10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(8):1591-1599. doi:10.7150/ijbs.34113 This issue Cite
Research Paper
miR-424-5p represses the metastasis and invasion of intrahepatic cholangiocarcinoma by targeting ARK5
1. Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery First Affiliated Hospital, School of Medicine, Zhejiang University
2. NHC Key Laboratory of Combined Multi-organ Transplantation
3. Key Laboratory of the diagnosis and treatment of organ Transplantation,CAMS
4. Key Laboratory of Organ Transplantation, Zhejiang Province, Hangzhou 310003,China
*These authors contributed equally to this work.
Received 2019-2-14; Accepted 2019-5-2; Published 2019-6-4
Abstract
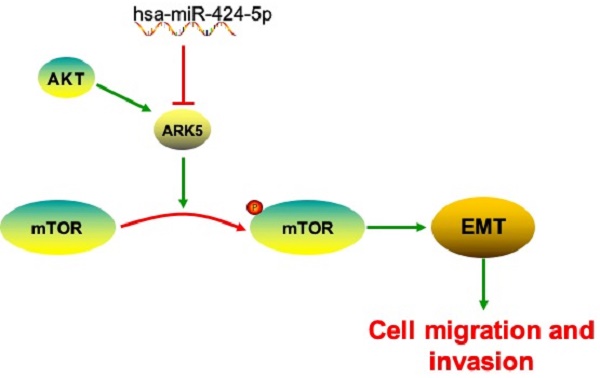
MicroRNAs (miRNAs) have been validated to play prominent roles in the occurrence and development of many kinds of malignant cancer. MiR-424-5p has been reported to participate in various tumors proliferation and metastasis as a suppressor. On the contrary, miR-424-5p would promote cell proliferation in some tumors. However, the expression of miR-424-5p in intrahepatic cholangiocarcinoma (ICC) is rarely reported and its mechanism remains unclear. Here, we discover that miR-424-5p is frequently downregulated in ICC tissues compared with adjacent normal tissues and in ICC cells. Over-expression of miR-424-5p significantly inhibits the invasion and migration of ICC cells in vitro. Importantly, miR-424-5p is found to be a suppressor of ARK5, by binding to 3'-UTR of ARK5 mRNA and then inhibiting mTOR phosphorylated, thus deregulating epithelial-mesenchymal transition (EMT) of ICC. Furthermore, ARK5 is found to play a role in ICC metastasis and regulating EMT. Knockdown of ARK5 inhibits invasion and migration of ICC, while the over-expression gives an opposite effect. Besides, high-expression of ARK5 is also associated with poor prognosis. In conclusion, our study reveals that miR-424-5p is critical to the invasion, migration and EMT progression in ICC cells. Targeting the pathway described here may be a novel approach to inhibit metastasis of ICC and the restoration of miR-424-5p expression may be a promising strategy for ICC therapy.
Keywords: intrahepatic cholangiocarcinoma (ICC), miR-424-5p, ARK5, EMT, mTOR
Introduction
Cholangiocarcinoma is an epithelial cell malignancy arising from varying locations within the biliary tree showing markers of cholangiocyte differentiation[1]. It is commonly divided into three subtypes according to the location of tumor, which are named intrahepatic, perihilar and distal cholangiocarcinoma. Intrahepatic cholangio-carcinoma (ICC) is believed to originate from liver progenitor cell in some recent studies. In recent years, ICC has become the second most common liver primary tumor following hepatocellular carcinoma[2], and its incidence and mortality grow up in the last three decades[3]. Unfortunately, there is still no effective target for ICC and the potential molecular mechanism of its oncogenicity remains unclear. Therefore, it is necessary to search for new biomarkers as valid therapeutic targets of ICC, by digging out their potential mechanism.
MicroRNAs (miRNAs) are short non-coding RNA molecules that usually repress gene expression by binding to the 3′-untranslated region (3′-UTR) of their target mRNAs[4]. Hsa-miR-424-5p has been reported to down-regulate in many kinds of tumors such as esophageal squamous cell carcinoma[5] and non-small cell carcinoma[6]. J. Wang et al. reported miR-424-5p as a tumor suppressive microRNA to regulate basal-like breast cancer proliferation, migration and invasion[7]. However, miR-424-5p is reported to up-regulate in colon cancer[8] and pancreatic carcinoma[9]. It is also reported that miR-424-5p is up-regulated in gastric cancer and promotes gastric cancer proliferation[10]. Therefore, there is no coincident conclusion of miR-424-5p function in malignant tumors.
ARK5 is a novel member of the human AMP-activated protein kinase (AMPK) family, which was discovered to be key molecules mediating migration activity of human pancreatic cancer cells[11]. Besides, the activation of ARK5 is directly mediated by AKT-dependent phosphorylation at serine residue on the regulatory domain[12]. Recently, a few studies elucidate that the expression of ARK5 plays an important role in tumor metastasis and invasion, such as glioma[13], human breast cancer[11], gastric cancer[14] and hepatocellular carcinoma[15]. Furthermore, some studies convince that ARK5 promotes cell epithelial-mesenchymal transition (EMT) in non-small cell lung cancers[16], while it enhances drug resistance in pancreatic carcinoma[17].
In our study, we firstly found that miR-424-5p down-regulated in intrahepatic cholangiocarcinoma (ICC). Then we tried to give evidence for the potential role miR-424-5p plays in ICC. Fortunately, we found that over-expression of miR-424-5p inhibited ICC migration and invasion by inhibiting epithelial-mesenchymal transition (EMT). Furthermore, we successfully demonstrated that miR-424-5p inhibits ICC metastasis and invasion by directly targeting ARK5 thus suppressing phosphorylated mTOR, which indicates that miR-424-5p/ARK5 might serve as a therapeutic target for intrahepatic cholangiocarcinoma.
Materials and Methods
Research involving human participants and animals
In this research, informed consent was obtained from all patients at the original time of collection (2009-2012) for the storage and use of their tissues. The Clinical Specimens Ethics Committee of the First Affiliated Hospital of Zhejiang University School of Medicine (Hangzhou, China) approved the present research.
Cell lines and culture
Three ICC cell lines (CCLP-1, RBE and HuCCT-1) and a normal human intrahepatic biliary epithelial cell line (HIBEC) were purchased from Cell Bank of Type Culture Collection of Chinese Academy of Sciences, (Shanghai, China). Cells were cultivated according to the protocols from their supplier. All cell lines were grown in RPMI-1640 complete medium (Biological Industries, Kibbutz Beit-Haemek, Israel) supplemented with 10% fetal bovine serum (FBS; Moregate Biotech, Brisbane, Australia), and were cultured in an incubator of 37°C and 5% CO2.
RNA Oligoribonucleotides construction and transfection
The hsa-miR-424-5p mimics (named miR-424-5p mimics; miRBase accession MIMAT0001341; sense: 5′-CAGCAGCAAUUCAUGUUUUGAA-3′) and the negative control duplex (named micrONTM miRNA mimic Ncontrol) were bought from Ribobio (Guangdong, PR China). ARK5 short hairpin (sh) RNA (sequence: TGGCCGAGTGGTTGCTATAAA) and negative control shRNA were purchased from BioLink Technologies (Shanghai, PR China). MicroRNA transfection was performed using Lipofectamine 3000 reagents (Invitrogen, Carlsbad, CA, USA) in accordance with the protocol of manufacturer.
Dual-luciferase reporter assay
Dual-luciferase assays were performed using 2 × 104 293T cells per well in a 48-well plate (Corning/Costar, Acton, MA, USA). After the cells attached for 24h, they were co-transfected with 50 ng of respective reporter constructs with either 50 nM of miRNA mimics or control miRNA using Lipofectamine 3000 reagents according to the protocol of manufacturer. After 48 h, a Reporter Assay System Kit (Promega E1910, Beijing, China) was used to measure the luciferase activity. There were three replicates for each transfect ant. Firefly luciferase activity was normalized to constitutive Renilla luciferase activity. The tests were repeated in three independent experiments.
Western blotting and antibodies
ICC tissues and cells were lysed using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) and sonicated. Lysates containing soluble proteins were collected and stored at -80°C. Protein concentration was determined by the Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Equal amounts (30 µg) of proteins were separated by 4-20% SDS-PAGE and transferred onto polyvinylidene difluoride membranes for 1h. The membranes were washed one time with TBS/0.1% Tween-20 (TBST) buffer, and incubated with a solution containing the primary antibody (1:1,000) at 4°C overnight. Then, the membranes were washed three times with TBST, and incubated with a solution containing the horseradish peroxidase (HRP)-conjugated secondary antibody (1:3,000) for 1-2h at room temperature. Following incubation, the membranes were washed three times with TBST. Enhanced chemiluminescence (ECL) (Guge Biotechnology, China) was used to detect the immunoreactive bands, according to the recommendations of manufacturer.
The antibodies used in our research for western blotting: anti-ARK5 (4458S, CST, 1:1000), beta-catenin (8480, CST, 1:1000), N-cadherin (13116, CST, 1:1000), E-cadherin (3195, CST, 1:1000), Claudin-1 (13255, CST, 1:1000), ZO-1 (8193, CST, 1:1000), mTOR (2983, CST, 1:1000), p-mTOR (5536, CST, 1:1000), GAPDH (Epitomics, Burlingame, CA, USA, 1:4000), beta-actin (A5441, Sigma-Aldrich, 1:2000). The antibodies used of immunofluorescence: beta-catenin (8480, CST, 1:200), N-cadherin (13116, CST, 1:200), E-cadherin (3195, CST, 1:200). The antibody used for immunohistochemistry: anti-ARK5 (22723-1-AP, Proteintech, 1:200).
Cell growth/cell viability assay (Cell Count Kit-8 assay)
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to assess cell viability. Cells (4×103 cells/well) were seeded in 96-well plates, and incubated in a humidified incubator for 24, 48 or 72h. The supernatant in each well was then replaced with 90 µl medium and 10 µl CCK-8 solution, and the cells were incubated at 37°C for 1 h. The absorbance was detected at 450 nm using a microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Colony formation assay
Cells (1,000 cells/well) were seeded onto 6-well plates and incubated at 37°C in a humidified incubator. The medium was changed every 4 days. After two weeks, the cells were washed with PBS, fixed with 100% methanol for 30 min at room temperature, and stained with 0.2% crystal violet for 15 min at room temperature. Following staining, the cells were washed with PBS three times and colonies were observed under a light microscope and counted.
Wound-healing assays
Cells (4×105/well) were seeded in 24-well plates, and wounds were generated by making a scratch on the plate using a sterile tip. Cells were washed with PBS and incubated in culture medium without serum. After the indicated time, the distance between the two margins of the wound was measured. The data for the HuCCT-1 cells and RBE cells were measured at 0, 24, 48 and 72h.
Cell migration and invasion assays
Cell migration and invasion assays were performed using Millicell Cell Culture Inserts (24-well plates; 8 µm pore size; Merck KGaA, Darmstadt, Germany). Stably transduced cells were used for these assays. For the migration assay, 5×104 HuCCT-1 cells or 5×104 RBE cells in serum-free medium were seeded on the upper chambers. For the invasion assays, the membranes of the upper chambers were coated with 4 µl Matrigel (BD Biosciences) in 36 µl RPMI-1640 medium for 2h in a humidified incubator. The cells were then seeded in the coated upper chambers. RPMI-1640 medium containing 10% FBS was added into the lower chambers. HuCCT-1 cells and RBE cells were incubated for 24 or 48h for the migration assays, and 48 or 72h for the invasion assays, respectively. Then, the cells on the lower membranes were stained using a Wright-Giemsa Stain kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) and observed at ×100 magnification. Five fields were randomly chosen and cells were counted upon observation under a light microscope, the number of cells of average field was calculated finally.
ICC tissue samples
In our research, 19 samples of ICC tissues with matching peritumoral tissues were collected between 2009 and 2012 at the First Affiliated Hospital, Zhejiang University School of Medicine (Hangzhou, China). Ten samples of ICC tissues with paired peritumoral tissues were randomly selected to measure the expression of miR-424-5p by RT-PCR. Immuno-histochemistry was performed as previously described[18]. The assessment of the staining was based on the following formula: Total score = the score of the percentage of positively stained cells over total × the score of the staining intensity. The scores of the percentage of positively stained cells were as follows: 0, ≤5%; 1, 5-25%; 2, 25-50%; 3, 50-75%; 4, >75%. The staining intensity scores were as follows: 1, low intensity; 2, medium intensity; 3, high intensity.
Results
The expression of hsa-miR-424-5p in ICC tissues and cell lines
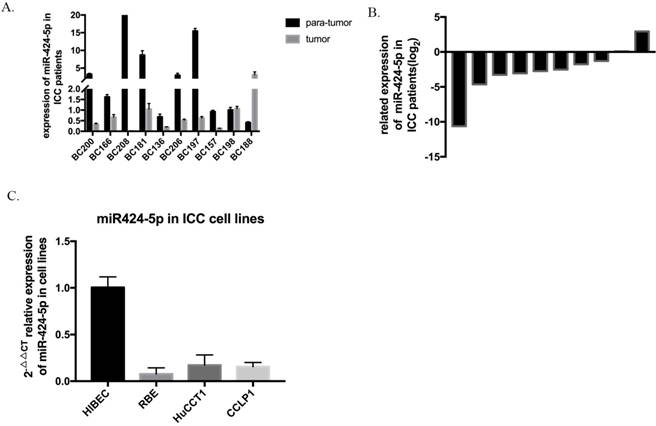
The expression level of hsa-miR-424-5p was analyzed by qRT-PCR and normalized to an endogenous control (U6 RNA). In our 10 pairs of intrahepatic cholangiocarcinoma patients' tissues, 8 pairs show that hsa-miR-424-5p expresses much lower in ICC tissue than in normal hepatic tissue (Figure 1A&B). Then we measured miR-424-5p expression in a panel of human ICC cell lines as wells as normal bile duct cell line HIBEC. Similarly, the expression of miR-424-5p is much lower in every cancer cell line (Figure 1C). These data suggest that hsa-miR-424-5p decreases in both intrahepatic cholangiocarcinoma tissues and cells.
Hsa-miR-424-5p inhibits tumor metastasis and invasion in vitro
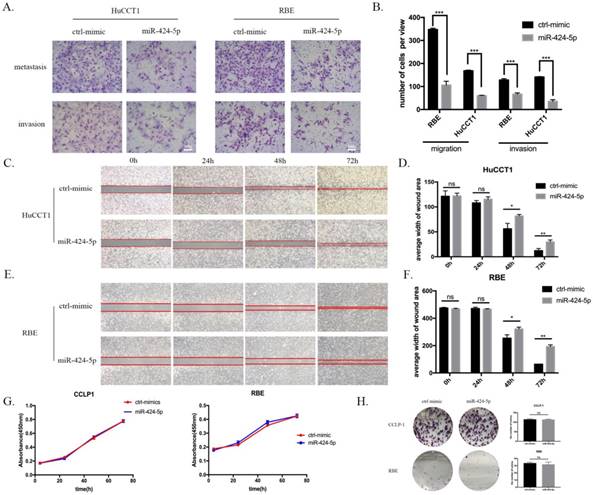
To explore the potential role of miR-424-5p in intrahepatic cholangiocarcinoma cells, we transfected ICC cells with miR-424-5p mimics (from Ribobio. China) and performed in vitro transwell assays and wound-healing assays in HuCCT1 and RBE cell lines. Compared to negative-control group, both HuCCT1 and RBE, after being transfected by miR-424-5p mimics in transwell assays, significantly decreased the number of cells invading or migrating the chamber membrane with or without Matrigel (Figure 2 A&B). What's more, in our wound-healing assay, the ability of migration of HuCCT1 cells, transfected by miR-424-5p mimics, was inferior to those transfected by negative-control mimics (Figure 2 C&D). We detected the similar situation in the assay of RBE cells (Figure 2 E&F). In transwell and wound-healing assays, we similarly found that miR-424-5p inhibits the motility of CCLP-1 cells (Supplementary Figure 1&2).
At the same time, we tested the proliferating ability of these two cell lines using in vitro CCK-8 assay and colony formation assay. As a result, none of CCLP-1, RBE and HuCCT-1 cell viability was inhibited by miR-424-5p mimics transfecting in CCK-8 assay, compared to the negative-control (Figure 2 G and Supplementary Figure 3). Besides, the colony formation assay showed that the colony dots of miR-424-5p mimics groups quite resembled to the negative-control groups in the number and diameter (Figure 2 H and Supplementary Figure 4).
To sum up, these data suggested that hsa-miR-424-5p can inhibit the migration and invasion of ICC cells in vitro, exerting no influences on the proliferation.
miR-424-5p inhibits metastasis of ICC cells by inhibiting EMT
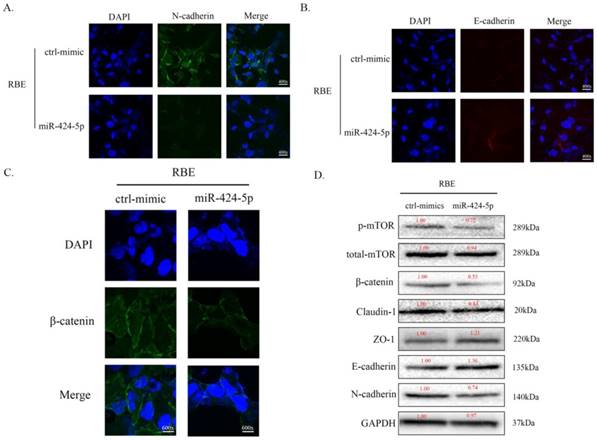
To investigate the potential mechanism of miR-424-5p in ICC metastasis, we transfected RBE cells with miR-424-5p mimics or negative-control (NC) mimics. In cell immunofluorescence assays, we found that the expression of N-cadherin in miR-424-5p group was downregulated compared to the NC group (Figure 3A) while the E-cadherin showed an opposite situation (Figure 3B). Meanwhile, the expression of beta-catenin in miR-424-5p group was also downregulated (Figure 3C). In addition, the western blot assays showed that, with miR-424-5p transfection, the expression of Claudin-1 reduced while the expression of ZO-1 increased (Figure 3D). Being taken together, these data indicate that miR-424-5p inhibits ICC cells metastasis by inhibiting their epithelial-mesenchymal transition.
The expression of miR-424-5p in intrahepatic cholangiocarcinoma. A. The expression of miR-424-5p in ten pairs of ICC tissues and adjacent normal tissues from patients, testing by qRT-PCR. B. Related expression of miR-424-5p in ICC patients were presented as the fold change to the corresponding adjacent normal tissues (T/N). C. The expression of miR-424-5p in different kinds of ICC cells (RBE, HuCCT-1, CCLP-1) and a normal human intrahepatic biliary epithelial cell line (HIBEC).
miR-424-5p inhibits invasion and migration of ICC in vitro. (A & B) Representative images of transwell assays of HuCCT-1 and RBE cells transfected with miR-424-5p mimics or negative control. (C-F) HuCCT-1 and RBE cells were pretreated with NC mimics or miR-424-5p mimics for 24h respectively. Wound-healing assay was performed to explore the motility of each cell line. Similar effect was observed in both HuCCT-1 and RBE cells. G. Cell Counting Kit-8 (CCK-8) assay results show no difference in cell proliferation and cell viability after transfected with NC mimic or miR-424-5p mimic for 24h. H. Representative images of colony formation assay give similar results in cell proliferation. Every experiment above was repeated 3 times independently. *:p<0.05, **:p<0.01, ***:p<0.001 compared to NC group.
miR-424-5p inhibits ICC metastasis by inhibiting EMT. A. Representative immunofluorescence images of expression of N-cadherin (green signal) in RBE cells after transfected by NC mimic or miR-424-5p mimic for 48h. DAPI (blue signal) was used to counterstain the cell nuclei. B. Representative images of expression of E-cadherin (red signal) in RBE cells after transfected by NC mimic or miR-424-5p mimic for 48h. C. Representative immunofluorescence images of expression of beta-catenin (green signal) in RBE cells. D. The cell lysates of RBE cells transfected with NC mimic or miR-424-5p mimic were used to perform western blot assays with indicated antibodies and GAPDH was used to be the loading control. Every experiment above was repeated 3 times independently.
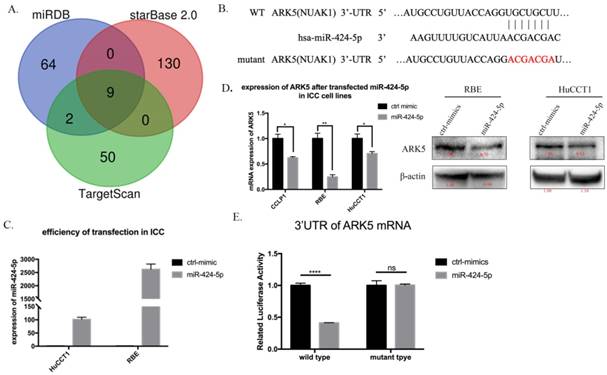
miR-424-5p directly target ARK5 by binding to its 3'UTR. A. Venn diagrams of calculating the intersections of the four target prediction engines (miRDB, starBase v2.0 and TargetScan; analyzed by R package Venn Diagram). B. miR-424-5p and its putative binding sequence in the 3′UTR of ARK5. The mutant miR-424-5p binding site was generated in the complementary site for the seed region of miR-424-5p. C. Expression of miR-424-5p in HuCCT-1 and RBE cells after transfected miR-424-5p mimic testing by qRT-PCR. D. Expression of ARK5 in mRNA level (left) and protein level (right) in ICC cell lines after transfected NC mimic or miR-424-5p mimic. E. 293T cells were co-transfected with miR-424-5p mimics or NC and wild-type or mutant-type of 3'UTR of ARK5. And the luciferase activities were examined. The firefly luciferase activity of each sample was normalized to the Renilla luciferase activity. ****:p<0;0001.
miR-424-5p inhibits ICC metastasis by directly targeting ARK5
Next, we searched for a candidate target gene responsible for suppression of miR-424-5p in ICC cells. We comprehensively analyzed TargetScan, miRDB and starBase database. ARK5(NUAK1) was chosen as a potential target of miR-424-5p (Figure 4A, Supplementary Table 1 and Supplementary Figure 5). Then we transfected HuCCT1 cells and RBE cells with negative-control mimics or miR-424-5p mimics. Figure 3C presents the qRT-PCR result of transfection efficiency. qRT-PCR and western blot assay indicated that over-expression of miR-424-5p, in HuCCT1 cells and RBE cells, occurred down-regulation of ARK5 expression (Figure 4D). Further, we explored whether miR-424-5p directly targets ARK5 by dual-luciferase reporter assays. The results revealed that co-transfection of miR-424-5p mimics significantly decreased the activity of firefly luciferase reporter with wild-type 3'UTR of ARK5 rather than mutant one (Figure 4E). After all, these data suggest that miR-424-5p could directly bind to the 3'UTR of ARK5, thereby down-regulating its expression.
ARK5 promotes metastasis of ICC cells in vitro
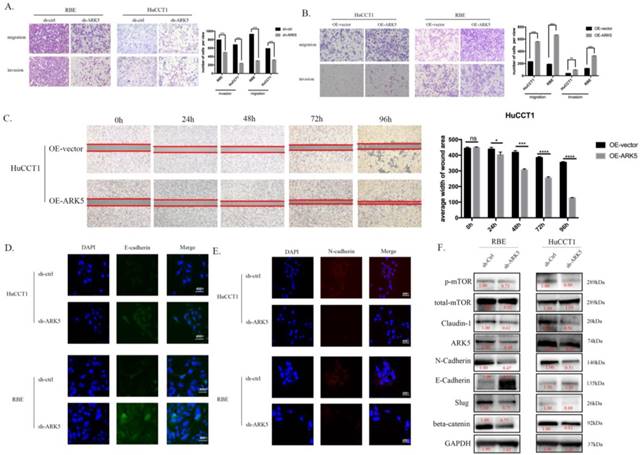
We detected the expression of ARK5 in HIBEC and all 3 ICC cell lines by western blot and found that the expression of ARK5 is higher in ICC cell lines (Supplementary Figure 6). We decreased the expression of ARK5 in HuCCT1 cells and RBE cells using short hairpin RNA. In transwell assays, knockdown ARK5 significantly decreased the motility of HuCCT1 cells and RBE cells comparing with negative control (Figure 5A), while over-expressing ARK5 leads to an opposite result (Figure 5B). Similarly, over-expressing ARK5 in HuCCT1 cells occurred an acceleration of wound closure compared to negative-control treatment (Figure 5C). In cell immunofluorescence assay, knockdown ARK5 deregulated the expression of N-cadherin in HuCCT1 cells and RBE cells, while up-regulating E-cadherin expression (Figure 5D&E). What's more, western blot assays show the same results of N-cadherin decreasing and E-cadherin increasing (Figure 5F) in ARK5 knockdown.
ARK5 is high-expressing in ICC tissue and associated with poor prognosis
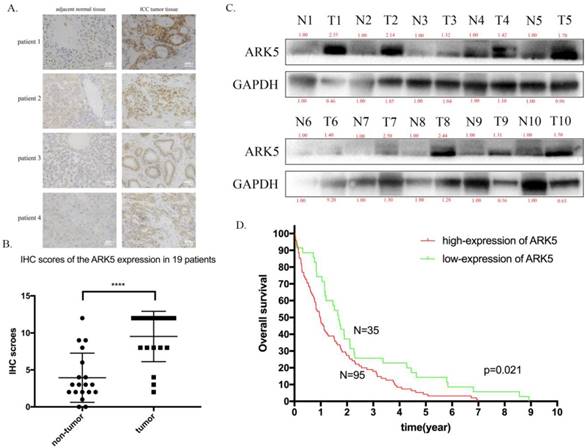
Then, we explored the expression of ARK5 in ICC tissues along with adjacent normal tissues by immunohistochemistry staining. As a result, the expression of ARK5 in ICC tissues is higher than their matched adjacent normal tissues (Figure 6A&B). Then we randomly chose 10 pairs of ICC samples to determine the protein expression level of ARK5 by western blotting. It turned out that the expression of ARK5 in 9 pairs is markedly higher in ICC tissues than their paired adjacent normal tissues (Figure 6C). To elucidate the association between ARK5 and overall survival, we studied the relevant information of an ICC cohort from The Cancer Genome Atlas (TCGA, n=130). The cut off for low/high expression of ARK5 is quarter. Kaplan-Meier survival analysis revealed that patients with low expression of ARK5 had a better overall survival than those with high expression of ARK5 (Figure 6D).
ARK5 promotes metastasis of ICC cells in vitro. (A&B) ARK5 knockdown significantly suppressed migration and invasion of HuCCT-1 and RBE cells, as detected by transwell assays. C. Over-expression ARK5 promotes HuCCT-1 cells motility by wound-healing assay. (D&E) Representative immunofluorescence images of the expression of E-cadherin (green signal) and N-cadherin (red signal) in RBE and HuCCT-1 cells after transfected by NC mimic or miR-424-5p mimic for 48h. DAPI (blue signal) was used to counterstain the cell nuclei. As expected, the expression of E-cadherin up-regulated while N-cadherin down-regulating. F. RBE and HuCCT-1 cells were transfected with ARK5 knockdown short hairpin RNA. And the lysates were used to perform western blot analysis with the indicated antibodies and GAPDH was used as a loading control. Error bars represent the S.D. obtained from three independent experiments. *:p<0.05, **p<0.01, ***:p<0.001, ****:p<0.0001.
miR-424-5p suppresses ICC metastasis by negative regulating ARK5 and mTOR
The potential molecular mechanism of the effect of miR-424-5p on ICC, has been revealed through the experiments above. The western blotting results showed that, after being transfected with miR-424-5p mimics, the expression of Claudin-1 and beta-catenin was down-regulated in RBE cells, so was the expression of phosphorylated mTOR (Figure 3D). Among the predicted targets of miR-424-5p, we chose ARK5 as the potential target, which has been reported to be along with the variation of phosphorylated mTOR[14]. In our study, ICC cells transfected with miR-424-5p mimics displayed a decrease of phosphorylated mTOR and ARK5 (Figure 4D) in western blotting assay. Meanwhile, ICC cells transfected with ARK5-knockdown short hairpin RNA also displayed a down-regulation of phosphorylated mTOR (Figure 5F).
Discussion
Intrahepatic cholangiocarcinoma (ICC) has become the focus of increasing concern in the few decades, largely due to its rising incidence and mortality worldwide. ICC is largely diagnosed at an advanced non-curable stage and usually occurs sporadically in patients without recognizable risk factors[3]. Unlike hepatocellular carcinoma (HCC), most ICCs develop in non-cirrhotic liver, although it has been reported that about 8-10% of all ICCs occur in patients with liver cirrhosis[19]. However, the molecular mechanism of oncogenesis in ICC remains complex.
MicroRNAs have been found to regulate the progress of various malignant tumor in recent years, which provides us a brand-new perspective to explore the potential mechanism. Recently, a number of evidences confirmed that some microRNAs were involved in this process[20]. miR-424-5p has been reported to inhibit tumor invasion and metastasis of esophageal squamous cell carcinoma by targeting Smad7[21]. Yet, there is a report showing that miR-424-5p would promote cell proliferation in gastric cancer by targeting Smad3 via TGF-beta pathway[10]. Therefore, the function of miR-424-5p remains complex and conflicting. Nonetheless, there were no reports on the expression and role of miR-424-5p in ICC though it is known as a kind of highly aggressive carcinoma with lymph nodes metastasis.
Accordingly, we tried to explore the potential role miR-424-5p playing in ICC, due to its reported relation between tumor invasion and migration. Our study, for the first time, reported that miR-424-5p is low expressed in ICC tissue compared to the adjacent normal tissue. The gain-of-function assays indicated that miR-424-5p plays an essential role in suppressing invasion and migration of RBE and HuCCT1 cells in vitro. The western blotting results revealed that ectopic expression of miR-424-5p increased the expression of E-cadherin and ZO-1, yet over-expression of miR-424-5p decreased the expression of N-cadherin, beta-catenin, Claudin-1 and phosphorylated mTOR. So did the cell immunofluorescence assays. These data suggest that miR-424-5p suppresses the epithelial-mesenchymal transition phenotype of ICC. Being taken together, these data indicate that miR-424-5p acts as a potential suppressor in ICC, especially in tumor invasion and migration via mTOR pathway.
In order to explore how miR-424-5p comes into play, we used TargetScan, miRBD and starBase databases to identify target gene. Our results firstly report AMPK-related kinase 5 (ARK5) as direct functional target of miR-424-5p in ICC. Depletion of ARK5, also known as NUAK1, induced the accumulation of MYC-expressing cells[22]. Besides, it is reported that AMPK inhibits the mTORC1 pathway that controls anabolic cell growth[23]. The dual-luciferase activity assay showed the decreasing of luciferase activity after co-transfection of miR-424-5p and CMT-ARK5-WT-UTR. These data reveal that miR-424-5p would directly bind to wild-type 3'-UTR of ARK5 rather than mutant one.
Moreover, silencing ARK5 by short hairpin RNA achieved the same effect of inhibiting invasion and migration in ICC cells. Western blotting results revealed the up-regulating of E-cadherin and down-regulating of N-cadherin, beta-catenin, Claudin-1, Slug and phosphorylated mTOR. Perumal, D., et al. reported a novel function of ARK5 in bridging the mTOR/Rb/MYC pathways[24], which supports our discoveries. What's more, we also uncover the expression in ICC tissues and discover its high expression relating to poor prognosis for the first time. Finally, we should take note that a single microRNA may regulate the mRNA transcripts of a few genes. We cannot deny other possibilities of other pathways mediated EMT[7, 21]. Therefore, the new miR-424-5p/ARK5/p-mTOR pathway might serve as a new orientation for us to know ICC, also as a promising therapeutic target, or at least an approach to reduce tumor metastasis and recurrence.
ARK5 is highly expressed in ICC tissues and associated with poor prognosis. A. Representative images of immunohistochemistry (IHC) staining of ICC patient. The deep brown staining demonstrated the high expression of ARK5, and is deeper in ICC tumor tissue than compared adjacent normal tissue. B. Total staining scores from the ARK5 IHC results in paired tumor and peritumoral tissues. C. ARK5 protein expression levels were examined by western blotting in randomly chosen 10 pairs of ICC and normal peritumoral tissues. D. Kaplan-Meier survival curves of ICC cohort from TCGA with different ARK5 expression levels. ****:p<0.0001.
In conclusion, our results show that miR-424-5p is low-expressed in both tissues and cells of intrahepatic cholangiocarcinoma. Exogenous expression of miR-424-5p could significantly inhibit invasion and migration of ICC in vitro. Under the investigation of the underlying mechanism, we found that miR-424-5p processes the effect by directly targeting ARK5 so as to inhibit mTOR phosphorylated, thus down-regulating epithelial-mesenchymal transition in ICC. Our study uncovers the potential role of miR-424-5p in ICC progression. We expect that our findings on miR-424-5p and its target ARK5 could provide a promising strategy for ICC therapy.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This research was supported by Innovative Research Groups of National Natural Science Foundation of China (No. 81721091) and National Natural Science Foundation of China (No. 81874228).
Author Contributions
Conception and design of the experiments: Jingbang Wu, Beng Yang, Bin He. Collection, analysis, and interpretation of all data: Jingbang Wu, Yanpeng Zhang, Xiaode Feng. Draft-ing and revising the paper critically for intellectual content: Haiyang Xie, Lin Zhou, Jian Wu, Shusen Zheng.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet (London, England). 2014;383:2168-79
2. Razumilava N, Gores GJ. Combination of gemcitabine and cisplatin for biliary tract cancer: a platform to build on. Journal of hepatology. 2011;54:577-8
3. Sirica AE, Gores GJ, Groopman JD, Selaru FM, Strazzabosco M, Wang XW. et al. Intrahepatic Cholangiocarcinoma: Continuing Challenges and Translational Advances. Hepatology (Baltimore, Md). 2018
4. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20
5. Chen X, Hu H, Guan X, Xiong G, Wang Y, Wang K. et al. CpG island methylation status of miRNAs in esophageal squamous cell carcinoma. International journal of cancer. 2012;130:1607-13
6. Donnem T, Fenton CG, Lonvik K, Berg T, Eklo K, Andersen S. et al. MicroRNA signatures in tumor tissue related to angiogenesis in non-small cell lung cancer. PloS one. 2012;7:e29671
7. Wang J, Wang S, Zhou J, Qian Q. miR-424-5p regulates cell proliferation, migration and invasion by targeting doublecortin-like kinase 1 in basal-like breast cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;102:147-52
8. Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. Journal of digestive diseases. 2010;11:50-4
9. Wu K, Hu G, He X, Zhou P, Li J, He B. et al. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathology oncology research: POR. 2013;19:739-48
10. Wei S, Li Q, Li Z, Wang L, Zhang L, Xu Z. miR-424-5p promotes proliferation of gastric cancer by targeting Smad3 through TGF-beta signaling pathway. Oncotarget. 2016;7:75185-96
11. Chang XZ, Yu J, Liu HY, Dong RH, Cao XC. ARK5 is associated with the invasive and metastatic potential of human breast cancer cells. Journal of cancer research and clinical oncology. 2012;138:247-54
12. Suzuki A, Kusakai G, Kishimoto A, Lu J, Ogura T, Lavin MF. et al. Identification of a novel protein kinase mediating Akt survival signaling to the ATM protein. The Journal of biological chemistry. 2003;278:48-53
13. Lu S, Niu N, Guo H, Tang J, Guo W, Liu Z. et al. ARK5 promotes glioma cell invasion, and its elevated expression is correlated with poor clinical outcome. European journal of cancer (Oxford, England: 1990). 2013;49:752-63
14. Chen D, Liu G, Xu N, You X, Zhou H, Zhao X. et al. Knockdown of ARK5 Expression Suppresses Invasion and Metastasis of Gastric Cancer. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;42:1025-36
15. Ye Z, Chen X, Chen X. ARK5 promotes invasion and migration in hepatocellular carcinoma cells by regulating epithelial-mesenchymal transition. Oncology letters. 2018;15:1511-6
16. Li M, Zheng C, Xu H, He W, Ruan Y, Ma J. et al. Inhibition of AMPK-related kinase 5 (ARK5) enhances cisplatin cytotoxicity in non-small cell lung cancer cells through regulation of epithelial-mesenchymal transition. American journal of translational research. 2017;9:1708-19
17. Wang X, Song Z, Chen F, Yang X, Wu B, Xie S. et al. AMPK-related kinase 5 (ARK5) enhances gemcitabine resistance in pancreatic carcinoma by inducing epithelial-mesenchymal transition. American journal of translational research. 2018;10:4095-106
18. Xiao H, Tong R, Cheng S, Lv Z, Ding C, Du C. et al. BAG3 and HIF-1 alpha coexpression detected by immunohistochemistry correlated with prognosis in hepatocellular carcinoma after liver transplantation. BioMed research international. 2014;2014:516518
19. Jesper D, Heyn SG, Schellhaas B, Pfeifer L, Goertz RS, Zopf S. et al. Effects of liver cirrhosis and patient condition on clinical outcomes in intrahepatic cholangiocarcinoma: a retrospective analysis of 156 cases in a single center. European journal of gastroenterology & hepatology. 2018;30:552-6
20. Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Current opinion in cell biology. 2013;25:200-7
21. Wang F, Wang J, Yang X, Chen D, Wang L. MiR-424-5p participates in esophageal squamous cell carcinoma invasion and metastasis via SMAD7 pathway mediated EMT. Diagnostic pathology. 2016;11:88
22. Liu L, Ulbrich J, Muller J, Wustefeld T, Aeberhard L, Kress TR. et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483:608-12
23. Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature reviews Cancer. 2009;9:563-75
24. Perumal D, Kuo PY, Leshchenko VV, Jiang Z, Divakar SK, Cho HJ. et al. Dual Targeting of CDK4 and ARK5 Using a Novel Kinase Inhibitor ON123300 Exerts Potent Anticancer Activity against Multiple Myeloma. Cancer research. 2016;76:1225-36
Author contact
![]() Corresponding author: Prof. Shusen Zheng, E-mail: shusenzhengedu.cn and Prof. Jian Wu, E-mail: drwujianedu.cn
Corresponding author: Prof. Shusen Zheng, E-mail: shusenzhengedu.cn and Prof. Jian Wu, E-mail: drwujianedu.cn

 Global reach, higher impact
Global reach, higher impact