10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(10):1321-1332. doi:10.7150/ijbs.26605 This issue Cite
Research Paper
Genome-wide Identification and Characterization of Enhancers Across 10 Human Tissues
School of Life Science and Engineering, Southwest Jiaotong University, Chengdu 610031, China.
* These authors contributed equally to this work.
Abstract
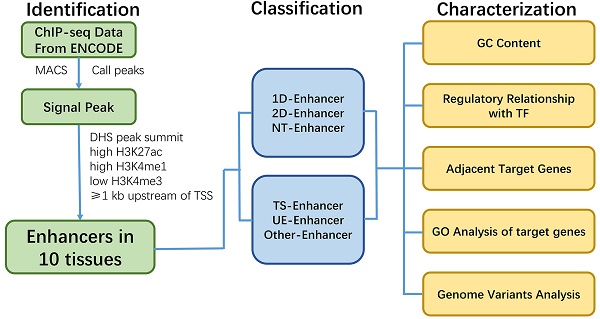
Background: Enhancers can act as cis-regulatory elements (CREs) to control development and cellular function by regulating gene expression in a tissue-specific and ubiquitous manner. However, the regulatory network and characteristic of different types of enhancers (e.g., transcribed/non-transcribed enhancers, tissue-specific/ubiquitous enhancers) across multiple tissues are still unclear.
Results: Here, a total of 53,924 active enhancers and 10,307 enhancer-associated RNAs (eRNAs) in 10 tissues (adrenal, brain, breast, heart, liver, lung, ovary, placenta, skeletal muscle and kidney) were identified through the integration of histone modifications (H3K4me1, H3K27ac and H3K4me3) and DNase I hypersensitive sites (DHSs) data. Moreover, 40,101 tissue-specific enhancers (TS-Enh), 1,241 ubiquitously expressed enhancers (UE-Enh) as well as transcribed enhancers (T-Enh), including 7,727 unidirectionally transcribed enhancers (1D-Enh) and 1,215 bidirectionally transcribed enhancers (2D-Enh) were defined in 10 tissues. The results show that enhancers exhibited high GC content, genomic variants and transcription factor binding sites (TFBS) enrichment in all tissues. These characteristics were significantly different between TS-Enh and UE-Enh, T-Enh and NT-Enh, 2D-Enh and 1D-Enh. Furt hermore, the results showed that enhancers obviously upregulate the expression of adjacent target genes which were remarkably correlated with the functions of corresponding tissues. Finally, a free user-friendly tissue-specific enhancer database, TiED (
Conclusion: Genome-wide analysis of the regulatory network and characteristic of various types of enhancers showed that enhancers associated with TFs, eRNAs and target genes appeared in tissue specificity and function across different tissues.
Keywords: Enhancer, eRNA, Histone modification, Tissue-specific, Transcription factor

 Global reach, higher impact
Global reach, higher impact