and higher yield and develop less apoptosis and slower senescence. Thus, the 3D differentiation system allows simple, cost-effective, and scalable production of high-quality EMSC and subsequently bone and cartilage tissues for therapeutic application.
Keywords: Mesenchymal stem cells, human pluripotent stem cells, 3D, spheroids, colitis
Introduction
Mesenchymal stem cells (MSC) are a unique stem cell type with not only multipotency as other somatic stem cells, but also the capability of immunomodulation and migration, and low immunogenicity. They have been used in clinical trials for the treatment of many degenerative, autoimmune, and inflammatory diseases [1]. MSC can be isolated from a variety of fetal and adult tissues such as amniotic membrane, amniotic fluid, placenta, umbilical cord, umbilical blood, dental pulp, adipose, and bone marrow. However, MSC isolated from these somatic tissues are subject to great variations of the source tissues, limitation of donors, and risk of disease transmission from donors.
In contrast, human pluripotent stem cells (hPSC) including embryonic stem cells (ESC) and induced pluripotent stem cells (iPSC) are unlimited and stable sources with the potential to differentiate into any cell type in the body. MSC have been previously derived from hESC via co-culturing with mouse OP9 cells, sorting, scraping or handpicking of cells [2-7]. However, these methods are limited by low MSC yield and purity. We have recently reported two methods for MSC derivation from hESC (EMSC). The first uses a multi-step protocol including embryoid body formation followed by hemangioblast enrichment [8]. Whereas, the second uses a simplified protocol with one intermediate step of trophoblast differentiation followed by multiple rounds of passaging in MSC medium [9]. Like somatic tissue-derived MSC, EMSC as well as iPSC-derived MSC also have immunomodulatory effect in vitro and can reduce the severity of an increasing list of degenerative, autoimmune, and inflammatory diseases in animal models such as osteochondral defects [5], allergic airway inflammation [10], lupus nephritis, uveitis [11], multiple sclerosis [8, 9], and inflammatory bowel disease [9]. Following intra-femoral transplantation into immunocompromised mice, EMSC can engraft and support hematopoiesis [12]. However, the above methods are all in two-dimensional (2D) systems, which demand tedious cell maintenance, passaging, and large culture spaces and supplies while having low EMSC yield.
It has been known that 3D spheroid formation grants somatic tissue-derived MSC with enhanced paracrine secretion of angiogenic, anti-tumorigenic, and pro- and anti-inflammatory factors, improved cell survival, increased differentiation potentials, and delayed replicative senescence [13]. Genes involved in hypoxia [14], apoptosis [15, 16], inflammation [15, 17], and mechanophysical changes [18, 19] are differentially expressed in MSC cultured in 2D versus 3D. Based on these studies, we recently demonstrated that spheroid formation allows hESC and MSC to tolerate ambient conditions for up 4 and 10 days, respectively, without loss of viability and functions including the efficacy of the spheropreserved MSC on mouse colitis models [20]. This method may replace cryopreservation to ship cells worldwide.
We have previously demonstrated that hESC-formed spheroids (hESCSp) can be passaged in 3D culture with sustained pluripotency, normal karyotype, and capability to efficiently differentiate to functional cardiac spheres [21, 22]. In this study, we further show that hESCSp could directly differentiate into MSC in spheroids (EMSCSp). Unlike the 2D differentiation systems, our method allows all the multi-step differentiation procedures to be completed in one vessel in 3D without a need for cell passaging. EMSCSp could further differentiate to osteocytes, chondrocytes, and adipocytes all in spheroids and in demineralized bone matrix. EMSCSp could also be reattached and passaged in monolayer (EMSCSp-ML) without compromising their multipotency. EMSCSp-ML retained immune-modulatory effects in vitro and therapeutic effects on mouse colitis models. Compared to EMSC without spheroid formation, EMSCSp-ML had faster proliferation and underwent less apoptosis and slower senescence.
Results
EMSCSp generation and characterization
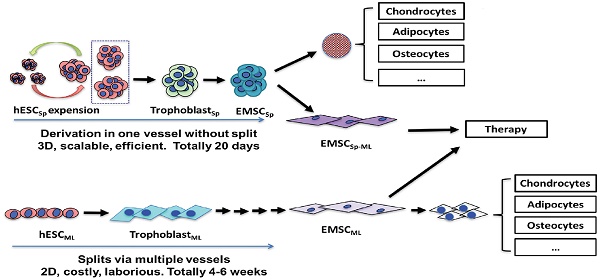
H9 and CT3 hESC were initially cultured in monolayer in the mTeSR1 medium (Fig. 1A and 1B). To generate hESCSp, we dislodged hESC colonies with dispase and passed cell clusters through a mesh (Fig. 1C) to form hESCSp (Fig. 1D), which were cultured in suspension and passaged via spheroid splitting as we reported recently [21]. Immunostaining and flow cytometric analysis confirmed that hESCSp retained pluripotency markers (Figs. S1A and S1B). Measured with ImageJ, the size of hESCSp was 108.2 ± 22.0 μm in diameter on day one after spheroid passaging (Fig. S1C). On day 2, hESCSp were treated with 10-ng/ml BMP4 and 1-μM A83-01 in mTeSR1 Minus Select Factors to initiate trophoblast-like (TB) differentiation in spheres. At day 3 of the treatment, the expression of several typical trophoblast-associated genes remarkably increased in the cells differentiating in spheres (TBSp/d3) as well as in monolayer (TBML/d3), indicating that the spheroid differentiation was also through a TB stage (Figs. S2A and S2B) as we reported before on hESC differentiation in monolayer [9]. On day 5, the consumed medium was replaced with MSC medium, followed by continuous culture for 15 days (Fig. 1E). To further determine the nature of the differentiated spheroids, we dissociated the spheroids, re-plated the single cells into a flask, and cultured them in 2D monolayer (EMSCSp-ML), which demonstrated spindle-like phenotype similar to that of typical MSC (Fig. 1F).
To track the multi-step differentiation, we analyzed cell surface markers TROP2 for trophoblast-like cells and CD73 for MSC on cells dissociated from spheroids at days 0, 5, 10, and 20 of the differentiation. On day 0, hESC (99.1%) were negative for either TROP2 or CD73. By day 5 of the differentiation, 98.2% of the cells became positive for TROP2. On day 10, 99.4% of the cells turned out positive for both markers. By day 20, 97.9% of the cells were positive for only CD73 (Fig. 2A). These results indicate a transition from hESC to trophoblast-like cells and then to MSC all in 3D, similar to that in 2D observed in our previous study [9]. Immunostaining of the day-20 spheroids shows that cells in the spheroids expressed typical MSC markers CD44, CD105, and CD90, but negative for the apoptotic marker activated caspase-3 (Fig. 2B), indicating that the 3D method allows generation of EMSCSp without noticeable apoptosis.
Flow cytometry demonstrates that EMSCSp-ML also expressed CD44, CD73, CD90, and CD105, but negative for a cocktail of hemangioblast and hematopoietic markers (Fig. 2C). Similar results were observed with EMSCSp-ML derived from three other hESC lines (CT2, CT3, and Envy) and one iPSC line derived from human peripheral blood mononucleocytes (PBY4) [23]. Human bone marrow MSC (BMSC) from two donors were tested as positive controls (Table S1). The ratio of CD105+ cells was lower among the cells that differentiated from PBY4 than that from hESC, which may be associated with different origins or epigenetic memory [24]. EMSCSp-ML also possessed the capability of differentiation into the three lineages of chondro-, osteo-, and adipocytes (Fig. 2D). These results suggest that EMSCSp and the subsequently derived EMSCSp-ML both possess typical features of MSC. In addition, G banding analysis shows that EMSCSp-ML derived from H9 and CT3 hESC lines both maintained normal keryotypes after prolonged culture (at p5 and p6, respectively) (Figs. S2C and S2D).
Scale-up production of EMSCSp and further differentiation in the demineralized bone matrix
Given the potential value of EMSCSp in clinical application, it is important to determine if our new method allows scale-up production of EMSCSp. Using a 100-ml plastic bag with an input for medium change and a filter for air exchange, we generated (7.0 ± 0.5) x 107 EMSC in spheres at (7.0 ± 0.3) x 105 cells/ml) from 3 x 106 hESC within 20 days (Fig. 3A and 3B). In contrast, using 21 T75 flasks we obtained (4.2 ± 0.3) x 107 EMSC in 2D monolayer (EMSCML), which took 5 passages and 39 days and consumed 555-ml MSC medium (Fig. 3B). Thus, compared to the 2D method, the 3D method produced 67% more EMSC, using half amount of medium in almost halftime in a single container without cell passaging. Moreover, following medium change EMSCSp in the same vessel could directly differentiate into spheroids of chondrocytes or adipocytes in 20 days, depending on the differentiation medium used. Staining shows that chondrocyte spheroids were positive for Alcian Blue, and adipocyte spheroids positive for Oil Red O with clear oil droplets (Fig. 3C).
Derivation of EMSCSp from hESCSp (A-D) At day 1, H9 hESC colonies (A) were dislodged and passed through a 50-μm strainer using mechanical pressure (B) to generate hESCSp of similar sizes (C). At day 2, the spheroid sizes were measured using ImageJ (D). (B) Scale bar is 100 μm. (D) Scale bar is 400 μm. (E) hESCSp were treated with 10-ng/ml BMP4 and 1-μM A83-01 in mTeSR1 Minus Select Factors for 3 days to induce differentiation into trophoblast-like cells also in spheroids. Scale bar is 400 μm. (F) The spent medium was replaced with the MSC medium to culture the trophoblast spheroids for 15 days with the medium refreshed every 5 days to form MSCSp, which were then dissociated and attached as a monolayer in a new dish. The resultant cells are termed EMSCSp-ML. Scale bar is 100 μm.
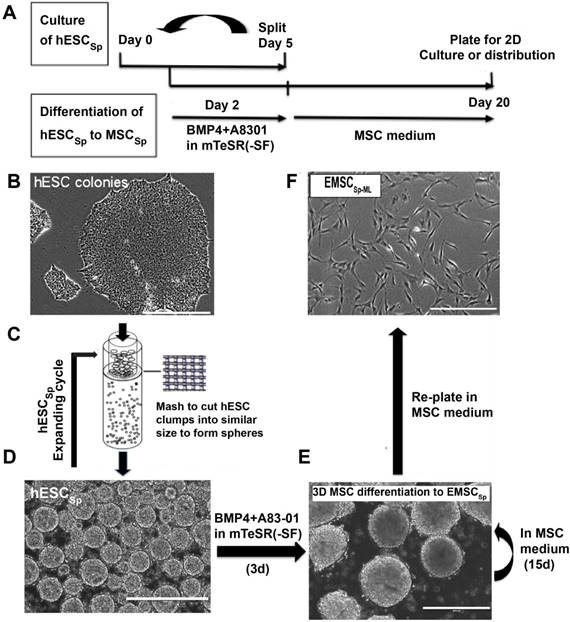
Characterization of EMSCSp and EMSCSp-ML (A) Stepwise differentiation from H9 hESC to trophoblasts and then MSC (EMSCSp) all in spheroids, reflected by transitional occurrence of markers of trophoblasts (TROP2) and MSC (CD73) via flow cytometry analysis. Blue dots stand for isotype control and red for tested markers. (B) Frozen sections of EMSCSp were immunostained positive for MSC markers CD90, CD105, and CD44, and negative for the apoptotic marker active caspase-3. Scale bars: 100 μm. (C) EMSCSp-ML were dissociated and quantified for the percentage of cells positive for four MSC markers and a mixture of negative control markers. Blue peaks stand for isotype control and red for tested markers. (D) Tri-lineage differentiation of EMSCSp-ML into chondrocytes, osteocytes, and adipocytes. Scale bars: 100 μm.
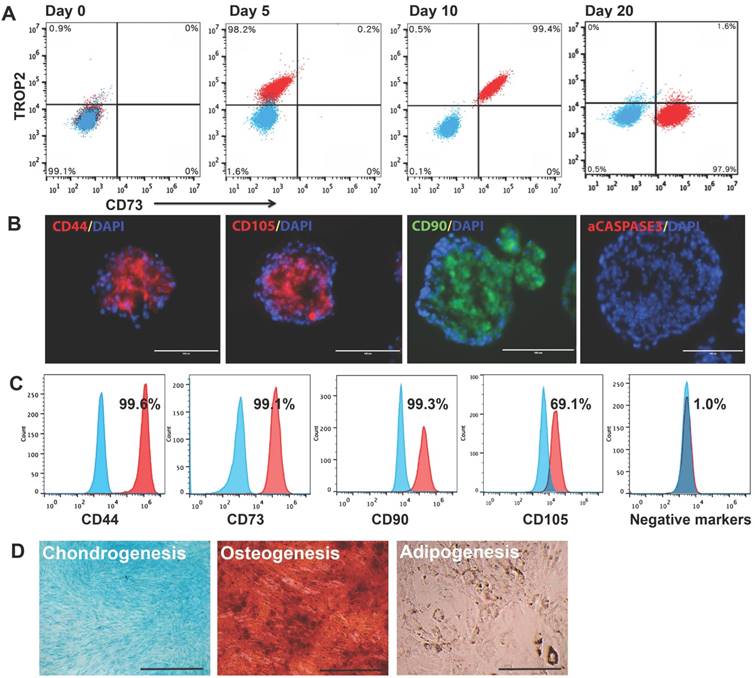
Demineralized bone matrix (DBM) is a widely accepted scaffold for bone and cartilage repair in clinic. Low attachment and viability of single cells filled into scaffolds are key problems that affect the outcomes of the cell-filled scaffolds in tissue engineering. Since spheroid formation of MSC enhances the cell viability and preserve the cell functions [20], we decided to fill DBM with EMSCSp and dissociated EMSC to enhance the cell survival and differentiation in the scaffold. We found that DBM could be recellularized with EMSC (Fig. 3D). Two cubes of DBM were aligned side by side, rinsed in osteo- or chondrogenic differentiation medium, covered with a mixture of EMSCSp and dissociated EMSC at a ratio of 2:1 in cell number (Fig. 3E), and cultured in a 37oC incubator for 20 days.
Both DBM cubes in each medium gradually fused with each other and became a whole piece of hard tissue (Fig. 3E). H&E staining shows that DBM alone contained lots of empty spaces, whereas the spaces inside EMSC-loaded DBM were filled with cells. Lineage-specific staining indicates that the cells in the spaces of the DBM cultured in the osteo- and chondrogenic differentiation media were positive for Alizarin Red S and Alician Blue, respectively. The Alcian Blue-stained cells were mostly aligned on the surface of the DBM (Fig. 3F). Together these data suggest that EMSCSp can differentiate into the skeletal tissues in DBM, which might be implanted for repair of bone fracture and cartilage injury.
Large-scale production of EMSCSp and continuous differentiation into chondrocytes and adipocytes all in spheroids. (A) Large-scale production of EMSCSp in a 100-ml plastic bag and EMSCML in multiple T75 flasks. Both were from 3 x 106 CT3 hESC. (B) Comparison of EMSCSp and EMSCML derivation for the time of production, cell yield (mean ± SE, n = 4), and consumption of culture supplies. *Medium change every 5 days. (C) EMSCSp were directed to differentiate into chondrocytes and adipocytes both in spheroids. Scale bars: 50 μm. (D) Sections of DBM alone or recellularized with EMSC were subjected to H&E staining. (E) Osteo- and chondrocyte differentiation of EMSC in spheres and single cells (at 2:1 in cell number ratio) loaded in two neighboring pieces of DBM. (F) H&E, Alizarin Red S, and Alcian Blue staining of the EMSC-loaded DBM cultured in the osteo- and chondrogenic differentiation media for 20 days. Scale bars: 100 μm.
Immune-responsive and -modulatory effects of EMSCSp-ML in comparison with authentic MSC from bone marrow
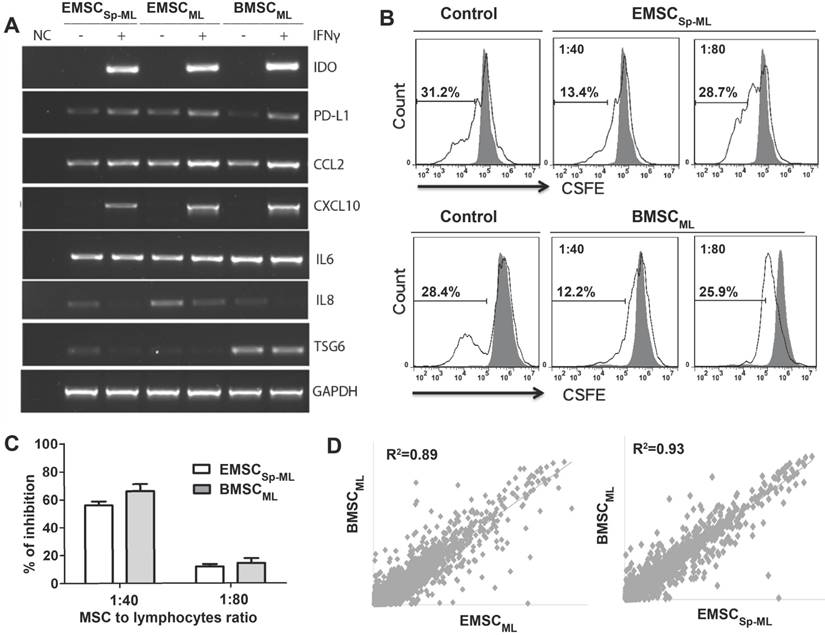
To further verify the therapeutic values of EMSCSp-ML, we tested the response of EMSCSp-ML to the inflammatory cytokine IFNγ, in comparison with BMSC cultured in monolayer (BMSCML) as a positive control. Following IFNγ treatment for 24 h, a series of anti- and pro-inflammatory genes including IDO, PD-L1, CCL2, and CXCL-10 were up-regulated in EMSCML, EMSCSp-ML and BMSCML compared to the untreated controls, whereas expression of the inflammatory cytokines IL6 changed little and IL8 decreased in all the three IFNγ treated samples (Fig. 4A).
Furthermore, the IFNγ-treated EMSCSp-ML and BMSCML cells dose-dependently inhibited proliferation of lymphocytes isolated from mouse lymph nodes and stimulated by antibodies against CD3 and CD28 (Fig. 4B). The percentages of inhibition are shown in Fig. 4C. These data suggest that EMSCSp-ML, like BMSCML, cells can respond to the inflammatory cytokine and repress lymphocyte proliferation.
Microarray assays show that the global gene expression profiles of both EMSCML (which had not experienced spheroid formation) and EMSCSp-ML are similar to that of BMSCML with the correlation coefficient as 0.89 and 0.93, respectively (Fig. 4D). The fact that EMSCSp-ML are more similar to BMSCML than EMSCML indicates that derivation via spheroidal formation may make EMSC to resemble the authentic BMSC more than derivation in monolayer culture.
Immune-responsive and -suppressive effects of EMSCSp-ML (A) EMSCML and EMSCSp-ML and BMSCML, pre-treated with or without 20 ng/ml IFNγ for 24 h, were subjected to RT-PCR assay for inflammation-related genes. GAPDH was detected for internal control. A mixture of all the samples was processed for the assay without reverse transcriptase to serve as a negative control (NC). (B and C) IFNγ-treated EMSCSp-ML as well as BMSCML were mixed at the cell ratio of 1:40 or 1:80 with CFSE-labeled mouse lymph node lymphocytes (2×105 cells/well) in a 96-well plate. The lymphocytes in the mixture were stimulated with anti-CD3 and -CD28 antibodies for 5 days. Lymphocyte proliferation was assayed via flow cytometry for CSFE-diluted cells (hollow peaks) against original CSFE-stained cells (the solid dark peak) as displayed in the histograms (C). The percentages of inhibition are shown displayed in D as mean ± SE (n = 3) (D) Correlation coefficients for comparisons of EMSCML and EMSCSp-ML with BMSCML based on microarray assay.
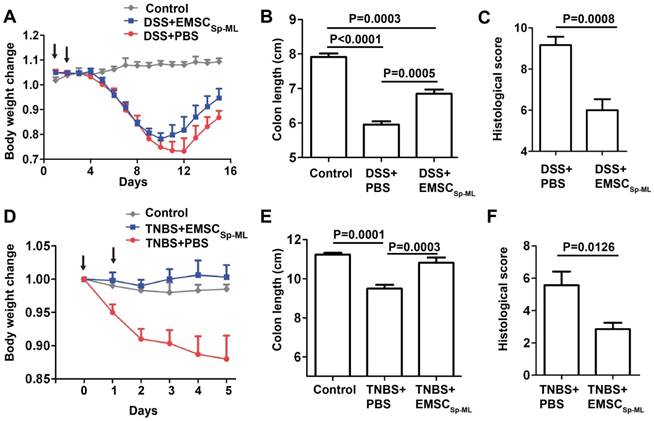
Efficacy of EMSCSp-ML on colitis model in mice
Given the immune responsive and modulatory effects of EMSCSp-ML cells, we decided to test whether the cells can treat mouse models of inflammatory colitis induced by DSS or TNBS. DSS involves immune responses driven by both Th1 and Th2, whereas TNBS involves only Th1-driven immune response [25]. DSS addition into the drinking water for 7 days caused dramatic decline of the body weight of C57BL/6 mice within 10 days or so. EMSCSp-ML treated with IFNγ, re-suspended in PBS at 1×106 cells/100 μl, and injected i.p. at days 1 and 2 post-DSS treatment reduced the loss of the body weight, compared to the control group that had been exposed to DSS water and injected with PBS (Fig. 5A). The length of the intestine was shortened the most in the DSS/PBS mice but moderately in the DSS/EMSCSp-ML mice, compared to the untreated mice (Figs. 5B and S3A). Histological analysis also supports the efficacy of EMSCSp-ML as the intestinal epithelium was less disrupted in the DSS/EMSCSp-ML group than the DSS/PBS group (Figs. 5C and S3C).
For the TNBS-induced colitis model, TNBS was administered into the anus at 2 mg/mouse twice at days 0 and 1. EMSCSp-ML cells were injected i.p. at 1x106 cells/100 μl/mouse also twice on days 0 and 1, respectively. These cells had even more obvious efficacy to prevent body weight loss than they did in the DSS-induced colitis model (Fig. 5D). Although EMSCSp-ML reduced colon length shortening in the TNBS-treated mice (Figs. 5E and S3B), it is much less obvious than in DSS-treated mice. EMSCSp-ML also reduced colon histology scores in the TNBS-treated mice compared to the vehicle control (Figs. 5F and S3D). These data together suggest that EMSCSp-ML have therapeutic effects on both colitis models.
Differential gene expression profiles in 2D- and 3D-derived EMSC
To elucidate differences between 2D- and 3D-derived EMSC at the molecular level, we analyzed the profiles of global gene expression using microarray in EMSCML and EMSCSp-ML at early (p3) and late (p8) passages. We also tested the response of the p3 EMSC samples to IFNγ treatment. First, we determined the quality of the dataset by comparing the probe intensity of all genes between the samples. As expected, the overall gene expression profiles of all the EMSC samples (no matter whether they were from 2D or 3D, and p3 or p8) are quite similar for comparisons between each other (R2 ≥ 0.8), whereas their profiles are very different from that of the parental hESC sample (R2 = 0.68-0.71) (Fig. S4A).
Alleviation of DSS-induced mouse colitis by IFNγ-treated EMSCSp-ML (A) Colitis was induced with DSS in C57BL/6 mice as described in Methods. IFNγ-treated EMSCSp-ML cells were injected i.p. on days 1 and 2 of the experiment. Body weight (mean ± SE) change of the mice was monitored everyday on days 0 through 15. The mice were sacrificed on day 15 and their colons were isolated. Control, n = 4; DSS+PBS, n = 6; DSS+EMSCSp-ML, n = 7. (B and C) The colon length (B) and histological scores (C) were measured on day 15. Data were analyzed via Student's t-test and presented as mean ± SE. (D) Colitis was induced with TNBS in C57BL/6 mice as described in Methods. IFNγ-treated EMSCSp-ML cells were injected i.p. on days 0 and 1 of the experiment. Body weight (mean ± SE) change of the mice was monitored every day on days 0 through 5. The mice were sacrificed on day 5 and their colons were isolated. Control, n = 4; TNBS+PBS, n = 7; TNBS+EMSC, n = 7. (E and F) The colon length (E) and histological scores (F) were measured on day 5. Data were analyzed via Student's t-test and presented as mean ± SE.
Next, the microarray dataset was normalized via Quantile Adjustment method, and filtered to remove probes with detection P value > 0.01 in at least 50% of the samples [26]. The rest of the data are shown in a heat-map to display expression of about 17,000 genes in each sample (Fig. S4B). Then, we compared the gene expression profiles of the samples in volcano plots. 190 (p3) and 115 (p8) genes were expressed 2-fold or higher, and 168 (p3) and 123 (p8) genes expressed 1/2-fold or lower in EMSCSp-ML compared to EMSCML (P < 0.001) (Figs. S4C and S4D).
Transcriptomic analysis of EMSCML and EMSCSp-ML Heat-maps are presented for differentially expressed genes associated to pluripotency and MSC (A), proliferation (B), inflammation (C), apoptosis and senescence (D).
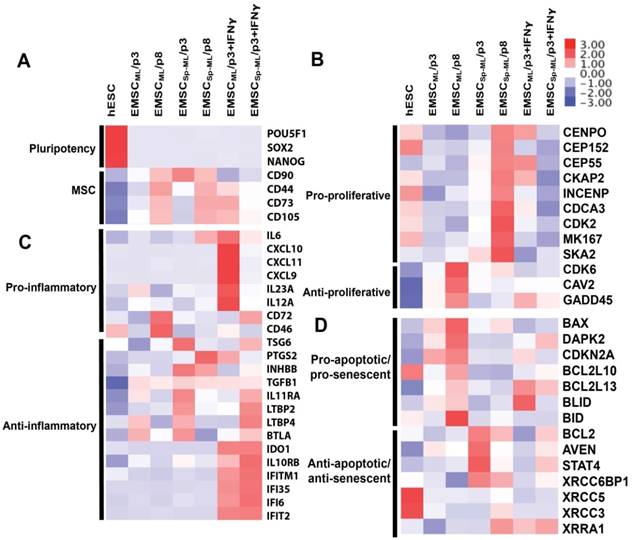
These genes were analyzed based on gene ontology using DAVID bioinformatics resources. First, pluripotency genes such as POU5F1 (OCT4), SOX2, and NANOG were down-regulated, and MSC marker genes including CD44, CD73, CD90, and CD105 up-regulated in all the MSC samples, which further increased at the later passage (Fig. 6A).
In addition, many of the differentially expressed genes are related to cell proliferation, inflammation, apoptosis, and senescence (Figs. 6B-D). For example, genes that positively regulate cell proliferation including CENPO, CEP152, CEP55, etc., were expressed higher in EMSCSp-ML than EMSCML, whereas genes that repress cell proliferation including CDK6, CAV2, and GADD45 expressed lower in EMSCSp-ML than EMSCML, which is more obvious at p8 than p3 (Fig. 6B). These data are consistent with the cell proliferation data in Figs. 3A-B. On the other hand, pro-inflammatory genes, e.g., CD72 and CD46 were expressed higher in EMSCML (p8) than EMSCML (p3) and EMSCSp-ML (both p3 and p8), and anti-inflammatory genes, e.g., TSG6, PTGS2, etc., expressed higher in EMSCSp-ML (p3) than EMSCML (p3) and in MSCs (either ML or Sp-ML) at p3 than p8 (Fig. 6C). Moreover, genes that promote apoptosis and senescence such as BAX, DAPK2, CDKN2A, etc., were expressed lower in EMSCSp-ML than EMSCML. In contrast, genes that repress apoptosis and senescence such as BCL2, AVEN, STAT4, etc., were expressed higher in EMSCSp-ML than EMSCML. Together, these results suggest that EMSCSp-ML, like MSC in spheroids derived from somatic issues [13], express higher levels of proliferative genes, and lower levels of inflammatory, apoptotic, and senescent genes than EMSCML, which is more obvious at the early stage than the late stage.
Upon IFNγ treatment, pro-inflammatory genes such as IL6, CXCL10, CXCL11, etc., were highly expressed in IFNγ-treated EMSCML but not EMSCSp-ML, whereas anti-inflammatory genes such as IDO1, IL10RB, and IFNγ-inducible genes (IFITM1, IFI35, IFI6, and IFIT2) were highly expressed in both IFNγ-treated EMSCML and EMSCSp-ML (Fig. 6C). These gene expression changes suggest that both EMSCML and EMSCSp-ML can respond to IFNγ, a feature important for their immune-modulatory and therapeutic effects. Together, the global gene expression profile of EMSCSp-ML is generally consistent with the features identified for MSC spheroids derived from other sources [13-19].
EMSCSp-ML have higher proliferation, less apoptosis, and delayed senescence than EMSCML
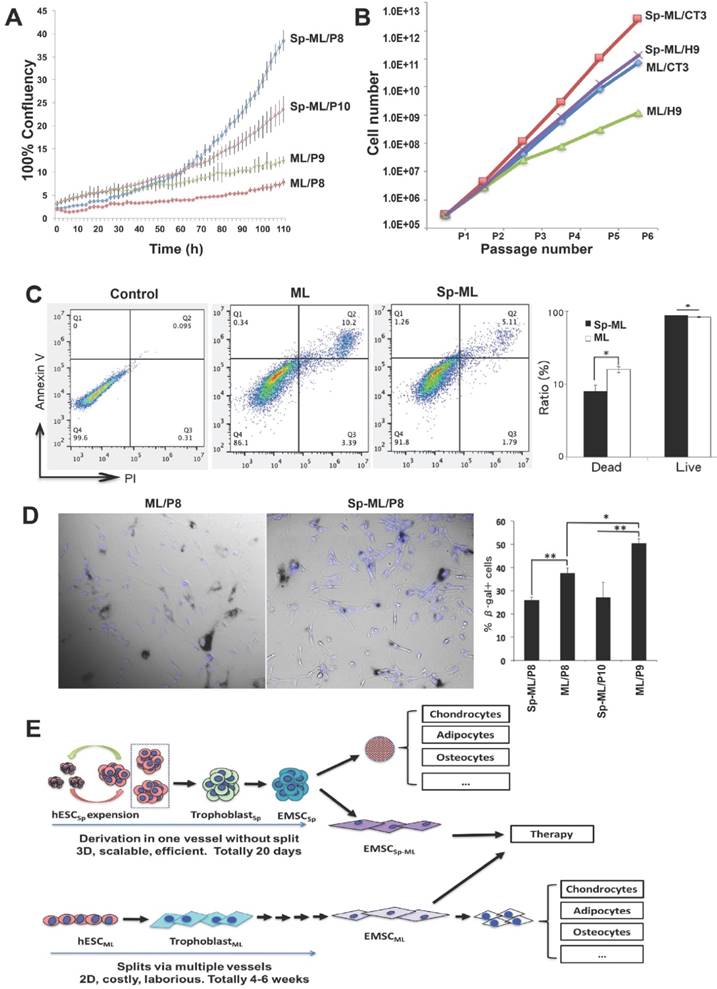
To obtain cellular evidence for the differences in the gene expression profile between the 2D- and 3D-derived EMSC, we compared the cells for some biological functions below. First, we monitored the proliferation of both EMSCML and EMSCSp-ML by tracking their confluence after being seeded in a 6-well plate in the IncuCyte Live-Cell Analysis System. As shown in Fig. 7A, while all groups starting from similar confluence at around 2.5%, p8 and p10 EMSCSp-ML reached 38.5% and 20.4% confluence, respectively, after being cultured for 110 h. In contrast, p8 and p9 EMSCML only reached 5.9% and 9.3% confluence, respectively, within the same period of time. This indicates that EMSCSp-ML proliferate faster than EMSCML, which was supported by the recording of cell numbers during the first five passages (derived from two hESC lines H9 and CT3). Higher cell numbers (about 50-100 fold) were obtained from EMSCSp-ML than EMSCML (Fig. 7B).
Since refrigeration is often used to protect viability of stored cells, we dissociated EMSCML and EMSCSp-ML and stored them at 4oC for 24 h followed by analysis of apoptosis. A higher ratio of apoptotic cells (about 2-fold) was found in culture of EMSCML (10.2%) than EMSCSp-ML (5.1%), based on staining for annexin V and propidium iodide (Fig. 7C). Again, higher viability was found in EMSCSp-ML than EMSCML. In addition, we tested cell senescence using staining for senescence-associated (SA) β-gal and found that up to 37.5% and 50.4% EMSCML were SA-β-gal+ at p8 and p9, respectively, where only 25.9% and 27.0% EMSCSp-ML were SA-β-gal+ at p8 and p10, respectively (Fig.7D). These results suggest that EMSCSp-ML have higher proliferation, less apoptosis, and delayed senescence than EMSCML, consistent with the gene expression profiles revealed via the microarray analysis (Fig. 6).
Discussion
Human MSC are a highly promising therapy for degenerative, inflammatory, and autoimmune diseases, which have been in preclinical studies and clinical trials worldwide. However, the great demand for a large number of MSC in clinic has proved to be a top challenge due to the limited number of MSC isolated from donated tissues and expanded in 2D monolayer cultures [27]. As alternative sources, hESC and iPSC can generate MSC theoretically for an unlimited time. Recently, substantial progress has been achieved to directly differentiate hESC spheroids into 3D structures such as cardiomyocytes [21], neural progenitors and hepatocytes [28, 29]. Without the need for culture splits (which are associated with repeat cell detachment, dissociation, and reattachment), these 3D direct differentiation methods have fewer cell loss, save space, time, supplies, and labor, thus having great advantages over 2D differentiation systems and favoring large-scale production of the therapeutic cells in a bioreactor. Although micro-beads, scaffold, and matrix have been used to generate MSC spheroids from adherent MSC culture [30-32], they demand trypsin treatment and cell dissociation. It has not yet been reported whether hESC spheroids can be directly differentiated into MSC spheroids, i.e., EMSCSp.
In this study, we derived EMSC completely in 3D with much higher efficiency and quality than EMSC derived in 2D, and show that EMSCSp could further differentiate into spheroids of chondrocytes and adipocytes in 3D in the same vessel. In addition, our method for EMSCSp generation does not require enzymatic treatment and culture split, thus it remarkably reduces cell damage and loss, and saves labor of experimenters. It also saves reagents and other culture consumables and enhances the differentiation efficiency and MSC yield. Moreover, we have shown that up to 70-million EMSC in spheroids could be generated in a 100-ml plastic bag. EMSCSp loaded into DBM differentiated into osteocytes or chondrocyte in the matrix, depending on the corresponding differentiation medium (Fig. 3).
Following re-plating of EMSCSp, the resultant EMSCSp-ML retained typical phenotypes, cell surface markers, and tri-lineage differentiation capability of MSCs. Interestingly, the spherical culture history enabled EMSCSp-ML to retain some unique features including faster proliferation, less apoptosis, and slower senescence than EMSCML. Consistently, gene expression profiles show that EMSCSp-ML had higher expression of pro-proliferative, anti-inflammatory, anti-apoptotic, and anti-senescent genes, and lower expression of pro-apoptotic, and pro-senescent genes than EMSCML. These features are also similar to those of MSC spheroids derived from somatic tissues [17, 33, 34], which can be retained even after 10 passages in monolayer culture [16].
Comparison of proliferation, apoptosis, and senescence between EMSCSp-ML and EMSCML (A) EMSCML and EMSCSp-ML at various passages were seeded at 3×104/well in a 12-well plate. Percentage of cell confluency in each well was monitored in the IncuCyte ZOOM Live-cell Analysis System. Results are presented as mean ± SD (n = 3). (B) EMSCML and EMSCSp-ML from two lines of hESC (H9 and CT3) were cultured for five consecutive passages and accumulated cell number was counted at each passage. (C) The concentrations of dissociated EMSCML and EMSCSp-ML were adjusted to 1×106 cells/ml. An Eppendorf tube containing 1 ml of the cell suspension was stored at 4oC for 24 h, and the cells were then subjected to flow cytometry analysis with Annexin V staining for apoptotic cells and PI staining for dead cells. The ratios of live and dead cells in the same samples from (C) was indicated in the histogram. Results are displayed as mean ± SD (n = 3). *P < 0.05 for comparison of EMSCSp-ML with EMSCML. (D) EMSCML and EMSCSp-ML were cultured at 37oC in a 12-well plate until about 50% confluency and stained for β-gal positive cells (dark blue). Cell nuclei were counterstained with DAPI (light blue). Results are displayed as mean ± SD (n = 3) in a bar chart, and representative images are shown beside. *P < 0.05. **P < 0.01. (E) Schematic to accommodate all the findings in this study.
It has been known that IFNγ-treated BMSC can enhance immunosuppressive factors, which then repress T cell activation, differentiation, and functions, and also have increased migratory and reparative capacities, which may aid tissue repair and prolong allograft survival [35]. We also found that IFNγ-treated EMSCSp-ML inhibited lymphocyte proliferation and reduced the severity of DSS-induced colitis in mice, suggesting that EMSCSp-ML are therapeutically effective similar to EMSCML [36] and MSC from other sources [1].
In spite of the remarkable functional changes in MSC following spherical culture, underlying molecular mechanisms and signaling pathways that trigger and sustain these changes are largely unknown. Cytoskeletal reorganization and drastic changes in cell morphology in MSC spheroids indicate a major difference in mechanophysical properties compared with 2D culture. It has been known that the self-aggregation process of BMSC initiates caspase-dependent IL1 autocrine signaling. IL1 then stimulates expression of early growth response gene-2 (EGR2), a zinc finger transcription factor that promotes synthesis of PGE2 an inhibitor of innate immunity via up-regulation of PTGS2/COX2 in MSCs [34, 35]. The IL1 autocrine signaling subsequently induces expression of chemokine receptors, such as CXCR4, or immunomodulatory mediators, such as TNFAIP6/TSG6 and IL6 [18, 20]. Although EGR2 was expressed low in all the MSC samples (data not shown), we found that transcripts for TGS6 and PTGS2 two anti-inflammatory genes were higher in EMSCSp-ML (p3) and EMSCML (p8), respectively, than the corresponding EMSCML samples (Fig. 6C), indicating that the IL1 autocrine signaling might be more active in EMSCSp-ML than EMSCML. Nevertheless, further investigations are warranted to illustrate the molecular mechanisms for how EMSCSp retain featured MSC capacity while gaining properties in cell functionalities different from EMSCML.
In summary, we have established a simple 3D method to efficiently differentiate hESC into MSC and then into chondrocytes and adipocytes, all in spheroids and in a single vessel. EMSCSp can recellularize DBM and differentiate in situ into osteocytes and chondrocytes. Following re-plating of EMSCSp, the resultant EMSCSp-ML share similarities in phenotypes and functionalities with BMSC, while possessing higher growth speed, lower apoptosis, and slower senescence than EMSCML (Fig. 7E). Since EMSCSp can be stored under ambient conditions for up to 10 days [20], EMSCSp generated using our method at any scale can be shipped worldwide without cryopreservation for research and therapeutic applications.
Methods
hESC culture and MSC generation from hESC spheroids and human bone marrow
All the hESC lines H9 [37], CT2, CT3 [38], and Envy (GFP+) derived from hES3 [39] used in this study had been registered with the National Institutes of Health, USA. For all these registered hESC lines, informed consents were taken from all the donors of leftover early-stage human embryos. hESC were cultured on Matrigel in E8 medium (StemCell Technologies) [40]. The PBY4 iPSC line was derived from donated human peripheral blood via Millipore [23]. For EMSCSp-ML generation, we first made hESC spheroids of homogenous sizes as described [21, 22]. Briefly, typical hESC colonies were digested for 5-10 min with 1 mg/ml dispase (StemCell Technologies). Once hESC colonies began to roll up, dispase was removed and cells were rinsed with Ca2- and Mg2-free PBS twice. The colonies were gently washed off the plate with mTeSR1 and collected with a 5-ml pipette. We then cut the colonies by passing them through a strainer with pore size at 50 μm in diameter (CellTrics) under pipette pressure to generate cell clusters of about 50-μm. The clusters were collected, re-suspended in 3 ml/well pre-warmed mTeSR1 medium (StemCell Technologies) supplemented with 10-μM Y-27632 (Sigma) to inhibit apoptosis, and cultured in suspension on Petri dish (Corning).
On day 2, hESC spheroids were treated with 10-ng/ml BMP4 and 1-μM A83-01 in mTeSR1 Minus Select Factors (StemCell Technologies) to induce differentiation into trophoblast-like cells, similar as we derived MSC from hESC cultured in monolayer via trophoblast differentiation in 2D [9]. On day 5, the spent medium was removed and replaced with the MSC medium, i.e., MEM basal medium supplemented with 20% FBS, 1% NEAA, and 1% L-Glutamine. The culture medium was refreshed with the MSC medium every 5 days. Initial analyses were conducted on cell spheroids collected at various days during the differentiation. For continuous analyses, some spheroids were dissociated at day 20 with 0.25% trypsin for 5 min. and then plated into a T75 flask for expansion and passaging in 2D in the MSC medium.
BMSC were derived from bone marrow aspirates donated by two healthy male adults under the approval of the University of Macau Panel of Research Ethics #APP004-FHS. The aspirates were washed with PBS twice followed by isolation of the mononucleocyte (MNC) fraction via centrifugation over Ficoll (Sigma) at 800 RCF. BMSCs were plated in T75 flasks in the MSC medium and fed every 3 days. When cells reached 80% confluence, they were detached with 0.05% trypsin and re-plated at 30 cells/cm2 (as passage 1 or p1) for further expansion. Resultant BMSC were passaged every 5-7 days.
Detection of cell surface markers for MSC
Cell surface markers were detected using BD StemflowTM Human MSC Analysis Kit (BD Biosciences). Briefly, cells were dissociated with 0.05% trypsin, washed and re-suspended in a staining buffer at 1×107 cells/ml. Cell suspensions were stained for typical MSC markers (CD44, CD73, CD90, and CD105) and a cocktail of negative markers (CD45, CD34, CD11b, CD19, and HLA-DR). After staining, cells were analyzed on a BD AccuriTM 6 flow cytometer (BD Biosciences).
Tri-lineage differentiation of MSC
MSC dissociated with 0.05% trypsin were seeded in a 12-well plate at 5×104 cells/well. After two days, the cells were induced to differentiate into osteogenic, adipogenic, and chondrogenic lineages, using the StemPro differentiation kits (Life Technologies), according to the manufacturer's instructions. Media were changed every three days. Resultant osteocytes and chondrocytes were fixed on day 21 post-differentiation, and stained with Alizarin Red S and Alcian Blue, respectively. Due to the slower adipogenic differentiation from EMSC than bone marrow derived MSC [41], we prolonged the differentiation time to 40 days. Resultant adipocytes were fixed on day 40 and stained with Oil Red O. Cells were photographed following the staining.
EMSC spheroidal differentiation into chondrocytes and adipocytes in 3D
Following precipitation of EMSCSp in an ultra-low attachment plate, the consumed medium was gently aspirated. The spheroids were rinsed with PBS twice and then re-suspended in the Cartilage or Adipocyte Differentiation Medium (StemPro differentiation kits, Life Technologies) for differentiation into chondrocytes and adipocytes, respectively. The medium was refreshed every four days. On day 20 the spheroids were collected and fixed in 4% paraformaldehyde for two days. Frozen sections of those spheroids were used for identification of chondrocytes and adipocytes using Alcian Blue and Oil Red O, respectively, followed by photography. EMSCSp without chondro- or adipogenic differentiation were used as a negative control. For EMSC differentiation to osteocytes and chondrocytes in DBM (Anjiu, BioTech), multiple cubes of DBM were placed in a well of a 6-well plate, aligned side by side, and rinsed in the MSC medium overnight. Next day, the medium was removed, a mixture of EMSCSp and dissociated EMSC at the ratio of 2:1 in cell number was loaded into the DBM overnight to fill all the spaces between the cell spheres and scaffold. The recellularized DBM was then rinsed in osteo- or chondrocyte differentiation medium that was added to the well until 75% full, and the plate was carefully placed in an incubator for differentiation with the medium refreshed every 4 days until day 20. The cell-loaded DBM blocks were fixed in 4% paraformaldehyde and sectioned for further analyses.
Detection of apoptotic and live/dead cells
EMSCSp were fixed in 4% paraformaldehyde for two days followed by immunostaining for the apoptotic marker active caspase-3 using corresponding antibody (SantaCruz, USA). To quantify live/dead cells, EMSC in spheroids or monolayer were dissociated with 0.25% trypsin and stained with Annexin V (for apoptotic cells), and acridine orange (AO, which labels live cells green), and propidium iodide (PI, which labels dead cells orange) (Cellometer), according the manufacturers' instructions. AO/PI-stained cells were photographed and quantified on Cellometer, and Annexin V-stained cells quantified on BD AccuriTM 6 flow cytometry.
Tracking of live cell proliferation
Cell growth was tracked using the IncuCyte ZOOMTM Live-Cell Analysis System (Essen BioScience). MSC with different treatments were plated on a 6-well plate at the same initial density, and the plate was placed in the IncuCyte ZOOM™ system. Phase-contrast images were captured at 2 hours intervals for a total duration of 110 hours. Percentage of confluence in each well, which is proportional to cell proliferation rate, was calculated using the IncuCyte ZOOM™ software.
Detection of cell senescence
Cell senescence was analyzed using the Senescence β-Galactosidase Staining Kit (Beyotime). Cells cultured as a monolayer until ~70% confluence were fixed and stained with the kit overnight, rinsed with 1× PBS, and then penetrated with 0.2% Triton X-100 for 10 min. Nuclei were counterstained with DAPI for the total cell number. Five different microscopic views of the stained cells were randomly photographed. The ratios of β-gal+ cells among the total cells in all the views were analyzed for statistics.
Reverse transcription- and quantitative polymerase chain reaction (RT-PCR and RT-qPCR)
Total RNA was extracted from cultured cells by using Trizol reagent (Life Technology) and cDNA was generated using PrimeScript RT Reagent kit (Clontech). PCR reactions were carried out by Quick-Load® Taq 2×Master Mix (NEB). PCR was performed under following condition: an initial denaturation at 95 °C for 30 sec.; followed by 28 cycles of 30 sec., denaturation at 95 °C, 20 sec., annealing at 53 °C, and 30 sec. extension at 68 °C; and with a final extension at 68 °C for 10 min.
qPCR was performed using iTaqTM Universal SYBR Green Supermix (Bio-rad), and detected with the CFX96TM Real-Time PCR Detection System (Bio-rad). qPCR cycling protocol was as follows: an initial denaturation and enzymatic activation at 95 °C for 5 sec; followed by 39 cycles of 5-sec. denaturation at 95 °C, 30-sec. annealing at 60 °C, and melt curve analysis by 0.5 increments at 5 sec./step from 65 °C to 95 °C; and the final incubation was at 95 °C for 30 sec. for polymerase activation and DNA denaturation. Data are normalized to the expressional level of GAPDH and represented as fold changes. Primers used to detect expression of specific genes are listed in Table S2.
Microarray
RNA was isolated from EMSCML and EMSCSp-ML treated with or without IFNγ for 24 h, using RNAeasy mini kit (Qiagen), and cDNA libraries were synthesized and processed by using HumanHT-12 v4 Expression BeadChip. Further analysis was performed using the BeadStudio Data Analysis software (Illumina) and Excel.
The microarray dataset was normalized and filtered to remove probes with detection P value > 0.01 in at least 50% of the samples [26]. Two normalization methods were utilized to analyze the dataset on the basis of their biological relevance. First, average probe intensity of all biological and technical replicates was used to make comparison of gene expression profile between test and control groups. Second, the probe intensity of analyzed genes in hESC was used as a baseline to identify the gene expression differences between EMSCML and EMSCSp-ML. The microarray data has been deposited to Gene Expression Omnibus, and its accession number is GSE93218.
Inhibition of T lymphocyte proliferation in vitro
Lymphocytes isolated from peripheral lymph nodes of 8 to 10 weeks-old C57BL/6 mice were labeled with 5-μM CFSE (Beyotime) for 10 min, at 37°C, seeded at 2×105 cells/100 μl/well in 1640 complete medium (RPMI-1640 + 10% FBS). MSC were seeded in a 6-well plate, cultured in the MSC medium for 7 days, dissociated with Trypsin-EDTA (StemCell Technologies), and added at the ratio of 1:40 or 1:80 (for cell number of MSC versus lymphocytes) per well into a 96-well plate containing lymphocytes at 100 μl/well. The lymphocytes in the mixture were stimulated with or without pre-coated 10-μg/ml anti-CD3 antibodies (BD Bioscience) and 2-μg/ml anti-CD28 antibodies (BD Bioscience). Suspended lymphocytes were collected 5 days after the stimulation and then subjected to flow cytometry analysis.
Mouse model of DSS/TNBS-induced colitis
Experiments were carried out on 8-week-old C57BL/6 mice, based on the animal use protocol #UMAEC-06-2015 approved by the University of Macau Panel of Research Ethics. The model was induced in mice as we recently reported [9] with minor modifications. Briefly, mice were administered with 2% (w/v) DSS (molecular weight 36,000-50,000, MP Biomedical) in the drinking water for 7 days. On days 1 and 2 after the start of the DSS treatment, mice were intraperitoneally (i.p.) injected with 1×106 MSC/mouse. MSC were suspended in 100 μl PBS. Some mice were injected with 100 μl PBS alone as a negative control. Body weight loss, rectal bleeding, and stool consistency were monitored daily.
All mice were sacrificed on day 15 after the start of the experiment. The colon was dissected from each mouse and measured for its length. The colons were flushed with sterile 1× DPBS and fixed in 4% paraformaldehyde at 4oC for 24 h. The distal part of the colon was embedded in paraffin wax and sectioned at 5 µm in thickness, mounted to glass slides for hematoxylin and eosin (H&E) staining. Images of the H&E stained sections were acquired on an Olympus CKX41 microscope. Histological score of the colon sections is calculated based on the percentage of area involved and crypt loss, erosions of the epithelium, and the severity of the other parameters [42].
The TNBS-induced colitis model was conducted on male BALB/C mice following the reported method [43] with some modifications. 100 mg/kg TNBS in 100 μl 45% ethanol was intra-rectally administrated to mice on days 0 and 1. 6 h after the administration, mice were i.p. injected with 1x106 MSC/mouse in 100-μl PBS or PBS alone. All mice were sacrificed on day 5 after the start of the experiment. The body weight loss, colon length, and histological score were documented as above.
Data analysis
Statistical analysis was performed with SPSS. Two-tailed Student's t-test was used to determine P values. Statistical significance was defined as *P < 0.05 and **P <0.01. Data obtained from flow cytometry were analyzed using FlowJo 7.6.
Abbreviations
hESC: human embryonic stem cell; hPSC: human pluripotent stem cell; iPSC: induced pluripotent sten cell; MSC: mesenchymal stem cell; BMSC: human bone marraw MSC; EMSC: hESC derived MSC; hESCSp: hESC spheroids; EMSCSp: EMSC spheroids; EMSCML: EMSC cultured in monolayer; EMSCSp-ML: EMSCSp reattached and passaged in monolayer; TB: trophlast; TBSp: TB spheroids; TBML: trophoblast cultured in monolayer; DBM: demineralized bone matrix; DSS: dextran sulfate sodium; TNBS: 2,4,6-trinitrobenzenesulfonic acid.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We appreciate the excellent services provided for this study by the Animal Facility, Single Cell and Gene Expression Analysis Core, and Histopathology Core of Faculty of Health Sciences, University of Macau. This work was supported by University of Macau Research Committee funds SRG #2014-00008-FHS and MYRG #2015-00169-FHS, 2016-00070-FHS, and 2017-00124-FHS, and Macau Science and Technology Development Fund (FDCT) #128-2014-A3, 028/2015/A1, and 095/2017/A2 to R.X.
Author Contributions
L.Y., B.J., E.H., and R.X. conceived and designed the experiments, and wrote the manuscript. L.Y., B.J., and Q.L. performed the experiments. X.D. and G.C. recruited bone marrow donors. E.L., X.W., and D.Z. assisted with the animal experiments and gene expression analyses. L.Y., B.J., J.W.P., Y.L., X.C., E.C., E.H. and R.X. analyzed the data. R.X. approved the manuscript.
Competing Interests
R.X. is a founder of ImStem Biotechnology, Inc., a stem cell company. He declares competing financial interests. No financial conflicts of interest exist for any of the authors.
References
1. Wang Y, Chen X, Cao W, Shi Y. Plasticity of mesenchymal stem cells in immunomodulation: pathological and therapeutic implications. Nat Immunol. 2014;15:1009-16
2. Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2005;2:e161
3. Brown SE, Tong W, Krebsbach PH. The derivation of mesenchymal stem cells from human embryonic stem cells. Cells Tissues Organs. 2009;189:256-60
4. Gruenloh W, Kambal A, Sondergaard C, McGee J, Nacey C, Kalomoiris S. et al. Characterization and in vivo testing of mesenchymal stem cells derived from human embryonic stem cells. Tissue engineering Part A. 2011;17:1517-25
5. Hwang NS, Varghese S, Lee HJ, Zhang Z, Ye Z, Bae J. et al. In vivo commitment and functional tissue regeneration using human embryonic stem cell-derived mesenchymal cells. Proc Natl Acad Sci U S A. 2008;105:20641-6
6. Olivier EN, Rybicki AC, Bouhassira EE. Differentiation of human embryonic stem cells into bipotent mesenchymal stem cells. Stem Cells. 2006;24:1914-22
7. Vodyanik MA, Yu J, Zhang X, Tian S, Stewart R, Thomson JA. et al. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell stem cell. 2010;7:718-29
8. Wang X, Kimbrel EA, Ijichi K, Paul D, Lazorchak AS, Chu J. et al. Human ESC-derived MSCs outperform bone marrow MSCs in the treatment of an EAE model of multiple sclerosis. Stem cell reports. 2014;3:115-30
9. Wang X, Lazorchak AS, Song L, Li E, Zhang Z, Jiang B. et al. Immune modulatory mesenchymal stem cells derived from human embryonic stem cells through a trophoblast-like stage. Stem cells. 2016;34:380-91
10. Sun YQ, Deng MX, He J, Zeng QX, Wen W, Wong DS. et al. Human pluripotent stem cell-derived mesenchymal stem cells prevent allergic airway inflammation in mice. Stem Cells. 2012;30:2692-9
11. Kimbrel EA, Kouris NA, Yavanian GJ, Chu J, Qin Y, Chan A. et al. Mesenchymal stem cell population derived from human pluripotent stem cells displays potent immunomodulatory and therapeutic properties. Stem cells and development. 2014;23:1611-24
12. Li O, Tormin A, Sundberg B, Hyllner J, Le Blanc K, Scheding S. Human embryonic stem cell-derived mesenchymal stroma cells (hES-MSCs) engraft in vivo and support hematopoiesis without suppressing immune function: implications for off-the shelf ES-MSC therapies. PloS one. 2013;8:e55319
13. Cesarz Z, Tamama K. Spheroid Culture of Mesenchymal Stem Cells. Stem Cells Int. 2016;2016:9176357
14. Song L, Webb NE, Song Y, Tuan RS. Identification and functional analysis of candidate genes regulating mesenchymal stem cell self-renewal and multipotency. Stem Cells. 2006;24:1707-18
15. Tsai AC, Liu Y, Yuan X, Ma T. Compaction, fusion, and functional activation of three-dimensional human mesenchymal stem cell aggregate. Tissue engineering Part A. 2015;21:1705-19
16. Cheng NC, Chen SY, Li JR, Young TH. Short-term spheroid formation enhances the regenerative capacity of adipose-derived stem cells by promoting stemness, angiogenesis, and chemotaxis. Stem Cells Transl Med. 2013;2:584-94
17. Bartosh TJ, Ylostalo JH, Bazhanov N, Kuhlman J, Prockop DJ. Dynamic compaction of human mesenchymal stem/precursor cells into spheres self-activates caspase-dependent IL1 signaling to enhance secretion of modulators of inflammation and immunity (PGE2, TSG6, and STC1). Stem Cells. 2013;31:2443-56
18. Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H. et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979-87
19. Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. 2014;13:547-57
20. Jiang B, Yan L, Miao Z, Li E, Wong KH, Xu RH. Spheroidal formation preserves human stem cells for prolonged time under ambient conditions for facile storage and transportation. Biomaterials. 2017;133:275-86
21. Jiang B, Xiang Z, Ai Z, Wang H, Li Y, Ji W. et al. Generation of cardiac spheres from primate pluripotent stem cells in a small molecule-based 3D system. Biomaterials. 2015;65:103-14
22. Otsuji TG, Bin J, Yoshimura A, Tomura M, Tateyama D, Minami I. et al. A 3D sphere culture system containing functional polymers for large-scale human pluripotent stem cell production. Stem Cell Reports. 2014;2:734-45
23. Martins-Taylor K, Nisler BS, Taapken SM, Compton T, Crandall L, Montgomery KD. et al. Recurrent copy number variations in human induced pluripotent stem cells. Nat Biotechnol. 2011;29:488-91
24. Jung Y, Bauer G, Nolta JA. Concise review: Induced pluripotent stem cell-derived mesenchymal stem cells: progress toward safe clinical products. Stem Cells. 2012;30:42-7
25. Randhawa PK, Singh K, Singh N, Jaggi AS. A review on chemical-induced inflammatory bowel disease models in rodents. Korean J Physiol Pharmacol. 2014;18:279-88
26. Schurmann C, Heim K, Schillert A, Blankenberg S, Carstensen M, Dorr M. et al. Analyzing illumina gene expression microarray data from different tissues: methodological aspects of data analysis in the metaxpress consortium. PLoS One. 2012;7:e50938
27. Sart S, Tsai AC, Li Y, Ma T. Three-dimensional aggregates of mesenchymal stem cells: cellular mechanisms, biological properties, and applications. Tissue Eng Part B Rev. 2014;20:365-80
28. Lin H, Li Q, Lei Y. An Integrated Miniature Bioprocessing for Personalized Human Induced Pluripotent Stem Cell Expansion and Differentiation into Neural Stem Cells. Sci Rep. 2017;7:40191
29. Shao Y, Sang J, Fu J. On human pluripotent stem cell control: The rise of 3D bioengineering and mechanobiology. Biomaterials. 2015;52:26-43
30. Chen AK, Reuveny S, Oh SK. Application of human mesenchymal and pluripotent stem cell microcarrier cultures in cellular therapy: achievements and future direction. Biotechnology advances. 2013;31:1032-46
31. Li WJ, Tuli R, Huang X, Laquerriere P, Tuan RS. Multilineage differentiation of human mesenchymal stem cells in a three-dimensional nanofibrous scaffold. Biomaterials. 2005;26:5158-66
32. Li WJ, Tuli R, Okafor C, Derfoul A, Danielson KG, Hall DJ. et al. A three-dimensional nanofibrous scaffold for cartilage tissue engineering using human mesenchymal stem cells. Biomaterials. 2005;26:599-609
33. Bartosh TJ, Ylostalo JH, Mohammadipoor A, Bazhanov N, Coble K, Claypool K. et al. Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiinflammatory properties. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13724-9
34. Ylostalo JH, Bartosh TJ, Coble K, Prockop DJ. Human mesenchymal stem/stromal cells cultured as spheroids are self-activated to produce prostaglandin E2 that directs stimulated macrophages into an anti-inflammatory phenotype. Stem Cells. 2012;30:2283-96
35. Sivanathan KN, Gronthos S, Rojas-Canales D, Thierry B, Coates PT. Interferon-gamma modification of mesenchymal stem cells: implications of autologous and allogeneic mesenchymal stem cell therapy in allotransplantation. Stem cell reviews. 2014;10:351-75
36. Wang X, Lazorchak AS, Song L, Li E, Zhang Z, Jiang B. et al. Immune Modulatory Mesenchymal Stem Cells Derived from Human Embryonic Stem Cells Through a Trophoblast-like Stage. Stem Cells. 2015
37. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS. et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-7
38. Lin G, Martins-Taylor K, Xu RH. Human embryonic stem cell derivation, maintenance, and differentiation to trophoblast. Methods in molecular biology. 2010;636:1-24
39. Costa M, Dottori M, Ng E, Hawes SM, Sourris K, Jamshidi P. et al. The hESC line Envy expresses high levels of GFP in all differentiated progeny. Nature methods. 2005;2:259-60
40. Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD. et al. Chemically defined conditions for human iPSC derivation and culture. Nature methods. 2011;8:424-9
41. Kang R, Zhou Y, Tan S, Zhou G, Aagaard L, Xie L. et al. Mesenchymal stem cells derived from human induced pluripotent stem cells retain adequate osteogenicity and chondrogenicity but less adipogenicity. Stem Cell Res Ther. 2015;6:144
42. Duijvestein M, Wildenberg ME, Welling MM, Hennink S, Molendijk I, van Zuylen VL. et al. Pretreatment with interferon-gamma enhances the therapeutic activity of mesenchymal stromal cells in animal models of colitis. Stem Cells. 2011;29:1549-58
43. Neurath MF, Fuss I, Kelsall BL, Stuber E, Strober W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J Exp Med. 1995;182:1281-90
Author contact
![]() Corresponding author: Ren-He Xu, Faculty of Health Sciences, University of Macau, Taipa, Macau, China. Email address: renhexumo
Corresponding author: Ren-He Xu, Faculty of Health Sciences, University of Macau, Taipa, Macau, China. Email address: renhexumo

 Global reach, higher impact
Global reach, higher impact