ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2023; 19(12):3640-3660. doi:10.7150/ijbs.85126 This issue Cite
Research Paper
Protection of c-Fos from autophagic degradation by PRMT1-mediated methylation fosters gastric tumorigenesis
1. Department of Integrative Biotechnology and Biomedical Institute for Convergence at SKKU (BICS), Sungkyunkwan University, Suwon 16419, Republic of Korea.
2. R&D Center, Yungjin Pharmaceutical Co, Suwon 16229, Republic of Korea.
3. Emergency Department, Hermina Hospital Tangkubanprahu, Malang 65119, Indonesia.
4. Pharmacy Program, Faculty of Science and Engineering, Universitas Ma Chung, Malang 65151, Indonesia.
5. Proteomic Analysis Team, Research Institute, National Cancer Center, Goyang 10408, Republic of Korea.
6. Division of Translational Science, Research Institute, National Cancer Center, Goyang 10408, Republic of Korea.
7. Department of Physiology and Division of Pathology, Michigan State University, East Lansing, Michigan 48824, USA.
8. Department of Molecular Cell Biology, Single Cell Network Research Center, Sungkyunkwan University School of Medicine, Suwon 16419, Republic of Korea.
9. Department of Surgery, Department of Biomedical Sciences, Ajou University School of Medicine, Suwon 16499, Republic of Korea.
*These authors contributed equally to this work.
Abstract
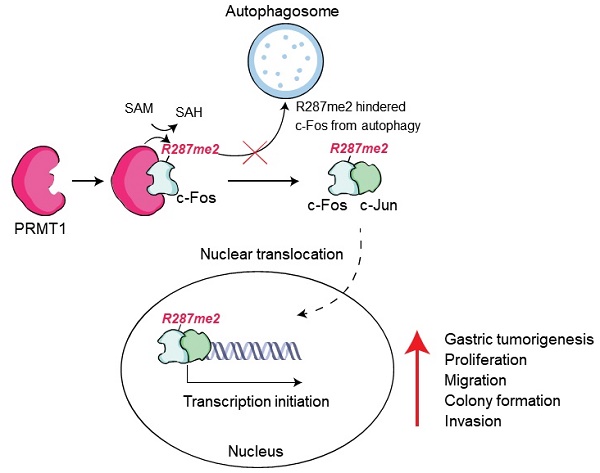
Both AP-1 and PRMT1 are vital molecules in variety of cellular progresssion, but the interaction between these proteins in the context of cellular functions is less clear. Gastric cancer (GC) is one of the pernicious diseases worldwide. An in-depth understanding of the molecular mode of action underlying gastric tumorigenesis is still elusive. In this study, we found that PRMT1 directly interacts with c-Fos and enhances AP-1 activation. PRMT1-mediated arginine methylation (mono- and dimethylation) of c-Fos synergistically enhances c-Fos-mediated AP-1 liveliness and consequently increases c-Fos protein stabilization. Consistent with this finding, PRMT1 knockdown decreases the protein level of c-Fos. We discovered that the c-Fos protein undergoes autophagic degradation and found that PRMT1-mediated methylation at R287 protects c-Fos from autophagosomal degradation and is linked to clinicopathologic variables as well as prognosis in stomach tumor. Together, our data demonstrate that PRMT1-mediated c-Fos protein stabilization promotes gastric tumorigenesis. We contend that targeting this modification could constitute a new therapeutic strategy in gastric cancer.
Keywords: autophagy, c-Fos, gastric cancer, PRMT1, protein methylation, tumorigenesis
Introduction
Protein arginine methyltransferases (PRMTs) modulate carrying a methyl group (CH3) to an arginine residue in proteinaceous substrates as a posttranslational modification [1,2]. In addition to being identified in histone substrates, PRMTs have been discovered in many nonhistone substrates, including those in the nucleus, cell membrane, and cytoplasm [3-5]. Members of the PRMTs are classified into 4 types derived from the type of arginine methylation catalyzed [6]. PRMT1 is a type I PRMT, which catalyzes the formation of monomethylated arginine (omega-NG-monomethylarginine [MME-R]) and asymmetric NG,NG-dimethylarginine (ADME-R). Among the types of PRMTs, PRMT1 is the predominant type in mammals and accounts for >80% of PRMT activity [3]. Hence, the various roles of PRMT1 in various processes and pathological conditions, such as inflammation, oxidative stress, cancer, and lymphocyte function, have been investigated [7-12].
The activator protein 1 (AP-1) contains homo- or hetero-dimers composed of different subunits from the Jun, Fos, and ATF families [13-16]. AP-1 activity is stimulated by a variety of ligands, including cytokines, chemokines, and oncogenic stimuli, to modulate numerous cellular processes. In addition, posttranslational modifications such as methylation and SUMOylation have been proved to alter the transcriptional capacity of AP-1 by affecting its subunits [17-21]. Fos proteins are basic region-leucine zipper (bZIP) proteins that interact with other bZIP or Jun proteins to build an AP-1 complex. Numerous tissues or types of cells constitutively express c-Fos, which regulates downstream genes by heterodimerizing with Jun proteins to mediate proliferation, angiogenesis, invasion, and metastasis [22-27]. Fos has been shown to be essential for autophagy induction and is expressed at truly lower levels in melanoma tissues than in the counterpart nontumor tissues [23]. In addition, c-Fos-mediated transcriptional activation is regulated by PRMT4/CARM1, which is involved in cancer and other diseases [28]. However, i whether the c-Fos protein is affected or methylated by other PRMTs and how c-Fos arginine methylation influences disease development are unknown.
Gastric cancer (GC) has the highest mortality rate among malignant tumors and is one of the central causations of cancer-pertinent death globally, with a five-year alive rate of 20% [29-31]. Despite advances in surgery and other therapies, patients with GC continue to have a poor prognosis and low chemosensitivity after surgical resection [29,32]. Suppression of metastasis is key to increasing the survival rate of stomach cancer patients; thereupon, it is crucial to fully comprehend the fundamental pathophysiological and molecular events of GC invasion and metastasis. Both c-Fos and PRMT1 are important in cellular processes, and an interesting question is whether their interaction participates in cellular functions. Research on the function of particular PRMT family members in GC development is scarce. Such information is essential for the reasonable development of PRMT modulators for the remedy of cancerous diseases.
In this study, we focused on the c-Fos AP-1 subunit and investigated the function of PRMT1 in regulating c-Fos activity. We also identified the methylated arginine sites of c-Fos and reported how this methylation affected underlying mechanisms in GC cells. Notably, PRMT1 methylated c-Fos to modulate its protein stability via autophagy blockade, which supported gastric tumorigenesis. Together, our studies support an important role for PRMT1 in c-Fos and AP-1 regulation; specifically, inhibiting this axis might be an approach to treat GC.
Materials and Methods
Chemicals and antibodies
MG132, 3-methyladenine (3-MA), bovine serum albumin (BSA), cycloheximide (CHX), polyethylenimine (PEI), and 4',6-diamidino-2-phenylindole (DAPI) (chemical identifiers: 1211877-36-9, 5142-23, C7698, 9002-98-6, and 11024-24-1, respectively) were procured from Sigma. Luciferase-encoded DNA constructs containing AP-1 response elements were attained from Promega (Shanghai, China). The Myc-c-Fos, Flag-c-Fos, Flag-PRMT1, and EGFP-PRMT5 plasmids were constructed in our laboratory; the EGFP-PRMT1 and EGFP-PRMT3 plasmids were kind gifts from Prof. Kim (Sookmyung Women's University); and the pLKO.1 vector was obtained from Addgene (10878; deposited by David Root). Proteinase K (KB-0111) was acquired from Bioneer (Daejeon, Republic of Korea). The following primary antibodies were used: anti-LC3B, anti-MME-R, anti-ADME-R, anti-Myc, and anti-Flag (3868, 8015, 13522, 2276, and 8146; Cell Signaling Technology, Beijing, China). Antibodies recognizing β-actin, c-Fos, and GFP (sc-166940, sc-47778, and sc-9996; Santa Cruz Biotechnology, Heidelberg, Germany) were obtained, and Alexa Fluor 405-labelled and Alexa Fluor 568-labelled secondary antibodies to mouse and rabbit immunoglobulin (Invitrogen, Carlsbad, CA, USA) (A-31553 and A-11011) were used for staining.
Cell culture
HEK293T (CRL-1573; ATCC, Manassas, VA, USA), MKN45, and MKN1 cells (80103 and 80101; KCLB, Seoul, Republic of Korea) were grown in RPMI 1640 medium or DMEM (SH30243.01 or SH30027.01; HyClone Laboratories, Sungnam, Republic of Korea) supplemented with antibiotics and fetal bovine serum (FBS) (16000-044; Gibco Laboratories, Tokyo, Japan). These cells were nourished in a five percent CO2 moisturized incubator (37 °C) following a previously reported protocol [33].
Constructs and mutagenesis
The expression constructs for c-Fos and PRMT5 were generated through PCR amplification, utilizing HEK293T cell cDNA as the template. A PRMT1 dominant negative (DN) mutant with mutation of 63VLD65 to 63AAA65 was also constructed with a Stratagene QuikChange mutagenesis kit (Stratagene, Seoul, Republic of Korea). Similarly, c-Fos methylated point mutants (R108K, R201K, R279K, R287K, and R287F) were generated using the same methodology. The nucleic acid primers employed for generating the site- or domain-targeted mutants are provided in Table 1.
List of primers used to generate the c-Fos mutant constructs.
| Target | Sequence (5' to 3') | |
|---|---|---|
| R108K | F | cgctggggcttactccaaggctggcgtt |
| R | aacgccagccttggagtaagccccagcg | |
| R201K | F | catcctggcagctcacaaacctgcctgcaagatc |
| R | gatcttgcaggcaggtttgtgagctgccaggatg | |
| R279K | F | ctgttcccagcatcatccaagcccagtggc |
| R | gccactgggcttggatgatgctgggaacag | |
| R287K | F | tggctctgagacagccaagtccgtgccagacatgg |
| R | ccatgtctggcacggacttggctgtctcagagcca | |
| R287F | F | ggctctgagacagccttctccgtgccagacat |
| R | atgtctggcacggagaaggctgtctcagagcc |
Lentivirus-mediated knockdown with short hairpin RNA (shRNA)
The plasmids containing shRNA sequences targeting PRMT1 were constructed following the protocols provided by Addgene (www.addgene.org). The pLKO.1 vector was used for constructing the plasmids containing the nontargeting scrambled shRNA sequence (TCCTAAGGTTAAGTCGCCCTCG) and the PRMT1 shRNA sequence (CCGGCAGTACAAAGACTACAA). Lentivirus production was carried out by transient transfection of HEK293T cells. The resulting lentiviruses were then utilized to infect cells, and subsequently, puromycin treatment was employed to select cells that were stably transduced with either shScramble or shPRMT1. The effectiveness of PRMT1 knockdown was verified by immunoblotting.
DNA transfection and luciferase reporter assay
It is generally accepted that cells with a high transfection efficiency are required for luciferase assays. Since HEK293T cells are known to have a higher transfection efficiency than other cell lines, these cells were chosen for this work, as reported previously [34,35]. To perform this assay, HEK293T cells were transfected in a 24-well plate with either empty vector or the specified plasmids (c-Fos, PRMT1, PRMT3, or PRMT5) at 0.25 μg per well. Additionally, Luc constructs were transfected at 0.25 μg per well along with β-galactosidase at 0.1 μg per well. Transfection was performed using polyethylenimine (PEI) following a previously reported method [36]. The next day, the cells were retreated with FBS-containing medium or the desired compound and further incubated for an additional day. Following this treatment, the cells transfected with DNA were harvested, and the activity of the luciferase reporter was quantified by employing a Promega enzyme determination kit (E1500; Promega, Beijing, China).
Preparation of whole-cell lysates and tissues; immunoblotting
We collected thirty pairs of stomach cancer tissues and normal adjacent tissues (NATs) from stomach cancer patients who received surgery at AUH (Ajou University Hospital, Suwon, Republic of Korea). The specimens were sourced from the AHBRB (Ajou Human Bio-Resource Bank). All patients agreed the use and storage of their samples. The project was orchestrated with strict adherence to the ethical guidelines and permission from the Institutional Review Board of Ajou University Hospital (AJIRB-BMRKSP-19-059), as previously mentioned [30,33]. The tumor tissue and NAT specimens obtained from GC patients were crushed under liquid nitrogen, while cells were washed with PBS (B2814; Samchun Pure Chemical, Pyeongtaek, Republic of Korea). The washed cells were collected, centrifuged, and lysed in buffer, as described in previous reports [37,38]. The upper layer obtained after centrifugation was collected and utilized for Western blot analysis. Antibodies specific for Myc, Flag, GFP, total c-Fos, LC3B, MME-R, ADME-R, and β-actin were used for immunoblotting [39].
Immunoprecipitation
Cell lysates containing 1,000 μg protein were prepared and reacted with 5 μL of the primary antibodies overnight with stirring in a refrigerator. After the incubation period, immune complexes were incubated with protein A- or G-coupled Sepharose beads (40 μL, 50% v/v) with rotation (4 h, 4 °C). Next, the immune complexes were boiled and subjected to immunoblotting to measure the protein levels following the methodology previously reported [40-42].
Immunofluorescence staining and image analysis
Adherent HEK293T cells expressing Myc-c-Fos and/or EGFP-PRMT1 were fixed with a solution of 3.7% paraformaldehyde, permeabilized by treating 1% Triton X-100, and incubated with 1% BSA. After staining with primary and secondary antibodies, the cells were immersed in Hoechst staining solution (diluted 1:1000). To counterstain nuclear DNA, DAPI was utilized [43]. Confocal imaging was conducted with a laser scanning microscope (LSM 700, Zeiss, Pendleton, IN, USA) [44,45].
mRNA analysis by semiquantitative RT‒PCR or quantitative RT-PCR
The mRNA expression status of c-Fos from Myc-c-Fos- and/or EGFP-PRMT1-transfected HEK293T cells was scrutinized by both semiquantitative reverse transcription polymerase chain reaction (RT‒PCR) [46] and quantitative real-time RT‒PCR (qRT‒PCR) in accordance with previously reported methods [47,48].
Protease protection assay
The protein level in proteasomes was measured by a protease protection assay in accordance with a previous report [49]. Myc-c-Fos- and/or EGFP-PRMT1-transfected HEK293T cells were incubated with digitonin solution (6.5 μg/mL, BN2006; ThermoFisher, Heysham, UK), mixed with protease (proteinase K) and Triton X-100, and were then scrutinized by electrophoresis.
LC‒MS/MS
Myc-c-Fos-overexpressing cells were harvested and prepared for immunoprecipitation using an anti-Myc antibody. Proteins in the immunoprecipitated samples were then separated by SDS‒PAGE. Subsequently, excised gel samples were digested with trypsin (37 °C, overnight). After digestion, the samples were lyophilized, reconstituted, and fractionated. The fractions were then analyzed using strong cation exchange liquid chromatography (SCXLC) in combination with mass spectrometry.
QuantSeq 3' mRNA sequencing
PRMT1-knockdown MKN45 cells and MKN45 cells with reconstitution of PRMT1 were harvested, and total RNA was isolated with TRIzol reagent. RNA sequencing and data interpretation were managed by E-Biogen (Daejeon, Republic of Korea) on a NextSeq 500 instrument from Illumina, Inc. (Seoul, Republic of Korea). The library was formulated with a QuantSeq 3' mRNA-Seq Library Prep Kit (Lexogen, Inc., Seoul, Korea). The sequencing data were analyzed with Excel-Based Differentially Expressed Gene Analysis (ExDEGA) GraphicPlus v2.0 by filtering DEGs based on a threshold fold change of greater than 2. In addition, gene set enrichment analysis (GSEA) of chosen genes was delved with GSEA software [33].
Analysis of microarray and RNA-seq data from a publicly available database
The GSE66229 [50], GSE26899 [50], GSE54129 (unpublished), and GSE79973 [51] datasets established with gene expression patterns of tumor and normal tissues were acquired from Gene Expression Omnibus (GEO). Unified STAD normal, STAD tumor, and healthy GTEx stomach tissue data were downloaded from Schultz et al. [52]. Nonpairwise comparisons of these data sets were evaluated with R version 4.2.1 (R Foundation for Statistical Computing, Indianapolis, IN, USA).
Multiple sequence alignment
The FASTA protein sequences of PRMT1 from various types of Genus and species were acquired from the UniProt database. Alignment of DNA sequences was carried out in Jalview 2.11.1.4 using ClustalW with default parameters.
Cell proliferation assay
DNA construct (c-Fos WT or its mutants)-transfected, PRMT1-overexpressing and PRMT1-knockdown MKN45 cells were incubated for indicated days. Cell proliferation was quantified by an MTT assay in accordance with previously reported protocols [30].
Wound healing assay
MKN45 cells transfected with empty vector or plasmids containing PRMT1 DNA together with the c-Fos WT or c-Fos R287K plasmid were seeded (5.5 × 105 cells/well) in a culture (12-well) plate. The cells were cultured overnight, scratched with a tip, and imaged after 0, 24, 48, and 72 h. The wound closure rate was determined with ImageJ of National Institute of Health (NIH, USA).
Colony formation assay
MKN45 cells were transfected with empty vector or the indicated plasmids (PRMT1 and c-Fos WT or c-Fos R287K) for 24 h using Lipofectamine 2000 (11668-019; Invitrogen) and thereafter seeded (0.5 × 103 cells per well) in a culture plate with six wells. After 10 days of culture, the cells were immersed in fixation solution (4% paraformaldehyde, 15 min). Subsequently, the cell aggregates were tinted with staining solution (0.5% crystal violet, C3886; Sigma). The stained cells in three random fields per well were counted and analyzed with a microscope connected to a camera and ImageJ.
Invasion assay
PRMT1-overexpressing MKN45 cells or c-Fos-transfected and PRMT1-knockdown MKN45 cells (5 × 104 cells/well) were plated in the top layer of a Transwell chamber with a membrane-permeable polycarbonate filter coated with BD Biosciences Matrigel (356237; San Diego, CA, USA) in Opti-MEM (11058021; Gibco Laboratories). The bottom compartment of the 24-well plate was filled with RPMI 1640 medium as a chemoattractant. Following 24 h of incubation, invaded cells from the upper compartment were fixed by 4% paraformaldehyde treatment and tinted with hematoxylin (ab220365; Abcam, Cambridge, US) and eosin Y solution (HT110116; Sigma‒Aldrich). The invaded cells in three random areas per well were counted and analyzed with a microscope connected to a camera and ImageJ.
Statistical analysis
Statistical values (P < 0.05) were figured with Student's t, the Mann-Whitney U, or R (version 4.2.1) tests, and data visualization was achieved with SigmaPlot 11.0. The Kaplan‒Meier method was specifically applied for cancer patient survival rate profile analysis.
Results
c-Fos directly interacts with PRMT1
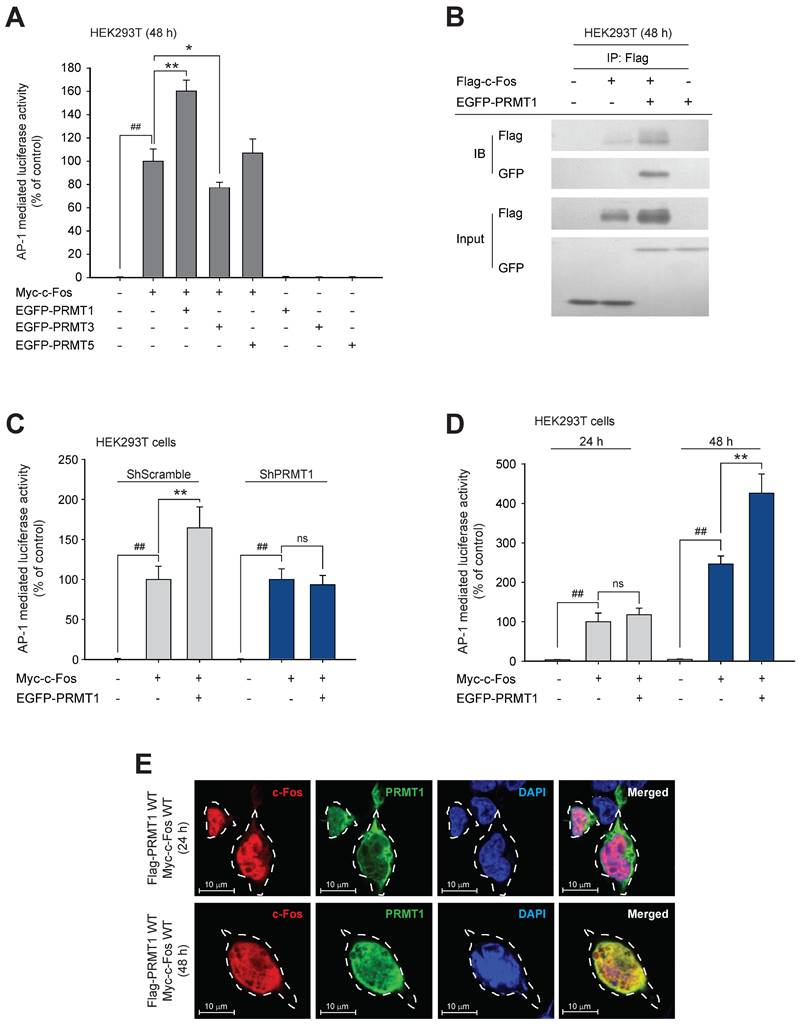
To identify whether c-Fos activity can be regulated by methyltransferases, we first performed mass spectrometry using c-Fos-immunoprecipitated protein samples (Fig. S1A). The results showed that RNA methyltransferases, DNA methyltransferases, and protein methyltransferases interacted with the c-Fos protein (Table 2). Eleven protein methyltransferases, including the protein arginine methyltransferase (PRMT) family and histone-lysine methyltransferases, were detected. Among the detected methyltransferases, we focused on PRMTs because they are located in both the nucleus and cytosol, which is similar to the cellular distribution of c-Fos. To study the effects of PRMTs on c-Fos function, we used a luciferase assay to ascertain whether c-Fos-mediated AP-1 activity is influenced by PRMTs. We observed that only PRMT1 dramatically and synergistically enhanced c-Fos-induced AP-1 activity, whereas transfection of PRMTs alone did not affect AP-1 activation (Fig. 1A). Unlike activity mediated by the other AP-1 subunits, c-Jun-mediated activity was not enhanced by PRMT1 (Fig. S1B). The increase in c-Fos-induced AP-1 activity was further evaluated by an immunoprecipitation assay, and we found that PRMT1 formed a complex with c-Fos (Fig. 1B), suggesting that PRMT1 interacts with the c-Fos transcription factor for specifically adjusting c-Fos-induced AP-1 activity. To determine whether activation of AP-1 is enhanced specifically by PRMT1, other protein methyltransferases were analyzed. Protein histidine methyltransferase (PHMT) and lysine methyltransferases (FAM86A and CAMKMT) did not affect c-Fos-mediated AP-1 activity (Fig. S1C). Interestingly, c-Fos-induced AP-1 liveliness was DNA-dose dependently upregulated by transfection of PRMT1 (Fig. S1C). Furthermore, knockdown of PRMT1 did not affect AP-1 activity induced by c-Fos (Fig. 1C). In a time-course assay, PRMT1-mediated c-Fos/AP-1 activation was observed after 48 h (but not after 24 h) of transfection (Fig. 1D). Concordant with this finding, in the immunohistochemistry and confocal microscopy experiments, colocalization of PRMT1 and c-Fos was verified in the nucleus 48 h post-transfection (Fig. 1E). This suggests that PRMT1 likely has a time-dependent synergistic effect on c-Fos activity.
Protein substrates interacting with the c-Fos protein.
| Putative substrate | Accession | MW (kDa) | Subcellular location |
|---|---|---|---|
| Protein arginine N-methyltransferase 1 | Q99873 | 41.5 | Nucleus, Cytoplasm, Cytosol |
| Protein arginine N-methyltransferase 5 | O14744 | 72.6 | Nucleus, Cytoplasm, Cytosol, Golgi apparatus, Chromosome |
| Histone-lysine N-methyltransferase SETD2 | Q9BYW2 | 287.4 | Nucleus, Chromosome |
| Histone-lysine N-methyltransferase NSD2 | O96028-1 | 152.2 | Nucleus, Chromosome |
| Probable methyltransferase-like protein 15 | A6NJ78 | 46.1 | Mitochondrion |
| Histone-lysine N-methyltransferase SETD1A | O15047 | 185.9 | Nucleus, Chromosome |
| Histone-lysine N-methyltransferase SMYD3 | Q9H7B4-1 | 49.1 | Nucleus, Cytoplasm, Cytosol |
| Methyltransferase-like protein 13 | Q8N6R0-5 | 78.7 | - |
| Histone-lysine N-methyltransferase EHMT2 | Q96KQ7 | 132.3 | Nucleus, Chromosome |
| Histone-lysine N-methyltransferase NSD3 | Q9BZ95 | 161.5 | Nucleus, Chromosome |
| Protein arginine N-methyltransferase 3 | O60678 | 59.8 | Cytoplasm |
PRMT1 methylates the c-Fos protein
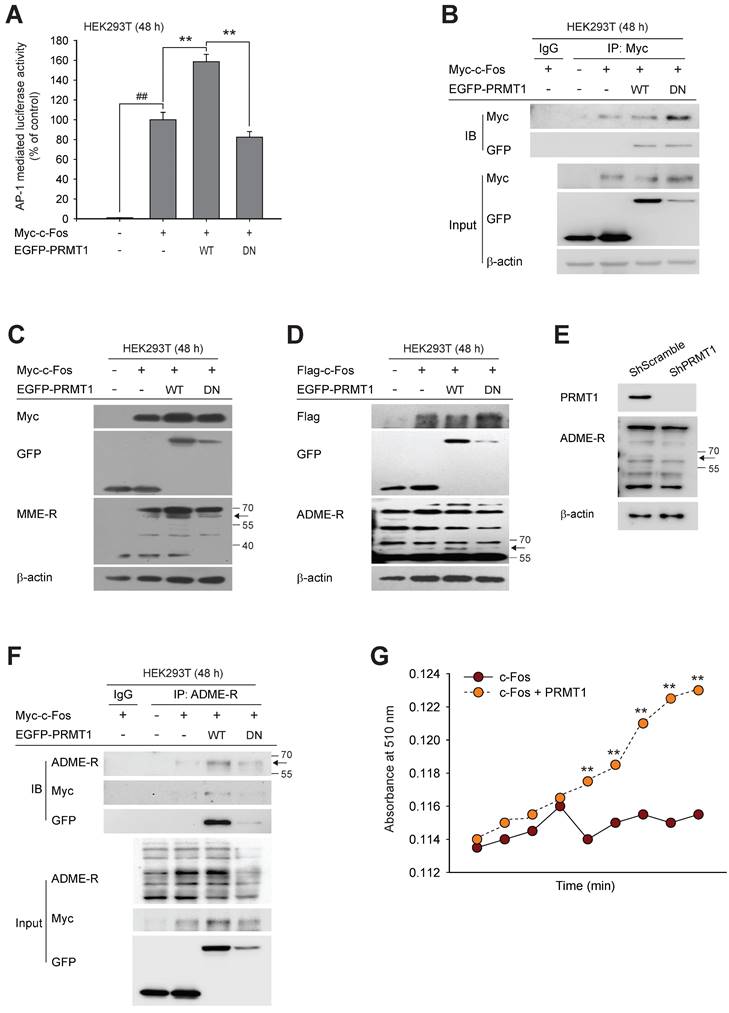
PRMT1 has been shown to catalyze the transfer of methyl groups to substrates, leading to diverse cellular processes [53,54]. However, there is no evidence that c-Fos is a methylation substrate for PRMT1. We hypothesized that the synergistic effect of PRMT1 on c-Fos activity is contingent on the methylation of c-Fos by PRMT1. Thus, we next evaluated whether PRMT1 can methylate c-Fos. A dominant negative (DN) mutant of PRMT1 without methyltransferase activity [55,56] was constructed (Fig. S2) and used to evaluate both the effect of PRMT1 on c-Fos-mediated AP1 activity via a luciferase assay and the interaction between PRMT1 and c-Fos using immunoprecipitation. As predicted, overexpression of the DN mutant did not enhance c-Fos-mediated AP-1 activation (Fig. 2A). However, the interaction between c-Fos and PRMT1 was not altered by the DN mutation (Fig. 2B). PRMT1 is classified as a type I PRMT that can generate both MME-R and ADME-R [63,39].
PRMT1 regulates c-Fos activity. (A) c-Fos and PRMT1, 3, or 5 were cotransfected into HEK293T cells for 48 h. c-Fos-induced AP-1-mediated luciferase reporter liveliness was quantified by a luminescence detector. (B) HEK293T cells overexpressing Flag-c-Fos and EGFP-PRMT1 were lysed for an immunoprecipitation assay. Flag and GFP levels were measured by immunoblotting of the anti-Flag immunoprecipitate obtained from whole-cell lysates of HEK293T cells. c-Fos-induced AP-1-mediated luciferase activity was measured in (C) shScramble- and shPRMT1-expressing cells and in (D) HEK293T cells transfected with Myc-c-Fos and EGFP-PRMT1 for the indicated times. Cells were lysed with buffer, and luminescence was measured. (E) HEK293T cells were transfected with Flag-PRMT1 and Myc-c-Fos for the indicated times. At the endpoint, cells were permeabilized and fixed for confocal analysis. Alexa Fluor 568 and Hoechst were used for recognition of c-Fos (indicated in red, c-Fos) and nuclei (indicated in blue, DAPI), respectively. ##P < 0.01 versus the normal and *P < 0.05 and **P < 0.01 versus the c-Fos alone.
To determine whether the interaction between PRMT1 and c-Fos leads to methylation of c-Fos, we sought to detect methylated arginine residues using antibodies specific for MME-R and ADME-R. When PRMT1 was cotransfected with c-Fos, both forms of methylated arginine residues were increased at the molecular weight corresponding to c-Fos (approximately 60 kDa) (seen in Fig. 2C and D). On the other hand, PRMT1 DN mutant-transfected cells showed very low levels of methylated c-Fos. In addition, a low level of methylated c-Fos was observed in PRMT1-knockdown cells (Fig. 2E). In the immunoprecipitation assay, we analyzed immunoprecipitated lysates using an anti-ADME-R antibody and detected a putative band corresponding to c-Fos at 55-70 kDa in PRMT1 wild-type (WT) and c-Fos-transfected cell lysates. The intensity of the ADME-R-modified c-Fos band was nearly zero in cells expressing the PRMT1 DN mutant that were cotransfected with c-Fos (Fig. 2F). These data demonstrate that c-Fos and PRMT1 were coimmunoprecipitated and that c-Fos was likely dimethylated asymmetrically by PRMT1. Next, we performed an in vitro methylation assay to confirm direct methylation of c-Fos by PRMT1. Fig. 2G shows that the presence of PRMT1 was able to elevate the methylation level of c-Fos. Collectively, such findings imply that c-Fos is undergoes arginine dimethylation catalyzed by PRMT1, which enhances AP-1 activity induced by c-Fos.
PRMT1 regulates c-Fos protein stability
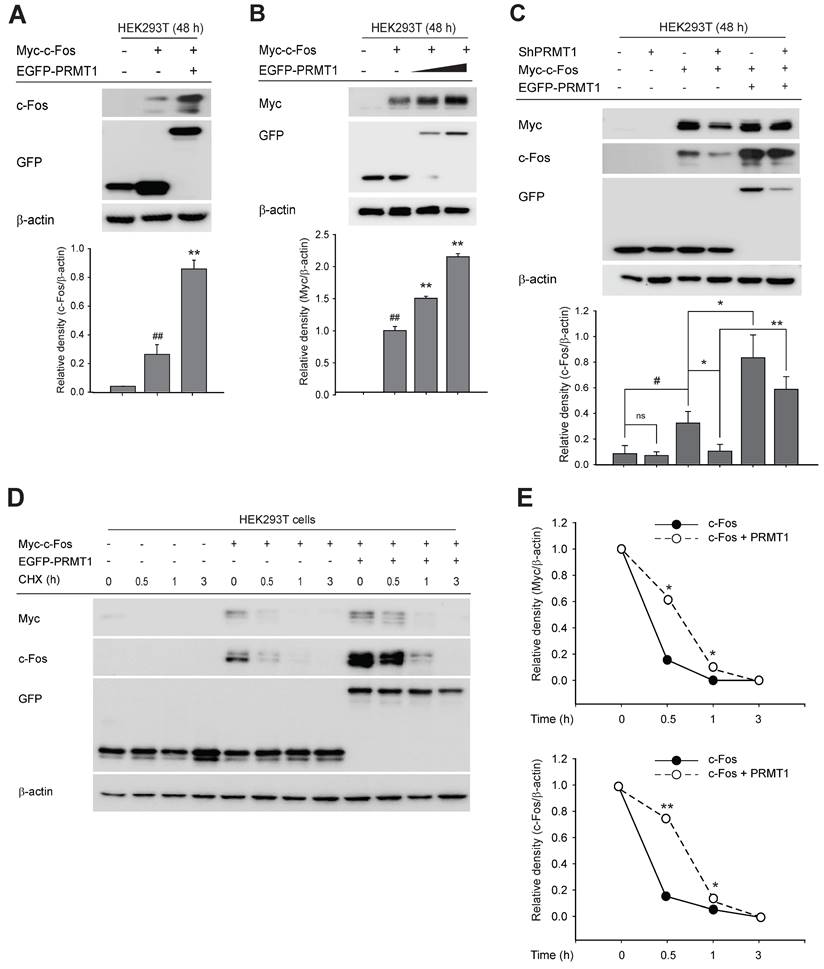
We next focused on the mechanism by which PRMT1 regulates c-Fos-mediated AP1 activity. Our initial experiments ruled out an effect of PRMT1 on c-Fos gene expression (Fig. S3A and S3B), phosphorylation of MAPK (Fig. S3C and S3D) or c-Fos (Fig. S3E), and dimerization of c-Fos and c-Jun (Fig. S3F). Although PRMT1 overexpression did not alter c-Fos gene expression (Fig. S3A and S3B), the protein level of the c-Fos, which shows a wide range of molecular weights due to its posttranslational modifications, such as ubiquitination, phosphorylation, and S-nitrosylation [57-59], was magnified by PRMT1 transfection (Fig. 3A) and was synergistically and concentration-dependently increased (Fig. 3B). Consistent with this finding, knockdown of PRMT1 blocked c-Fos expression, which was restored when the c-Fos expression plasmid was transfected (Fig. 3C). To examine whether PRMT1 regulates c-Fos protein stability, a cycloheximide (CHX) chase assay was carried out. CHX has been widely used for protein stability assays because it blocks protein translation [60-62]. In this experiment, CHX was added 48 h after transfection of the c-Fos and PRMT1 plasmids. As expected, the c-Fos protein level decreased over a period of 3 h. Interestingly, the PRMT1/c-Fos cotransfected group had a higher c-Fos level than the c-Fos control-transfected group (Fig. 3D and E). Together, these data demonstrate that PRMT1 regulates c-Fos activity by enhancing its protein stability.
c-Fos protein stability is modulated by autophagy
To decipher how PRMT1 regulates c-Fos stability, we used the protein degradation inhibitors MG132 (a proteasomal degradation inhibitor) and 3-MA (an autophagy inhibitor). Proteasomal degradation and autophagy are the two main mechanisms of protein degradation [63,64]. The addition of 3-MA enhanced PRMT1-mediated c-Fos/AP-1 activity, whereas MG132 inhibited AP1 activity (Fig. 4A). We subsequently confirmed that 3-MA treatment increased the c-Fos protein level (Fig. 4B). Conversely, treatment with rapamycin, an autophagy inducer, decreased the c-Fos protein level (Fig. 4C) and reduced c-Fos-induced AP-1 activity (Fig. S4A).
During autophagic degradation, autophagosomes and lysosomes are formed and envelop degradable proteins. In this study, we isolated autophagosomes by a protease protection assay [49]. The lysosomal location of c-Fos was identified (Fig. 4D), and we determined whether the concentration of c-Fos was affected by the expression level of PRMT1. Upon coexpression of PRMT1 and c-Fos, c-Fos accumulated in the proteasome, and methylated arginine was detected by immunoblotting (Fig. 4E). In contrast, when PRMT1 was knocked down, the c-Fos protein was rapidly degraded, and the intensity of the band corresponding to ADME-R-modified c-Fos was reduced (Fig. 4F). In addition, the LC3-II level was increased under PRMT1-knockdown conditions (Fig. S4B). Moreover, overexpression of c-Fos increased the number of puncta visualized by confocal microscopy (Fig. 4G). Moreover, the number of vacuoles was reduced in the presence of PRMT1 but was restored when PRMT1 DN was cotransfected. A similar pattern was observed in the LysoTracker assay (Fig. 4H). Collectively, these data demonstrate that c-Fos could be degraded via the autophagic manner and that PRMT1 could prevent c-Fos's autophagic degradation.
PRMT1 methylation-dependent regulation of c-Fos activity. (A) Myc-c-Fos and EGFP-PRMT1 WT or DN were cotransfected into HEK293T cells for 48 h; then, c-Fos-mediated AP-1 activity was measured by a luciferase assay. (B) Immunoprecipitation was performed with whole-cell lysates prepared from HEK293T cells transfected with Myc-c-Fos, EGFP-PRMT1 WT, or EGFP-PRMT1 DN. The levels of Myc and GFP were measured by immunoblotting. (C) Monomethylated arginine levels and (D) dimethylated arginine, Flag, and GFP levels were measured by immunoblotting in whole-cell lysates of HEK293T cells transfected with Myc-c-Fos and EGFP-PRMT1. (E) Asymmetric dimethylarginine (ADME-R) was detected in whole-cell lysates prepared from shScramble- or shPRMT1-expressing cells by immunoblotting. (F) ADME-R, Myc, and GFP levels were measured by immunoblotting in the ADME-R immunoprecipitate from whole-cell lysates of HEK293T cells transfected with Myc-c-Fos and EGFP-PRMT1 for 48 h. (G) Ectopic c-Fos or PRMT1 was immunoprecipitated, and these proteins were incubated with SAM following the manufacturer's instructions to measure the methyl-accepting capacity of c-Fos. ##P < 0.01 versus the normal and **P < 0.01 versus the c-Fos alone.
PRMT1 regulates c-Fos protein stability. (A) Myc-c-Fos and EGFP-PRMT1 were cotransfected into HEK293T cells for 48 h. Harvested cells were used for immunoblot analysis of c-Fos and GFP levels. (B) Myc-c-Fos and EGFP-PRMT1 were transfected at gradually increasing concentrations into HEK293T cells for 48 h. Harvested cells were used for immunoblot analysis of Myc and GFP levels. The relative density of the Myc protein band was determined by ImageJ. (C) Myc-c-Fos and EGFP-PRMT1 were transfected into HEK293T WT or PRMT1-knockdown cells. Harvested cells were used for immunoblot analysis of c-Fos, GFP, and Myc levels. A cycloheximide (CHX) chase assay was performed with Myc-c-Fos/EGFP-PRMT1-overexpressing HEK293T cells by treatment with 30 μg/mL CHX for the indicated times (0-3 h). (D) Whole-cell lysates were prepared for immunoblot analysis, and (E) the relative densities of the Myc and c-Fos protein bands were determined by ImageJ.
PRMT1 protects c-Fos from autophagic degradation. (A) HEK293T cells were transfected with Myc-c-Fos and EGFP-PRMT. A proteasome or autophagy inhibitor (MG132 or 3-MA, respectively) was added for an additional 24 h of incubation. AP-1-mediated luciferase reporter liveliness was quantified by a luminescence detector. (B and C) Myc-c-Fos- and EGFP-PRMT1-overexpressing HEK293T cells were treated with 3-MA (5 mM) or rapamycin (1 μM); then, the protein levels of Myc and GFP were measured by immunoblotting. (D) Autophagic degradation of c-Fos, ADME-R, and LC3B was detected in autophagosomes/lysosomes of HEK293T cells by a protease protection assay and immunoblot analysis. (E and F) Endosomes were isolated for a proteasome protection assay. With whole-cell lysates prepared from HEK293T cells (E) or PRMT1-knockdown cells (F), the protein levels of Myc, ADME-R, and GFP were measured by immunoblot analysis. (G) HEK293T cells were cotransfected with Myc-c-Fos and EGFP-PRMT1 WT or DN for 48 h and then immunostained with Alexa Fluor 568 (for c-Fos) and Hoechst (for nuclear DNA), and the formation of puncta was observed by confocal microscopy. (H) HEK293T cells transfected with c-Fos and PRMT1 were incubated with LysoTracker following the manufacturer's manual. Lysosomes were identified by fluorescence microscopy. ##P < 0.01 versus the normal and **P < 0.01 versus the c-Fos alone or c-Fos/PRMT1 cotransfection group. Arrows (H) indicate lysosomes.
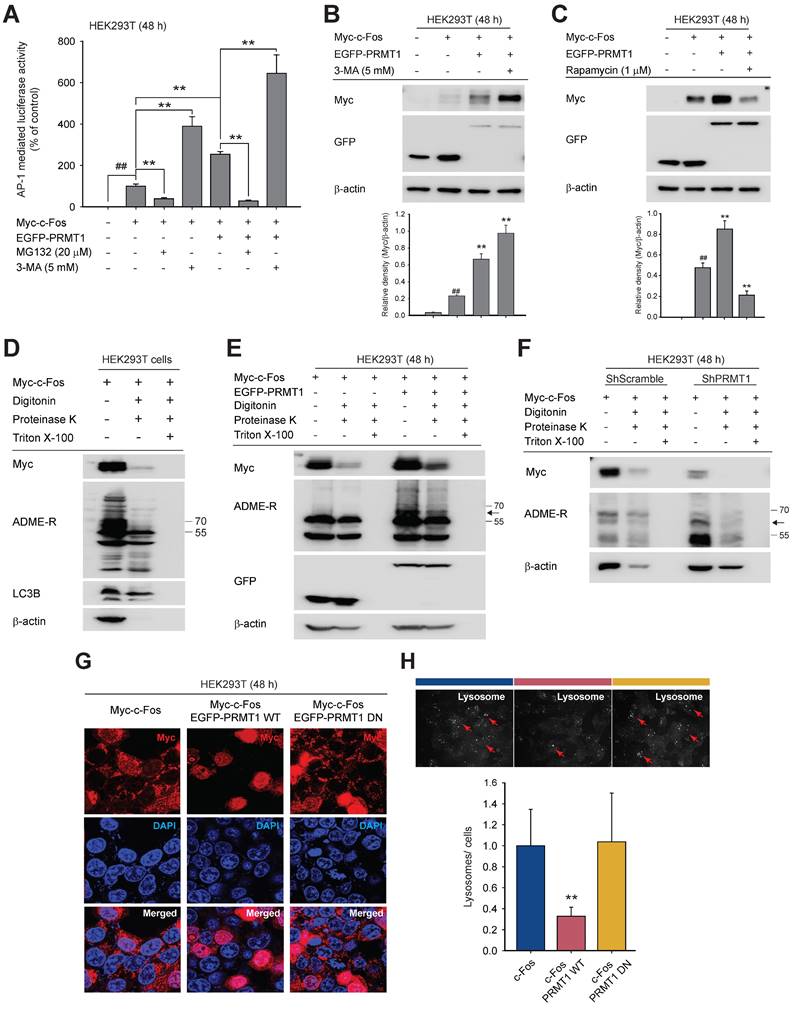
Methylation of c-Fos on R287 protects it from autophagic degradation
By mass spectrometry, we identified the methylated arginine sites of c-Fos as R108, R201, R279, and R287. Many methylated sites of proteins are conserved among species [65,66]. We compared these four sites among 10 species, including Rattus norvegicus, Mus musculus, and Homo sapiens, and found that all were highly conserved (Fig. S5 and Fig. 5A). To identify the site methylated directly by PRMT1, arginine (R)-to-lysine (K) substitution mutants were generated as shown in Fig. 5B, and asymmetric dimethylarginine levels were evaluated. The translational states of c-Fos R279K and R287K were slightly decreased, but a reduced methylated arginine level was observed only in the R287K mutant (Fig. 5C). Protein stability was examined with c-Fos WT and R287K under co-expression of PRMT1. As exhibited in Fig. 5D, PRMT1 protected c-Fos WT against degradation, but the c-Fos R287K mutant protein was rapidly lost. Consistent with the results of the protein stability assay, reduced c-Fos R287K-induced activation of AP-1 and weak synergistic AP-1-medited induction of PRMT1 were also observed (Fig. 5E). To further strengthen these findings, a c-Fos methylation mimetic mutant, R287F, was constructed. Compared to c-Fos WT, the R287F mutant significantly enhanced AP-1 activity (Fig. 5F). Finally, we measured the c-Fos R287K methylation level. In line with the previous result (Fig. 2G), PRMT1 overexpression increased the c-Fos WT methylation level. However, the c-Fos R287K methylation level was decreased even in the presence of PRMT1 overexpression, suggesting that c-Fos R287K was not methylated (Fig. 5G). In addition, using mass spectrometry, we further verified methylation of c-Fos at R287 (Fig. 5H) and, as revealed in Fig. 5A, showed that c-Fos R287 is conserved across species, indicating that this regulatory mechanism is also conserved across species. Together, these results show that PRMT1 methylates c-Fos at R287 and protects c-Fos from autophagic proteolysis.
PRMT1 is upregulated in GC and associated with clinicopathologic variables of GC
To study the biological roles of PRMT1 and c-Fos, we analyzed PRMT1 expression data for a variety of cancers obtained from The Cancer Genome Atlas (TCGA) by employing TIMER. These analysis results showed higher expression of PRMT1 in almost all cancer types, including GC, than in the corresponding normal tissues (Fig. S6A). PRMT1 upregulation has been linked to numerous cancer types [67-70]. However, the specific role of PRMT1 and c-Fos in GC remains to be elucidated. To expand the knowledge of the role that PRMT1 plays in GC pathogenesis, an RNA sequencing (RNA-seq) dataset from TCGA, including data from the Genotype-Tissue Expression (GTEx) database, and a number of human GC tissue microarray datasets were analyzed. PRMT1 expression was observed to be higher in stomach cancer tissues in GSE62254 (Fig. 6A), GSE26899 (Fig. 6B), GSE54129 (Fig. 6C), and GSE79973 (Fig. 6D) than in normal gastric tissues, normal samples in GTEx, or normal samples in TCGA (Fig. 6E), confirming that PRMT1 expression is high in patients with GC. Further profiling of PRMT1 expression was completed to characterize its expression in patients with various subtypes of GC. Intestinal-type GC categorized by Lauren classification showed significantly higher expression of PRMT1 mRNA than diffuse-type GC (Fig. S6B), suggesting that the expression of PRMT1 is more strictly associated with well-differentiated gastric tumor types than with poorly differentiated types. Importantly, the overall survival probability was compared between the groups of GC patients with low and high PRMT1 expression using Kaplan‒Meier Plotter (www.kmplot.com) with Affymetrix input ID 1565016_at, corresponding to the PRMT1 gene. The outcomes displayed that patients showing high PRMT1 expression had a lower survival probability (Fig. 6F and Fig. S6C). Interestingly, the distinction between the low and high PRMT1 groups in terms of survival was more pronounced in cohorts of patients with intestinal-type GC (hazard ratio, 2.08) (Fig. 6F). In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG) 2021 human pathway enrichment analysis of PRMT1-associated genes identified in our RNA-seq analysis indicated a significant regulatory role of PRMT1 in GC (Fig. S6D). Together, these data validated the potential role of PRMT1 as an oncogene in GC. Furthermore, GSEA of the RNA-seq data with PRMT1-knockdown MKN45 cells (shPRMT1) as a control and PRMT1-knockdown MKN45 cells with reconstitution of PRMT1 expression (recovery) (Fig. S7B) showed that PRMT1 is negatively associated with the regulation of autophagy genes (false discovery rate [FDR]-adjusted P = 0.009, normalized enrichment score [NES] = -1.36) (Fig. 6G). Moreover, positive correlations of PRMT1 with gene markers (FDR-adjusted P = 0.000, NES = 1.66) and FOS target genes (FDR-adjusted P = 0.030, NES = 1.43) were found in advanced GC (Fig. 6H and I). This suggests that the promoting effect of PRMT1on GC is related to c-Fos target genes and autophagy pathways. Among autophagy genes in the Reactome database, PRMT1 was negatively correlated with genes such as ATG9A, DYNC1H1, EPAS1, PINK1, TSC1, TUBB6, and ULK1 in gastric tumor tissues (STAD-TCGA) (Fig. 6J and S6E) and positively correlated with HMMR, ESCO2, CHAC2, and NUDT6 (STAD-TCGA) (Fig. 6K and S6F). Of interest, these genes are FOS target genes, based on analysis of chromatin immunoprecipitation sequencing datasets from the ENCODE Transcription Factor Targets database (ncbi.nlm.nih.gov/gene/2353). Thus, we focused on how PRMT1-methylated c-Fos contributes to gastric tumorigenesis.
PRMT1 methylates the R287 residue of c-Fos. (A) Amino acid sequence alignment of c-Fos arginine methylation sites across species. (B) Diagram of c-Fos and its mutants (WT, R108K, R201K, R279K, and R287K). (C) c-Fos WT and its mutants were cotransfected with PRMT1 into HEK293T cells. Myc, GFP, and ADME-R levels were measured by immunoblot analysis. (D) A CHX chase assay was conducted in HEK293T cells cotransfected with PRMT1 and c-Fos WT or c-Fos R287K by CHX treatment (30 μg/mL) at the indicated times. Myc and GFP levels were measured by immunoblot analysis. (E and F) Myc-c-Fos WT, R287K, or R287F was transfected into HEK293T cells in the presence or absence of Flag-PRMT1 with the AP-1-Luc construct. AP-1 activation was then measured by a luminometer. (G) Lysates of cells cotransfected with PRMT1 and c-Fos WT or c-Fos R287K were subjected to immunoprecipitation, and the immunoprecipitated proteins were incubated with SAM for measurement of the methyl-accepting capacity by spectrometry. (H) Methylation of the R287 residue of c-Fos was detected by LC‒MS/MS. ##P < 0.01 versus the normal, *P < 0.05 and **P < 0.01 versus the c-Fos alone or c-Fos/PRMT1 cotransfection group.
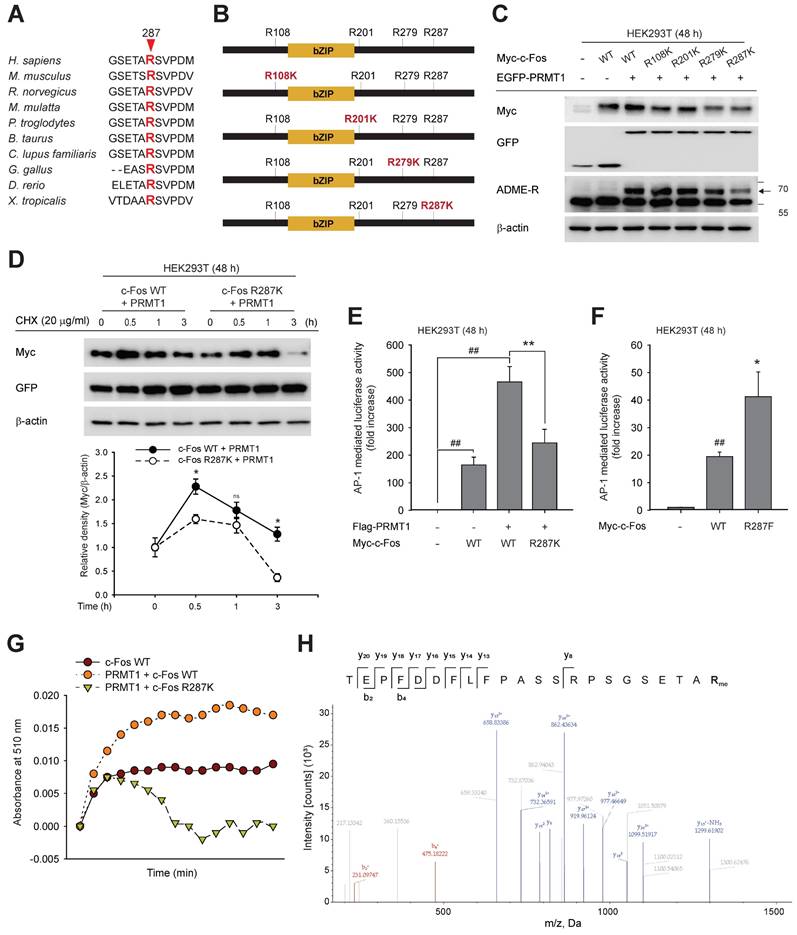
Gene expression profile of PRMT1 and survival rate of patients based on PRMT1 expression in GC. (A-D) PRMT1 gene expression in gastric normal vs. tumor tissues in several GEO datasets containing data from gastric cancer patients. (E) Comparison between PRMT1 gene expression in gastric normal tissues (STAD_TCGA+GTEx) vs. gastric tumor tissues (STAD_TCGA). (F) Overall survival analysis of gastric cancer patients based on the transcriptional level of PRMT1 was completed by employing Kaplan‒Meier Plotter (www.kmplot.com). (G-I) GSEA of autophagy genes, gene markers of advanced GC, and FOS target genes based on our RNA-seq data from PRMT1-reconstituted cells compared to shPRMT1 cells. (J and K) Visualization of the correlations of PRMT1 expression with that of several genes in the STAD tumor dataset.
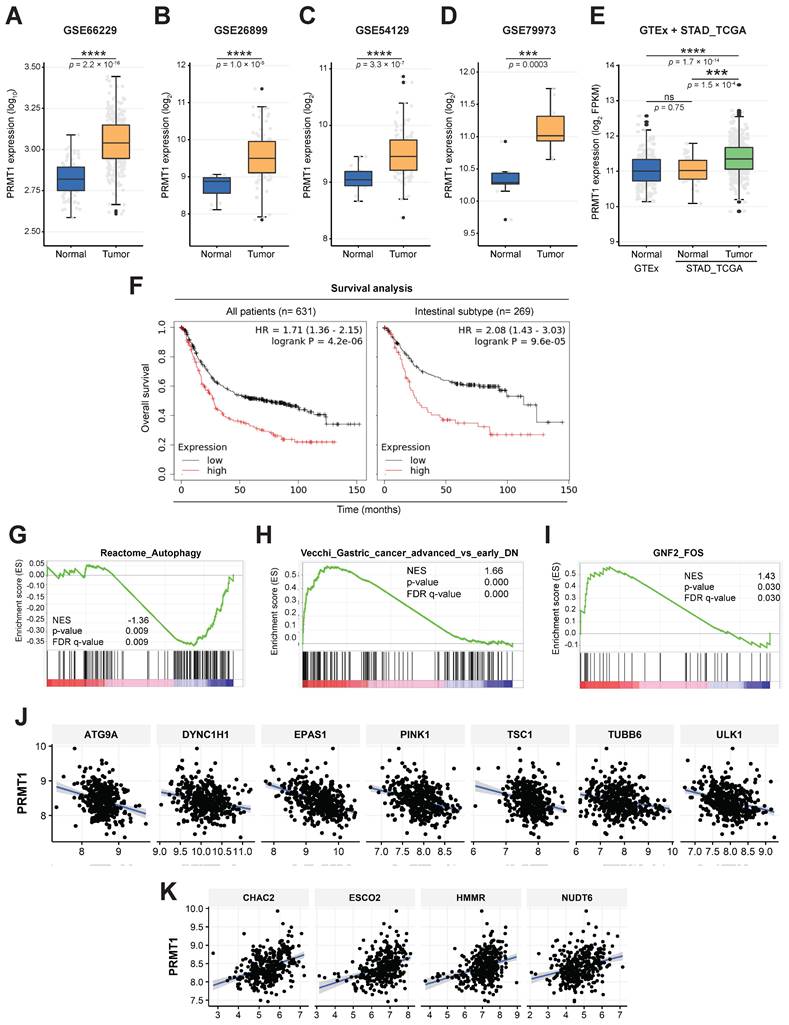
PRMT1-methylated c-Fos supports gastric tumorigenesis
We used cell lines to examine the molecular relationship between PRMT1 and c-Fos in the milieu of GC. Consistent with c-Fos protein, PRMT1 protein level was higher in human GC cells (MKN1 and MKN45) than in a normal gastric cell line (HFE-145) (Fig. S7A). In paired normal gastric tissues and GC samples from human patients, higher PRMT1 expression in GC samples was confirmed (Fig. 7A). In addition, c-Fos expression was increased and decreased with overexpression and knockdown of PRMT1, respectively, in both MKN45 and MKN1 cells, as revealed by immunoblotting (Fig. 7B, C, S7C, and S7D). These outcomes indicate that the c-Fos protein level can be positively affected by the PRMT1 expression level. Furthermore, in line with findings in HEK293T cells (Fig. 3A), the c-Fos expression level was higher in c-Fos/PRMT1-cotransfected MKN45 cells compared with c-Fos-control-transfected MKN45 cells (Fig. 7D). We hypothesized that PRMT1 promotes GC by regulating c-Fos stability. To confirm this hypothesis, we tested c-Fos degradation using a CHX chase assay in MKN45 cells. As appeared in Fig. 7E and F, the c-Fos level decreased over the 3-h period but remained higher in the c-Fos/PRMT1-cotransfected group than in the c-Fos-transfected group. Moreover, consistent with data shown earlier (Fig. 3D and 4B), 3-MA treatment enhanced the PRMT1-mediated enhancement of c-Fos, and both protein-translational pattern and stability of c-Fos were elevated by PRMT1 transfection (Fig. 7G). This might be the result of inhibition of LC3B.
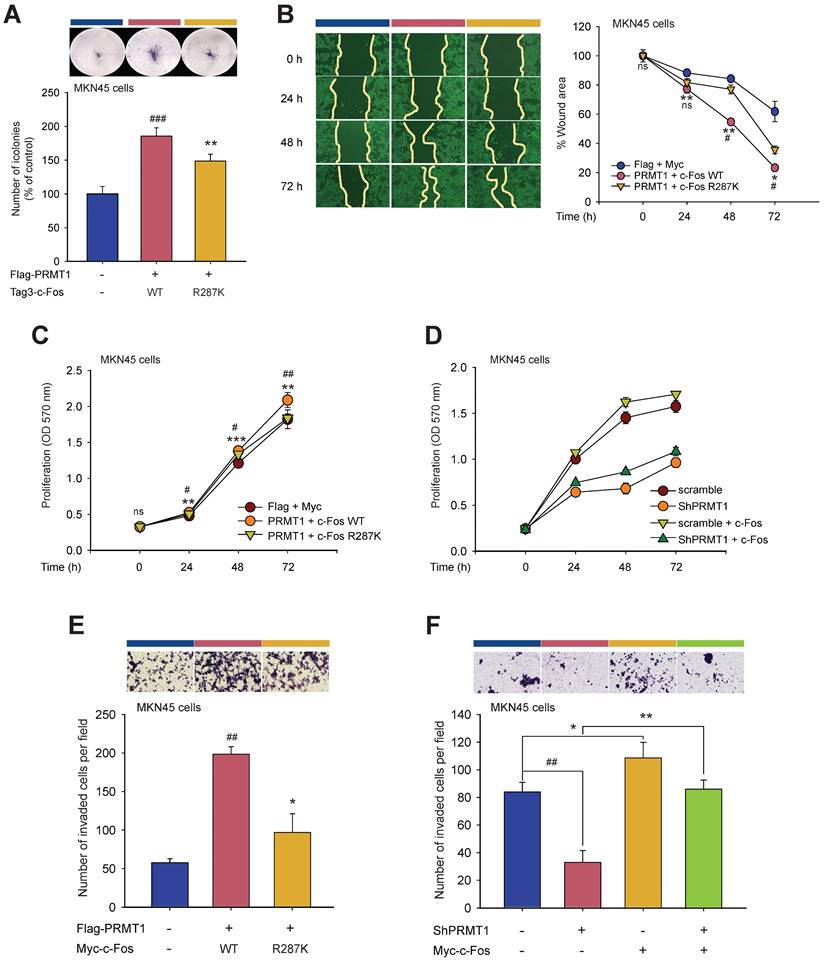
To determine whether PRMT1-mediated methylation of c-Fos fosters gastric tumorigenesis, the functional role of c-Fos was examined by comparing cellular responses in PRMT1-overexpressing and PRMT1-knockdown cells cotransfected with WT c-Fos or the c-Fos R287K mutant (nonmethylated form). Overexpression and silencing of PRMT1 were performed in MKN45 cells, and the PRMT1 level was confirmed through immunoblotting (Fig. S8A and S8B). In combination with PRMT1 overexpression, c-Fos WT overexpression promoted tumorigenic responses, including colony formation (Fig. 8A), migration (Fig. 8B, S8C, and S8D), accelerated cell growth (Fig. 8C and D), and invasion (Fig. 8E and F). On the other hand, overexpression of nonmethylated c-Fos significantly suppressed these responses compared to those in WT c-Fos-expressing MKN45 cells. Besides, knockdown of PRMT1 repressed cellular growth and invasion, but c-Fos overexpression restored these responses (Fig. 8D and F). Hence, the data demonstrated that PRMT1-methylated c-Fos plays an important role in gastric tumorigenesis.
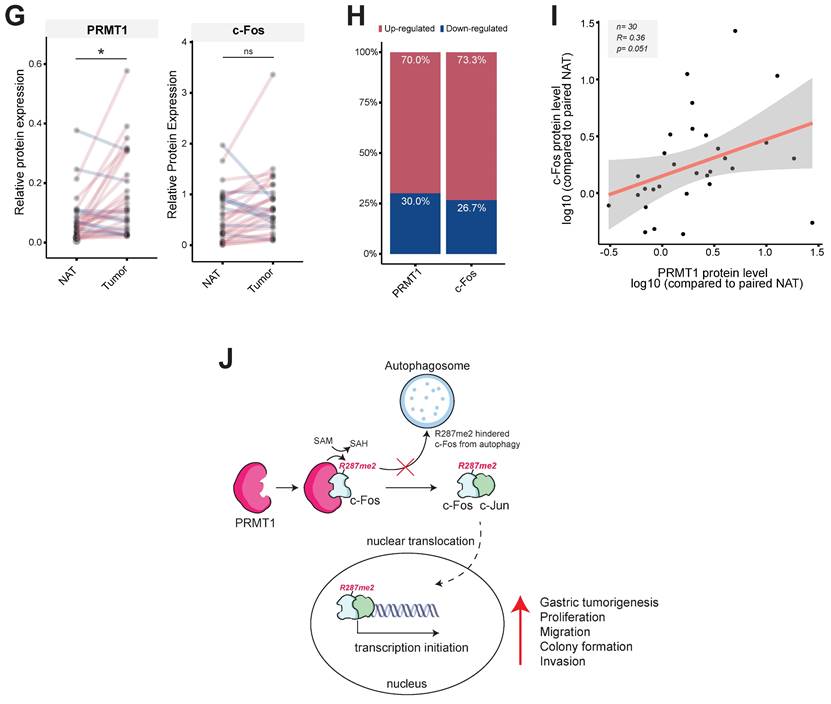
Moreover, using immunoblot analysis of samples in paired normal gastric tissues and GC samples from human patients, the correlation between the PRMT1 and c-Fos protein levels was evaluated (Fig. S8E). Pairwise analysis of PRMT1 and c-Fos protein expression, bar plot generation, and correlation analysis were performed to visualize differences in the normalized PRMT1 and c-Fos levels between NATs and GC samples. PRMT1 protein expression was significantly higher in GC samples than in NATs from human patients (n = 30, P = 0.010), with 70% of samples showing an increased PRMT1 level, while c-Fos was expressed in tumor tissue in approximately 73.3% of patients. Our findings suggest that the PRMT1 and c-Fos proteins are both highly expressed in GC samples compared to NATs from human patients (Fig. 8G and H). Moreover, correlation analysis showed a moderate positive correlation of PRMT1 and c-Fos expression in GC samples compared to NATs (R = 0.36) (Fig. 8I), suggesting that PRMT1 appears to have a synergistic effect on c-Fos expression in patients with GC.
Discussion
In our present research, the regulatory function of PRMT1 on the activation of c-Fos was delineated. Methylation of c-Fos protects it from autophagic degradation, which contributes to gastric tumorigenesis. We first observed a direct interaction between c-Fos and PRMT1, and that interaction synergistically enhanced c-Fos-mediated AP-1 activity. This phenomenon was dependent on PRMT1-mediated c-Fos methylation. Moreover, our study revealed c-Fos R287 as a new substrate of PRMT1 for arginine methylation. In addition, PRMT1 protected the c-Fos protein from autophagic degradation, enhanced c-Fos protein stability, and enhanced c-Fos activity, impacting human gastric tumorigenesis. Hence, our study established an important function of PRMT1 in c-Fos/AP-1 regulation, and targeted inhibition of this axis might be another approach for treating GC.
PRMT1 is a methyltransferase that regulates various cellular processes via methylation or another mechanism. AP-1 can contain various subunits (c-Fos, ATF, and c-Jun family members), and its activation is induced by various ligands. As a result, AP-1 mediates diverse cellular reactions, including differentiation, proliferation, and tumorigenesis. Other studies have demonstrated that posttranslational modifications such as SUMOylation, ubiquitination, and phosphorylation can regulate c-Fos activity.
PRMT1 expression in GC tissues and the role of PRMT1 in c-Fos protein expression in GC cell lines. (A) PRMT1 expression in paired normal stomach tissues (N) and GC samples (T) from human patients was evaluated by immunoblot analysis. (B and C) c-Fos level was evaluated in MKN45 cells with overexpression (B) or knockdown (C) of PRMT1 together with LC3B levels. (D) Myc-c-Fos- and Myc-c-Fos/Flag-PRMT1-overexpressing MKN45 cells were harvested and used for immunoblot analysis of Myc, c-Fos, and Flag levels. (E) MKN45 cells overexpressed Myc-c-Fos and Flag-PRMT1. CHX was further applied for the indicated times, and the translational levels of c-Fos and Flag were measured in whole-cell lysates by immunoblotting; then, (F) the relative density of the c-Fos protein band was measured by ImageJ. (G) Myc-c-Fos and Flag-PRMT1 were cotransfected into MKN45 cells for 2 days, and 3-MA (5 mM) was added. The translational levels of Myc, c-Fos, Flag, and LC3B were measured by immunoblotting.
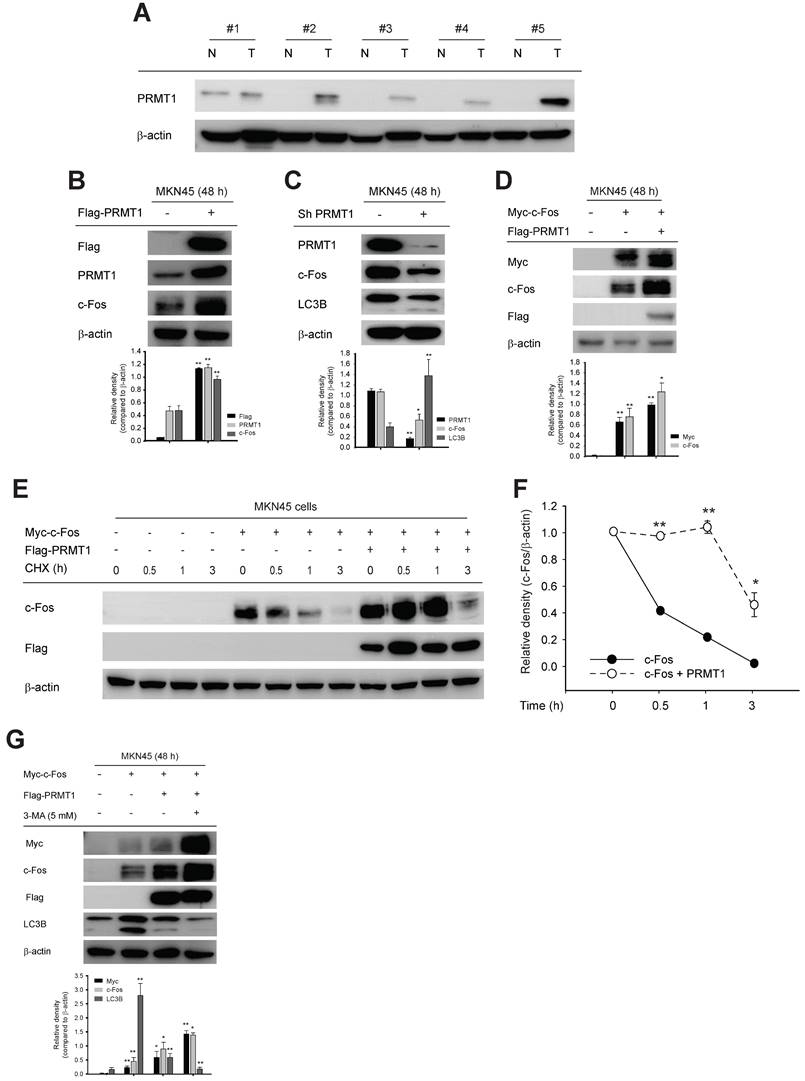
PRMT1-mediated methylation of c-Fos at R287 supports gastric tumorigenesis. Cellular responses were examined in c-Fos WT-overexpressing or R287K mutant (nonmethylated)-overexpressing MKN45 cells. (A) A clonogenic assay was performed in a six-well plate (1 x 103 cells/well) by incubation for ≥10 days. The culture medium was taken away, the cells were fixed, stained, and imaged. (B) A wound healing effect was utilized to evaluate the migrative capability of MKN45 cells. Cells were imaged after indicated times, and the wound closure rate was determined by ImageJ. (C and D) Proliferation was evaluated at the indicated days by an MTT assay in (C) WT c-Fos- or nonmethylated c-Fos-cotransfected MKN45 cells and (D) c-Fos-overexpressing MKN45 cells expressing Scramble or shPRMT1. (E and F) An invasion assay was performed with c-Fos WT/Flag-PRMT1 or c-Fos R287K/Flag-PRMT1-cotransfected MKN45 cells (E) or c-Fos-overexpressing MKN45 cells expressing Scramble or ShPRMT1 (F). Invaded cells were tinted with hematoxylin and eosin Y solution, imaged using a camera connected to a microscope, and counted across three random areas of each well. (G) Pairwise analysis and (H) bar plot of the relative protein expression of PRMT1 and c-Fos in paired normal gastric tissue and GC samples from human patients. The relative expression of PRMT1 and c-Fos in tissues compared to MKN45 cells (control) was measured and normalized to that of β-actin determined by immunoblotting. Statistical significance was evaluated using the paired t test. (I) The correlation between PRMT1 and c-Fos expression in paired normal gastric tissue and GC samples from human patients was evaluated by immunoblotting and plotted. The correlation coefficient (R) between PRMT1 and c-Fos expression was calculated using Pearson correlation analysis. (J) A schematic diagram showing how PRMT1 regulates c-Fos activity by methylation. PRMT1 methylates the R287 residue of c-Fos and protects c-Fos from autophagic degradation. The increased protein stability of c-Fos leads to an increase in the functionality of AP-1 to foster gastric tumorigenesis and tumorigenic behaviors in GC cell lines.
c-Fos is commonly phosphorylated at several sites by p70S6K, MAPKs, protein kinases A and C, or 90-kDa ribosomal S6 kinase [71,72], c-Fos complexes on target gene promoters can be SUMOylated to promote transcription [20]. However, c-Fos protein methylation is not well understood. Some reports have examined the ability of c-Fos DNA methylation to regulate its gene expression [73-75]. Our data showed that PRMT1 specifically increases c-Fos-mediated AP-1 luciferase activity. We detected the interaction of c-Fos and PRMT1 and identified an increased level of methylated arginine on c-Fos. We discovered four arginine-methylated sites on the c-Fos protein, among which PRMT1-methylated c-Fos R287 appears to specifically regulate c-Fos protein stability. It was evident that the c-Fos transcription factor is methylated and expression levels of AP-1-regulated genes are altered via PRMT4. However, the methylated sites of c-Fos have not been identified [28]. In summary, we demonstrated that c-Fos is posttranslationally methylated to regulate its stability and activity and identified the methylated arginine residues in c-Fos.
c-Fos proteins are short-lived proteins whose expression is tightly regulated by proteolytic pathways [76-79]. c-Fos can be degraded by proteasomes via ubiquitin-dependent or ubiquitin-independent pathways. c-Fos degradation is differentially regulated based on the involvement of various destabilizers (kinases, enzymes, or c-Jun) [80,81]. For example, ERK5 phosphorylation simultaneously regulates c-Fos nuclear translocation and stability [82]. The interaction between c-Fos and NAD(P)H:quinone oxidoreductase 1 prevents c-Fos degradation through a ubiquitin-independent proteasomal pathway [77]. Previous reports have shown that c-Fos can undergo proteasomal degradation. We demonstrated here that c-Fos degradation is activated via autophagy, which is an important intracellular degradation process mediated by lysosomes that is designed to reutilize cellular components. We found that c-Fos was located in autophagosomes and that knockdown of PRMT1 increased the protein level of the autophagic degradation marker LC3B. Autophagy is usually triggered by nutrient starvation [83,84], and 48 h was a sufficient length of time for induction of starvation. This indicates that PRMT1 plays a pivotal role in nutrient starvation by modulating c-Fos degradation. Although further studies are needed to understand how PRMT1-methylated c-Fos is protected from degradation, our research documents a new posttranslational mechanism that regulates c-Fos stability and activity.
Previous studies have demonstrated that transcription factors can be modulated by autophagic degradation. PU.1 undergoes autophagic degradation dependent on p62 (a selective autophagy adaptor) to suppress T helper 9 (Th9) cell differentiation [85]. Transcriptional adjustment by hypoxia-inducible factor-1 (HIF-1) was shown to be modulated by autophagy [86]. HIF-1α, a major subunit of HIF-1, is targeted by chaperone-mediated autophagy, resulting in altered expression of HIF-1 target genes. Thus, transcription factors could be regulated by degradation pathways (especially autophagy). Similar to these studies, our study shows that c-Fos-mediated AP1 activity is regulated by autophagic degradation of c-Fos.
The pathophysiological role of c-Fos is deeply studied, especially because c-Fos is a proto-oncogene that plays diverse roles in cells [25,26,28,87]. In addition, various types of human cancers have been reported to exhibit PRMT1 upregulation [8,11,53,55]. However, the roles and underlying mechanisms of PRMT1 and c-Fos in GC are not clearly understood. Here, we found upregulation of PRMT1 in patient tissues and established human gastric cancer cells. Notably, an intensive mRNA expression level of PRMT1 has been suggested to be an unfavorable marker in GC. Many biological processes, including cancer progression, involve arginine methylation of various substrate proteins [70]. Interestingly, our research revealed that c-Fos expression and activity are regulated by PRMT1. Our findings demonstrate that arginine methylation of c-Fos at R287 increases the stability of the c-Fos protein and protects it from autophagic degradation.
This study also elucidates a pro-oncogenic function for PRMT1 in the regulation of gastric tumorigenesis. Analysis of PRMT1 gene expression was conducted in multiple GC cohorts, and the results confirmed the upregulation of PRMT1 in GC tissues in comparison with the NATs. Furthermore, enrichment analysis verified that PRMT1 positively and significantly regulates GC progression. These results support a previous finding about the important roles of PRMT1 in the progression of GC [31]. Mechanistically, by increasing the stability of c-Fos, PRMT1-mediated c-Fos methylation accelerates the growth and enhances the invasion and migration of human GC cells. Conversely, the R287K mutation at the arginine methylation site of c-Fos reduced its stability and attenuated these tumorigenic phenotypes. In addition, the PRMT1-c-Fos signaling axis in GC was revealed by correlation analysis of PRMT1 and c-Fos protein levels in the Ajou cohort, indicating that the interaction of PRMT1 and c-Fos contributes to GC. Therefore, combined inhibition of c-Fos and PRMT1 enzymatic activity could inhibit tumorigenesis.
Conclusions
In conclusion, our study suggests that the regulatory mechanism of c-Fos/AP-1 activity could be altered by PRMT1 (Fig. 8J). PRMT1 is a specific regulator of c-Fos and protects it from autophagic degradation by regulating its methylation. Notably, the correlation in the levels of these two proteins greatly contributes to an important role in gastric tumorigenic responses, and specific targeting of PRMT1-catalyzed arginine methylation of c-Fos could become a potential new therapeutic strategy for GC.
Abbreviations
PRMT1: Protein arginine methyltransferase 1
AP-1: Activator protein-1
MAPKs: Mitogen-associated protein kinases
GC: Gastric cancer
WT: Wild-type
3-MA: 3-Methyladenine
CHX: Cycloheximide
NES: Normalized enrichment score
NATs: Normal adjacent tissues
GSEA: Gene set enrichment analysis
GEO: Gene Expression Omnibus
ShRNA: Short hairpin RNA
MS: Mass spectrometry
IP: Immunoprecipitation
RT‒PCR: Real-time polymerase chain reaction
Supplementary Material
Supplementary methods and figures.
Acknowledgements
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1A6A1A03015642 and 2020R1F1A1073219), Republic of Korea.
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Author contributions
EK, LR NA, BM, JL, and JYC conceptualized the research. EK, LR NA, BM, JL, and JYC developed the methodology. EK, LR, NA, HGK, JHK, KHK, and BCY performed the experiments and data analysis. EK, LR, and NA visualized the results. EK, LR NA, BM, JL, NP, JSK, H.H., and JYC assisted in data interpretation and provided valuable comments. EK, LR NA, BM, JL, and JYC wrote and revised the manuscript. EK, LR NA, BM, JL, and JYC reviewed and edited the manuscript. All authors discussed and commented on the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li S, Yang P, Tian E, Zhang H. Arginine methylation modulates autophagic degradation of PGL granules in C. elegans. Mol Cell. 2013;52:421-33
2. Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1-13
3. Wei H, Mundade R, Lange K, Lu T. Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle. 2014;13:32-41
4. Bedford MT, Richard S. Arginine methylation: an emerging regulatorof protein function. Mol Cell. 2005;18:263-72
5. Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425-33
6. Bachand F. Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot Cell. 2007;6:889-98
7. Kim D-I, Park M-J, Choi J-H, Kim I-S, Han H-J, Yoon K-C. et al. PRMT1 and PRMT4 regulate oxidative stress-induced retinal pigment epithelial cell damage in SIRT1-dependent and SIRT1-independent manners. Oxid Med Cell Longev. 2015. 2015
8. Li L, Zhang Z, Ma T, Huo R. PRMT1 regulates tumor growth and metastasis of human melanoma via targeting ALCAM. Mol Med Rep. 2016;14:521-8
9. Hata K, Yanase N, Sudo K, Kiyonari H, Mukumoto Y, Mizuguchi J. et al. Differential regulation of T-cell dependent and T-cell independent antibody responses through arginine methyltransferase PRMT 1 in vivo. FEBS Lett. 2016;590:1200-10
10. Mathioudaki K, Scorilas A, Ardavanis A, Lymberi P, Tsiambas E, Devetzi M. et al. Clinical evaluation of PRMT1 gene expression in breast cancer. Tumor Biol. 2011;32:575-82
11. Liu L, Sun Q, Bao R, Roth M, Zhong B, Lan X. et al. Specific regulation of PRMT1 expression by PIAS1 and RKIP in BEAS-2B epithelia cells and HFL-1 fibroblasts in lung inflammation. Sci Rep. 2016;6:21810
12. Sun Q, Yang X, Zhong B, Jiao F, Li C, Li D. et al. Upregulated protein arginine methyltransferase 1 by IL-4 increases eotaxin-1 expression in airway epithelial cells and participates in antigen-induced pulmonary inflammation in rats. J Immunol. 2012: 1102635.
13. Wurm S, Zhang J, Guinea-Viniegra J, García F, Muñoz J, Bakiri L. et al. Terminal epidermal differentiation is regulated by the interaction of Fra-2/AP-1 with Ezh2 and ERK1/2. Genes Dev. 2015;29:144-56
14. Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859
15. Karin M, Liu Z-g, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240-6
16. Gazon H, Barbeau B, Mesnard J-M, Peloponese Jr J-M. Hijacking of the AP-1 signaling pathway during development of ATL. Front Microbiol. 2018;8:2686
17. Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965-73
18. Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483-6
19. Savickienë J, Pivoriûnas A, Treigytë G, Navakauskienë R. AP-1 binding activity is modulated by post-translational modifications during granulocytic differentiation of HL-60 cells. Biologija. 2003;3:10-2
20. Tempe D, Vives E, Brockly F, Brooks H, De Rossi S, Piechaczyk M. et al. SUMOylation of the inducible (c-Fos: c-Jun)/AP-1 transcription complex occurs on target promoters to limit transcriptional activation. Oncogene. 2014;33:921
21. Garaude J, Farrás R, Bossis G, Charni S, Piechaczyk M, Hipskind RA. et al. SUMOylation regulates the transcriptional activity of JunB in T lymphocytes. J Immunol. 2008;180:5983-90
22. Saez E, Rutberg SE, Mueller E, Oppenheim H, Smoluk J, Yuspa SH. et al. c-Fos is required for malignant progression of skin tumors. Cell. 1995;82:721-32
23. Shao J, Jiang F, Hu M, Mei E, Pan Z, Chen C. et al. The role of FOS-mediated autophagy activation in the indocyanine green-based photodynamic therapy for treating melanoma. J Photochem Photobiol B. 2021;214:112101
24. Mahner S, Baasch C, Schwarz J, Hein S, Wölber L, Jänicke F. et al. C-Fos expression is a molecular predictor of progression and survival in epithelial ovarian carcinoma. Br J Cancer. 2008;99:1269
25. Liu ZG, Jiang G, Tang J, Wang H, Feng G, Chen F. et al. c-Fos over-expression promotes radioresistance and predicts poor prognosis in malignant glioma. Oncotarget. 2016;7:65946-56
26. You L, Ren X, Du Y, Zhao W, Cui M, Chen G. et al. c-Fos/ERK promotes the progression from pancreatic intraepithelial neoplasia to pancreatic ductal adenocarcinoma. Oncol Rep. 2016;36:3413-20
27. Lu C, Shen Q, DuPré E, Kim H, Hilsenbeck S, Brown PH. cFos is critical for MCF-7 breast cancer cell growth. Oncogene. 2005;24:6516-24
28. Fauquier L, Duboé C, Joré C, Trouche D, Vandel L. Dual role of the arginine methyltransferase CARM1 in the regulation of c-Fos target genes. FASEB J. 2008;22:3337-47
29. Wadhwa R, Song S, Lee J-S, Yao Y, Wei Q, Ajani JA. Gastric cancer—molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643-55
30. Aziz N, Hong YH, Jo MK, Kim JK, Kim K-H, Ashktorab H. et al. Molecular signatures of JMJD10/MINA53 in gastric cancer. Cancers. 2020;12:1141
31. Zhang Y, Wang D, Zhang M, Wei H, Lu Y, Sun Y. et al. Protein arginine methyltransferase 1 coordinates the epithelial-mesenchymal transition/proliferation dichotomy in gastric cancer cells. Exp Cell Res. 2018;362:43-50
32. Gao Y, Qi W, Sun L, Lv J, Qiu W, Liu S. FOXO3 inhibits human gastric adenocarcinoma (AGS) cell growth by promoting autophagy in an acidic microenvironment. Cell Physiol Biochem. 2018;49:335-48
33. Hong YH, Aziz N, Park JG, Lee D, Kim JK, Kim SA. et al. The EEF1AKMT3/MAP2K7/TP53 axis suppresses tumor invasiveness and metastasis in gastric cancer. Cancer Lett. 2022;544:215803
34. Yang WS, Kim HG, Kim E, Han SY, Aziz N, Yi YS. et al. Isoprenylcysteine carboxyl methyltransferase and its substrate Ras are critical players regulating TLR-mediated inflammatory responses. Cells. 2020 9
35. Park JG, Son YJ, Yoo BC, Yang WS, Kim JH, Kim JH. et al. Syk plays a critical role in the expression and activation of IRAK1 in LPS-treated macrophages. Mediators Inflamm. 2017;2017:1506248
36. Shen T, Lee J, Park MH, Lee YG, Rho HS, Kwak Y-S. et al. Ginsenoside Rp1, a ginsenoside derivative, blocks promoter activation of iNOS and COX-2 genes by suppression of an IKKβ-mediated NF-кB pathway in HEK293 cells. J Ginseng Res. 2011;35:200
37. Yi YS, Kim HG, Kim JH, Yang WS, Kim E, Jeong D. et al. Syk-MyD88 axis is a critical determinant of inflammatory-response in activated macrophages. Front Immunol. 2021;12:767366
38. Sutopo NC, Kim JH, Cho JY. Role of histone methylation in skin cancers: Histone methylation-modifying enzymes as a new class of targets for skin cancer treatment. Biochim Biophys Acta Rev Cancer. 2023 1878):188865
39. Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195-203
40. Beitz LO, Fruman DA, Kurosaki T, Cantley LC, Scharenberg AM. SYK is upstream of phosphoinositide 3-kinase in B cell receptor signaling. J Biol Chem. 1999;274:32662-6
41. Kim E, Yoon KD, Lee W-S, Yang WS, Kim SH, Sung NY. et al. Syk/Src-targeted anti-inflammatory activity of Codariocalyx motorius ethanolic extract. J Ethnopharmacol. 2014;155:185-93
42. Kim E, Son Y-J, Yang Y, Shen T, Kim I, Aravinthan A. et al. 1-(2, 3-dibenzimidazol-2-ylpropyl)-2-methoxybenzene is a Syk inhibitor with anti-inflammatory properties. Molecules. 2016;21:508
43. Latt SA, Stetten G. Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. J Histochem Cytochem. 1976;24:24-33
44. Yoo S, Kim M-Y, Cho JY. Beauvericin, a cyclic peptide, inhibits inflammatory responses in macrophages by inhibiting the NF-κB pathway. Korean J Physiol Pharmacol. 2017;21:449-56
45. Sung NY, Kim SC, Kim YH, Kim G, Lee Y, Sung G-H. et al. Anti-proliferative and pro-apoptotic activities of 4-methyl-2, 6-bis (1-phenylethyl) phenol in cancer Cells. Biomol Ther. 2016;24:402
46. Shim MK, Lee YJ. Estrogen receptor is activated by Korean red ginseng in vitro but not in vivo. Journal of ginseng research. 2012;36:169
47. Kim Y-O, Lee SW. Microarray analysis of gene expression by ginseng water extracts in a mouse adrenal cortex after immobilization stress. J 717 Ginseng Res. 2011;35:111-23
48. Sun H, Lee O-R, Kim Y-J, Jeong S-K, In J-G, Kwon W-S. et al. Identification of'Chunpoong'among Panax ginseng cultivars using real time PCR and SNP marker. J 717 Ginseng Res. 2010;34:47-50
49. Albrecht LV, Ploper D, Tejeda-Muñoz N, De Robertis EM. Arginine methylation is required for canonical Wnt signaling and endolysosomal trafficking. Proc Natl Acad Sci USA. 2018;115:E5317-E25
50. Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE, Park KC. et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun. 2018;9:1777
51. He J, Jin Y, Chen Y, Yao HB, Xia YJ, Ma YY. et al. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. OncoTargets Ther. 2016;9:6099-109
52. Wang Q, Armenia J, Zhang C, Penson AV, Reznik E, Zhang L. et al. Unifying cancer and normal RNA sequencing data from different sources. Sci Data. 2018;5:180061
53. Herrmann F, Lee J, Bedford MT, Fackelmayer FO. Dynamics of human protein arginine methyltransferase 1 (PRMT1) in vivo. J Biol Chem. 2005
54. Nicholson TB, Chen T, Richard S. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol Res. 2009;60:466-74
55. Yamaguchi A, Kitajo K. The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS One. 2012;7:e49267
56. Wada K, Inoue K, Hagiwara M. Identification of methylated proteins by protein arginine N-methyltransferase 1, PRMT1, with a new expression cloning strategy. Biochim Biophys Acta Mol Cell Res. 2002;1591:1-10
57. Piechaczyk M, Blanchard JM. c-fos proto-oncogene regulation and function. Crit Rev Oncol Hematol. 1994;17:93-131
58. Curran T, Morgan JI. Barium modulates c-fos expression and post-translational modification. Proc Natl Acad Sci USA. 1986;83:8521-4
59. Plenchette S, Romagny S, Laurens V, Bettaieb A. S-Nitrosylation in TNF superfamily signaling pathway: Implication in cancer. Redox Biol. 2015;6:507-15
60. Kao S-H, Wang W-L, Chen C-Y, Chang Y-L, Wu Y-Y, Wang Y-T. et al. Analysis of protein stability by the cycloheximide chase assay. Bio Protoc. 2015 5, e1374
61. Yeh P-Y, Lu Y-S, Ou D-L, Cheng A-L. IκB kinases increase Myc protein stability and enhance progression of breast cancer cells. Mol Cancer. 2011;10:53
62. Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091-7
63. Mayer RJ. The meteoric rise of regulated intracellular proteolysis. Nat Rev Mol Cell Biol. 2000;1:145
64. Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349
65. Kumar P, Joy J, Pandey A, Gupta D. PRmePRed: A protein arginine methylation prediction tool. PLoS One. 2017;12:e0183318
66. Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS. et al. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. 2010: jbc. M109. 089821.
67. Kim JH, Yoo BC, Yang WS, Kim E, Hong S, Cho JY. The role of protein arginine methyltransferases in inflammatory responses. Mediators Inflamm. 2016;2016:4028353
68. Hwang JW, Cho Y, Bae GU, Kim SN, Kim YK. Protein arginine methyltransferases: promising targets for cancer therapy. Exp Mol Med. 2021;53:788-808
69. Pyun JH, Ahn BY, Vuong TA, Kim SW, Jo Y, Jeon J. et al. Inducible Prmt1 ablation in adult vascular smooth muscle leads to contractile dysfunction and aortic dissection. Exp Mol Med. 2021;53:1569-79
70. Wang L, Jia Z, Xie D, Zhao T, Tan Z, Zhang S. et al. Methylation of HSP70 orchestrates its binding to and stabilization of BCL2 mRNA and renders pancreatic cancer cells resistant to therapeutics. Cancer Res. 2020;80:4500-13
71. Chen R-H, Abate C, Blenis J. Phosphorylation of the c-Fos transrepression domain by mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase. Proc Natl Acad Sci USA. 1993;90:10952-6
72. Yokoyama K, Hiyama A, Arai F, Nukaga T, Sakai D, Mochida J. C-Fos regulation by the MAPK and PKC pathways in intervertebral disc cells. PLoS One. 2013;8:e73210
73. Choi E, Uyeno S, Nishida N, Okumoto T, Fujimura S, Aoki Y. et al. Alterations of c-fos gene methylation in the processes of aging and tumorigenesis in human liver. M Mutat Res. 1996;354:123-8
74. Niu Q, Liu H, Guan Z, Zeng Q, Guo S, He P. et al. The effect of c-Fos demethylation on sodium fluoride-induced apoptosis in L-02 cells. Biol Trace Elem Res. 2012;149:102-9
75. Saunderson EA, Spiers H, Mifsud KR, Gutierrez-Mecinas M, Trollope AF, Shaikh A. et al. Stress-induced gene expression and behavior are controlled by DNA methylation and methyl donor availability in the dentate gyrus. Proc Natl Acad Sci USA. 2016;113:4830-5
76. Monje P, Hernández-Losa J, Lyons RJ, Castellone MD, Gutkind JS. Regulation of the transcriptional activity of c-Fos by ERK: A novel role for the prolyl-isomerase Pin1. J Biol Chem. 2005
77. Adler J, Reuven N, Kahana C, Shaul Y. c-Fos proteasomal degradation is activated by a default mechanism, and its regulation by NAD (P) H: quinone oxidoreductase 1 determines c-Fos serum response kinetics. Mol Cell Biol. 2010;30:3767-78
78. Alfonso Pecchio AR, Cardozo Gizzi AM, Renner ML, Molina-Calavita M, Caputto BL. c-Fos activates and physically interacts with specific enzymes of the pathway of synthesis of polyphosphoinositides. Mol Biol Cell. 2011;22:4716-25
79. Gomard T, Jariel-Encontre I, Basbous J. et al. Fos family protein degradation by the proteasome. In.: Portland Press Limited. 2008
80. Papavassiliou AG, Treier M, Chavrier C, Bohmann D. Targeted degradation of c-Fos, but not v-Fos, by a phosphorylation-dependent signal on c-Jun. Science. 1995;258:1941-4
81. Tsurumi C, Ishida N, Tamura T, Kakizuka A, Nishida E, Okumura E. et al. Degradation of c-Fos by the 26S proteasome is accelerated by c-Jun and multiple protein kinases. Mol Cell Biol. 1995;15:5682-7
82. Sasaki T, Kojima H, Kishimoto R, Ikeda A, Kunimoto H, Nakajima K. Spatiotemporal regulation of c-Fos by ERK5 and the E3 ubiquitin ligase UBR1, and its biological role. Mol Cell. 2006;24:63-75
83. Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861-73
84. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3-12
85. Vargas TR, Cai Z, Shen Y, Dosset M, Benoit-Lizon I, Martin T. et al. Selective degradation of PU. 1 during autophagy represses the differentiation and antitumour activity of TH 9 cells. Nat Commun. 2017;8:559
86. Hubbi ME, Hu H, Ahmed I, Levchenko A, Semenza GL. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J Biol Chem. 2013;288:10703-14
87. Muhammad N, Bhattacharya S, Steele R, Phillips N, Ray RB. Involvement of c-Fos in the promotion of cancer stem-like cell properties in head and neck squamous cell carcinoma. Clin Cancer Res. 2017;23:3120-8
Author contact
![]() Corresponding authors: Jae Youl Cho; jaechoedu, Jongsung Lee: bioneeredu, and Balachandran Manavalan: bala2022edu.
Corresponding authors: Jae Youl Cho; jaechoedu, Jongsung Lee: bioneeredu, and Balachandran Manavalan: bala2022edu.
Received 2023-4-9
Accepted 2023-7-8
Published 2023-7-15
