ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2023; 19(13):4052-4060. doi:10.7150/ijbs.80468 This issue Cite
Review
Seeing the T cell Immunity of SARS-CoV-2 and SARS-CoV: Believing the Epitope-Oriented Vaccines
1. CAS Key Laboratory of Infection and Immunity, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences (CAS), Beijing, China.
2. Department of Pathogen Biology & Microbiology, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China.
3. NHC Key Laboratory of Biosafety, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention (China CDC), Beijing, China.
4. School of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, Wenzhou, Zhejiang, China.
5. CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences (CAS), Beijing, China.
6. Shenzhen Children's Hospital, Shenzhen, Guangdong, China.
†These authors contributed equally to this work.
Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at the end of 2019 stimulated vigorous research efforts in immunology and vaccinology. In addition to innate immune responses, both virus-specific humoral and cellular immune responses are of importance for viral clearance. T cell epitopes play a central role in T cell-based immune responses. Herein, we summarized the peptide/major histocompatibility complex (pMHC) structures of the SARS-CoV-2-derived T cell epitopes available in the Protein Data Bank (PDB) and proposed the challenge and opportunities for using of T cell epitopes in future vaccine development efforts. A total of 27 SARS-CoV-2 related pMHC structures and five complexes with T cell receptors were retrieved. The peptides are mainly distributed on spike (S), nucleocapsid (N), and ORF1ab proteins. Most peptides are conserved among variants of concerns (VOCs) for SARS-CoV-2, except for several mutated peptides located in the S protein. The structures of human leukocyte antigen (HLA) complexed with seven epitopes derived from SARS-CoV were also retrieved, which showed a potential cross T cell immunity with SARS-CoV-2. Structural studies of antigenic peptides from SARS-CoV-2 and SARS-CoV help to visualize the processes and the mechanisms of cross T cell immunity. T cell epitope-oriented vaccines are potential next-generation vaccines for SARS-CoV-2, which are worthy of further investigation.
Keywords: Sarbecoviruses, COVID-19, SARS-CoV-2, pMHC, Epitope, TCR, Vaccine
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic [1], with more than 757 million confirmed cases and more than 6.8 million deaths reported worldwide as of February, 2023 [2, 3]. COVID-19 presents with a variety of clinical symptoms, and it is currently estimated that approximately 80% of COVID-19 cases are mild to moderate [4, 5]. In addition to innate immune responses, virus-specific humoral and cellular immunity play a pivotal role in viral clearance. Cell immunity is a vital aspect of adaptive immunity and an effective method to combat viruses. Although some COVID-19 patients may not develop strong antibody responses, particularly in mild cases, T cell responses can still provide significant protection [6]. T cell-mediated immunity plays a crucial role in clearing SARS-CoV-2, generating long-term memory responses to the virus, and identifying SARS-CoV-2 variants [7-12]. Furthermore, the strength of T cell responses in patients is strongly linked to the severity of COVID-19 symptoms, with weaker T cell responses observed in those with milder symptoms [13]. Numerous studies also have shown that T cell responses can serve as a highly sensitive indicator of exposure to SARS-CoV-2, and can accurately predict the status and prognosis of COVID-19 patients [14-17]. Thus, it is essential to conduct a thorough examination of T cell immune properties against SARS-CoV-2, as this may lead to more effective design strategies for antiviral drugs and vaccines [18].
T cell immunity of the SARS-CoV-2 infection
T cell immunity is essential for the control and clearance of viral infections, with virtually all individuals who contract the virus exhibiting T cell responses [19]. T cell immunity is comprised of two types: CD4+ T cell-mediated immunity and CD8+ T cell-mediated immunity, each activated by specific peptide-HLA (pHLA) molecule interactions. CD4+ T cells recognize 13-17 peptides presented by HLA class II molecules, while CD8+ T cells require the recognition of 8-10 amino acid peptides presented in combination with HLA class I molecules, known as T cell epitopes [20, 21]. Moderate and severe symptoms in COVID-19 patients are associated with significantly reduced numbers of CD4+ and CD8+ T cells [22, 23]. The level of CD8+ T cells reflects the severity of the patient's disease, while reduced levels of CD4+ T cells are only independently associated with increased in-hospital mortality in COVID-19 patients [24]. Both CD4+ and CD8+ T cells have been observed to respond robustly to epitopes derived from the spike (S), nucleocapsid (N), and membrane (M) proteins of SARS-CoV-2. However, CD4+ T cells displayed more significant response to SARS-CoV-2 than CD8+ T cells [19, 25]. CD4+ T cells possess a unique ability to differentiate into various helper and effector cell types. This enables them to provide guidance to B cells, assist CD8+ T cells, and exhibit direct antiviral activity. In contrast, CD8+ T cells play a crucial role in eradicating viral infections by directly eliminating infected cells [26]. Therefore, studying the immune response characteristics of CD4+ and CD8+ T cells, and their interactions in antiviral immunity, is crucial in designing effective COVID-19 therapeutic and vaccine strategies.
COVID-19 vaccines need to boost the breadth and strength of T cell immunity
Many types of vaccines are currently used to fight COVID-19, such as inactivated vaccines, nucleic acid-based vaccines, viral vector-based vaccines, and subunit (recombinant protein) vaccines [27, 28]. Generally, each vaccine type has its characteristics, and vaccination possesses a relatively safe profile and is fairly well tolerated. Vaccines can trigger robust humoral, Th1- dominated, and/or Th2-dominated cellular immune responses after vaccination to protect against severe and fatal infections [29-33].
According to recent research, existing vaccines are mostly based on the study of humoral immunity, and how long SARS-CoV-2-specific antibodies can provide protective immunity has not been well determined [34-40]. Although the target antigen spectrum of vaccines is different, except for inactivated vaccines, vaccine design based on the S protein is the most common strategy to fight the COVID-19 pandemic [41]. The receptor-binding motif (RBM) in the receptor binding domain (RBD) is a primary target for neutralizing antibodies, and most of the substitutions of variants of concern (VOCs) occur in the antibody-recognized regions [42]. The currently authorized COVID-19 vaccines remain effective against mutant strains, but vaccinated sera show decreased neutralization titers against different SARS-CoV-2 VOCs compared to the ancestral strain [43-47]. The virus can mutate to escape under the pressure of immunity raised by both natural infection and vaccination and produces a variety of mutant viruses in immunocompromised hosts, which also poses a huge challenge to existing vaccines [43]. It has been demonstrated that even in the absence of antibodies, cellular responses mediated by T cell can maintain sufficient immune responses against SARS-CoV-2 [8, 48-50]. For example, in the study by Soresina et al., they found that two patients with X-linked aglobulinemia (XLA) (congenital immunodeficiency (IEI)) had no B cells in their peripheral blood but could recover from infection, suggesting that the B-cell response may not be the only key factor in defeating SARS-CoV-2, and T cell mediated immunity may contribute to the antiviral process [48].
The ability of existing vaccines to stimulate T cell immunity also remains limited. For example, mRNA vaccines can induce CD8+ cells to mediate T cell immune responses. However, the SARS-CoV-2 mRNA vaccines that have been approved for clinical use are S protein-based vaccines, which cannot generate immune responses against other SARS-CoV-2 proteins. The gene encoding the S protein is more prone to mutations than other regions of the SARS-CoV-2 genome. This may be why Omicron can escape the full repertoire of S-specific T cells induced by mRNA vaccines in some people [51]. Similarly, other S protein-based vaccines such as adenovirus vaccines and DNA vaccines also have the same problem [52]. Even the inactivated vaccine containing the full sequence of the virus only induces a low number of S-specific T cell responses, mostly inducing CD4+ T cell responses and lacking virus-specific CD8+ T cell immune responses [53]. Vaccines targeting T cell epitopes are already in development, and CoVac-1 is a peptide vaccine candidate consisting of multiple T cell epitopes from various SARS-CoV-2 viral proteins and contains toll-like receptor 1/2 agonist XS15. In its phase I open-label trial, CoVac-1 displayed good security and induced a potent SARS-CoV-2 T cell immunity. Considering the conservation of the selected T cell epitopes, it may also contribute to cross-reactivity to SARS-CoV-2 VOCs [54, 55]. Given the emergence of mutant strains of SARS-CoV-2 and the heightened vulnerability of elderly and immunocompromised individuals to this virus, there remains an urgent need to develop next-generation SARS-CoV-2 vaccines. In addition to inducing antibody responses, an ideal SARS-CoV-2 vaccine needs to contain conserved T cell epitopes to induce broad-spectrum and persistent cellular immunity.
MHC presentation of SARS-COV-2-derived epitopes
The molecular mechanism of HLA presenting virus-derived epitopes provides useful information for the development of T cell-related vaccines. In the course of T cell immunity, the stable binding of peptides to HLA is crucial for the presentation process, which both affects the way the peptides are presented and affects the T cell receptor recognition of pHLA complexes to activate T cells [20, 56, 57]. Current bioinformatics approaches, including AlphaFold, cannot precisely predict protein-protein interactions. Thus, crystallography-based structural studies are still a direct way to provide detailed insights into the peptide conformations in pHLA complexes. The pHLA complex structures can help to visualize the typical anchoring residues and peptide conformation [58], define the minimal (optimal) epitopes [59], confirm the conformations of immunodominant epitopes, and aid in the design of altered peptides with higher immunogenicity [60, 61].
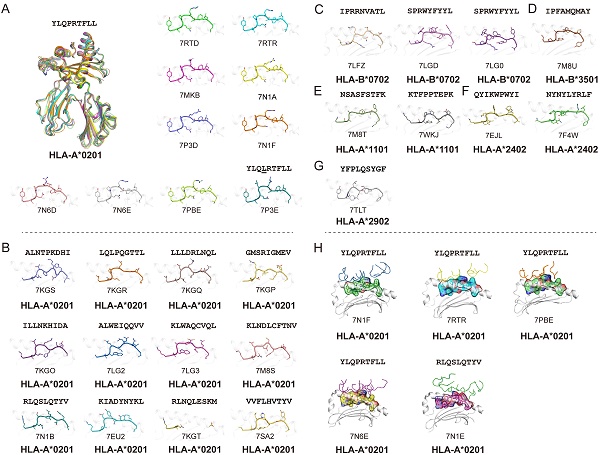
Using the Protein Database Bank (PDB), we searched for pMHC structures with the keywords “MHC” and “SARS”. A total of 27 structures of HLA class I molecules with peptides derived from SARS-CoV-2 S, N, and ORF 1ab proteins were found (Table 1) [1, 15, 56, 62-68], while no HLA class II complexes were found. These HLA I complexes are derived from six HLA allele types, HLA-A*0201, HLA-A*1101, HLA-A*2402, HLA-A*2902, HLA-B*0702, and HLA-B*3501, and belong to the five supertypes A02 (HLA-A*0201), A03 (HLA-A*1101), A24 (HLA-A*2402), A01A24 (HLA-A*2902), and B07 (HLA-B*0702 and HLA-B*3501). These supertypes are more common in humans, and the combination of three supertypes (A02, A03, and B07) covers 86% of the population [69]. HLA-A*0201, as one of the most common HLAs in the global population [70], has 18 pMHC structures and five pMHC/TCR complexes, involving a total of 14 different SARS-CoV-2-derived peptides (Fig. 1). Among the 14 peptides, the dominant peptide S269-277 and its variants represent a total of 10 structures (including one mutant) (Fig. 1A). Like other HLA-A*0201-presented peptides, the peptides derived from SARS-CoV-2 in all of these HLA-A*0201 structures use typical P2-Met/Leu and PΩ-Val/Leu as primary anchors [69], with residues P3 and P5 or P7 as secondary anchors. Other HLA alleles involved in the structural investigations of SARS-CoV-2-derived peptide presentation include three HLA-B*0702, one HLA-B*3501, two HLA-A*1101, two HLA-A*2402, and one HLA-A*2902 structures, respectively, with typical anchoring characteristics for all of the peptides in the corresponding HLA alleles (Fig. 1C-G).
As the viral receptor binding protein and the dominant antibody target, the S protein is the main location for the mutations related to immune escape. Compared to the N protein, the S protein is more prone to mutation. There are more than 30 mutations in the S protein of the Omicron variant of SARS-CoV-2 [71]. We analyzed the conservation of peptides utilized in these available structures among the SARS-CoV-2 VOCs (Alpha, Beta, Gamma, Delta, and Omicron) and found four peptides in the S protein, S417-425, S370-378, S448-456 and S489-497, were mutated among VOCs. Peptide S269-277 is included in nine structures and is conserved in all of the current VOCs. This suggests that peptide S269-277 is a highly conserved epitope. However, the appearance of the mutant peptide (YLQLRTFLL) also indicated that peptide S269-277 had a low level of variation among the SARS-CoV-2 variants (Fig. 1A and Table 1) [63].
In contrast, the N protein is more conserved compared to the RBD of the S protein. All of the currently structurally available N protein-derived peptides are conserved among the VOCs. Our recent study identified one HLA-A*1101-restricted peptide N25. This peptide is both conserved among SARS-CoV-2 VOCs and within SARS-CoV [15, 18]. Furthermore, Szeto et al. found that the stability of the peptide on the N protein affects its immunogenic potential [66]. The N219-227, N222-230, and N316-324 peptides, which have higher Tm values (as determined by circular dichroism), are immunogenic in recovered patients with COVID-19, while N138-146 and N159-167, with lower Tm values, are not immunogenic [72-74].
The TCR recognition of SARS-CoV-2-derived epitopes
Five pMHC-TCR complex structures have been determined to demonstrate the T cell recognition of the SARS-CoV-2-derived epitopes, four of which are different pMHC-TCRs based on the dominant epitope YLQPRTFLL [62]. In the two studies by Szeto et al. and Wu et al., they determined the structure of the immunodominant peptide S269-277 bound to the HLA-A*02:01 molecular complex and performed a ternary structure resolution of the pHLA-TCR in response to the observation of a public TCR in multiple unrelated individuals, which is essential for a thorough understanding of the CD8+ T cell response to specific epitopes (Fig. 1H). Moreover, another structure of HLA-A*02:01/S269-277 distinct from the public TCR was identified by Chaurasia et al. [75]. It was found that the HLA-A*02:01/S269-277 restricted TCR library does not efficiently cross-react with S269-277 epitope variants or homologous epitopes of other β-coronaviruses, and the resolution of this complex provides information about potential mechanisms by which the virus evades SARS-CoV-2-specific CD8+ T cell responses. Another pMHC-TCR complex based on the peptide RLQ (RLQSLQTYV) and a private T cell receptor relies heavily on the CDR3α and CDR3β loops produced by somatic cells to recognize the pMHC complex, implying the optimal activation of CD8+ T cell responses [62]. The determination of these pMHC-TCR complex structures provides useful information and insights for the rational optimization and design of effective T cell vaccines capable of durability and cross-protection.
The pMHC/pMHC-TCR structures of SARS-CoV-2 and SARS-CoV derived peptides in the Protein Data Bank (PDB).
| Peptide | PDB | Sequence | Protein | Position | MHC allele | Reference |
|---|---|---|---|---|---|---|
| SARS-CoV-2 | ||||||
| YLQ | 7RTD | YLQPRTFLL | S | 269-277 | HLA-A*0201 | (Szeto et al., 2021) |
| YLQ | 7RTRb | HLA-A*0201 | (Szeto et al., 2021) | |||
| / | 7MKB | HLA-A*0201 | To be published | |||
| YLQ | 7N1A | HLA-A*0201 | (Wu et al., 2021) | |||
| / | 7P3D | HLA-A*0201 | (Dolton et al., 2021) | |||
| YLQ | 7N1Fb | HLA-A*0201 | (Wu et al., 2021) | |||
| S269-277 | 7N6D | HLA-A*0201 | (Chaurasia et al., 2021) | |||
| S269-277 | 7N6Eb | HLA-A*0201 | (Chaurasia et al., 2021) | |||
| / | 7PBEb | HLA-A*0201 | (Dolton et al., 2021) | |||
| / | 7P3E | YLQLRTFLL | S | 269-277 | HLA-A*0201 | (Dolton et al., 2021) |
| S370-378 | 7M8T | NSASFSTFKc | S | 370-378 | HLA-A*1101 | (Nguyen et al., 2021) |
| S386-395 | 7M8S | KLNDLCFTNV | S | 386-395 | HLA-A*0201 | (Nguyen et al., 2021) |
| KIA S | 7EU2 | KIADYNYKLc | S | 417-425 | HLA-A*0201 | (Zhang et al., 2021) |
| NYN S | 7F4W | NYNYLYRLFc | S | 448-456 | HLA-A*2402 | (Zhang et al., 2021) |
| / | 7TLT | YFPLQSYGFc | S | 489-497 | HLA-A*2902 | To be published |
| S896-904 | 7M8U | IPFAMQMAY | S | 896-904 | HLA-B*3501 | (Nguyen et al., 2021) |
| RLQ | 7N1Eb | RLQSLQTYV | S | 1000-1008 | HLA-A*0201 | (Wu et al., 2021) |
| RLQ | 7N1B | HLA-A*0201 | (Wu et al., 2021) | |||
| / | 7SA2 | VVFLHVTYV | S | 1060-1068 | HLA-A*0201 | To be published |
| CoV-2 | 7EJL | QYIKWPWYI | S | 1208-1216 | HLA-A*2402 | (Shimizu, K., 2021) |
| / | 7LG0 | SPRWYFYYL | N | 105-113 | HLA-B*0702 | To be published |
| SPR | 7LGD | HLA-B*0702 | (Lineburg et al., 2021) | |||
| N138-146 | 7KGS | ALNTPKDHI | N | 138-146 | HLA-A*0201 | (Szeto et al., 2021) |
| N159-167 | 7KGR | LQLPQGTTL | N | 159-167 | HLA-A*0201 | (Szeto et al., 2021) |
| N222-230 | 7KGQ | LLLDRLNQL | N | 222-230 | HLA-A*0201 | (Szeto et al., 2021) |
| N226-234 | 7KGT | RLNQLESKM | N | 226-234 | HLA-A*0201 | (Szeto et al., 2021) |
| N316-324 | 7KGP | GMSRIGMEV | N | 316-324 | HLA-A*0201 | (Szeto et al., 2021) |
| N351-359 | 7KGO | ILLNKHIDA | N | 351-359 | HLA-A*0201 | (Szeto et al., 2021) |
| N25 | 7WKJ | KTFPPTEPK | N | 361-369 | HLA-A*1101 | (Zhang et al., 2021) |
| / | 7LG3 | KLWAQCVQL | ORF 1ab | 3886-3894 | HLA-A*0201 | To be published |
| / | 7LG2 | ALWEIQQVV | ORF 1ab | 4094-4102 | HLA-A*0201 | To be published |
| / | 7LFZ | IPRRNVATL | ORF 1ab | 5916-5924 | HLA-B*0702 | To be published |
| SARS-CoV | ||||||
| / | 3C9N | VQQESSFVMd | ORF 1ab | 1775-1783 | HLA-B*1501 | (Røder et al., 2008) |
| N216-225 | 6IEX | GETALALLLLd | N | 216-225 | HLA-B*4001 | (Ji et al., 2019) |
| N1 | 3I6L | QFKDNVILLd | N | 346-354 | HLA-A*2402 | (Liu et al., 2010) |
| SNP362-370 | 1X7Q | KTFPPTEPK | N | 362-370 | HLA-A*1101 | (Blicher et al., 2005) |
| Md3 | 3I6K | TLACFVLAAVd | M | 60-69 | HLA-A*0201 | (Liu et al., 2010) |
| Md3-C9 | 3TO2 | LACFVLAAVd | M | 61-69 | HLA-A*0201 | (Liu et al., 2011) |
| Mn2 | 3I6G | GLMWLSYFVd | M | 88-96 | HLA-A*0201 | (Liu et al., 2010) |
aThe conservation of SARS-CoV-2 (NCBI reference sequence: NC_045512.2) is based on alignment with variants of concern (VOCs), and conservation of SARS-CoV-1 (GenBank: AY654624.1) is based on alignment with SARS-CoV-2 and VOCs. VOCs: Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529). The accession numbers from GISAID are as follows: EPI_ISL_683466, EPI_ISL_6693552, EPI_ISL_833172, EPI_ISL_3473618, and EPI_ISL_6640916. Mutated positions of peptides are highlighted in bold and underlined. bTCR structures. cPeptides with substitutions among SARS-CoV-2 variants of concern (VOCs). S370-378, NSASFSTFK: NLAPFFTFK (Omicron); KIA S, KIADYNYKL: NIADYNYKL (Beta), TIADYNYKL (Gamma), NIADYNYKL (Omicron); NYN_S, NYNYLYRLF: NYNYRYRLF (Delta); and YFPLQSYGF: YFPLRSYSF (Omicron). dPeptides with substitutions among SARS-CoV-2 and SARS-CoV. VQQESSFVM: VQQESPFVM (SARS-CoV-2 and five VOCs); N216-225, GETALALLLL: GDAALALLLL (SARS-CoV-2 and five VOCs); N1, QFKDNVILL: NFKDQVILL (SARS-CoV-2 and five VOCs); Md3, TLACFVLAAV: TLTCFVLAAV (Omicron); Md3-C9, LACFVLAAV: LTCFVLAAV (Omicron); and Mn2, GLMWLSYFV: GLMWLSYFI (SARS-CoV-2 and five VOCs).
Summary of pMHC/pMHC-TCR structures of SARS-CoV-2-derived peptides in the Protein Data Bank (PDB). (A) The structure of HLA-A*0201 presentation of SARS-derived dominant epitope peptide (YLQPRTFLL) and mutant peptide (YLQLRTFLL). (B) HLA-A*0201 presents other SARS-CoV-2-derived peptides in the PDB (https://www.rcsb.org/). Note: for peptide RLQSLQTYV, we only show one pMHC structure (PDB: 7N1B), and for another TCR-pMHC structure (PDB: 7N1E) not shown separately in Figure 1B. (C-G) Structural summary of the five alleles of HLA-B*0702, HLA-B*3501, HLA-A*1101, HLA-A*2401, and HLA-A*2902 in the PDB bound to the SARS-CoV-2-derived peptides. (H) The pHLA-TCR structures of SARS-CoV-2-derived peptides.
Potential for cross-T cell recognition of SARS-CoV-2 and SARS-CoV
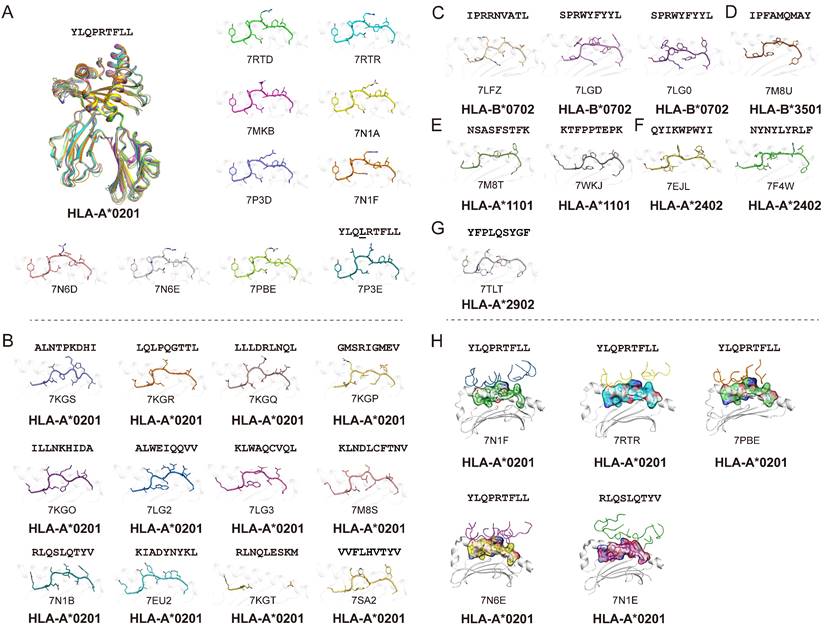
In general, T cells can cross-recognize some mutant epitopes, and mutations at certain sites on the peptide may not be detrimental to TCR recognition [75, 76]. Homologous epitopes from other coronaviruses have the potential to elicit cross T cell responses to SARS-CoV-2 infection, so we also summarized the structures of HLA complexes loaded with SARS-CoV-derived peptides (Table 1 and Fig. 2A) [60, 77-81]. Three peptides are based on the HLA-A*0201-restricted type, and there is one for each of the other HLA alleles, i.e., HLA-A*2402, HLA-B*1501, HLA-A*1101, and HLA-B*4001. The peptides in these structures have high conservation when aligned to the corresponding peptides in SARS-CoV-2. The peptide SNP362-370 (KTFPPTEPK) has the same sequence between SARS-CoV-2 and SARS-CoV, while only one or two amino acid mutations were found in ORF 1775-1783, N216-225, N 346-354, M 60-69, M 61-69, and M 88-96 fragments (Table 1 and Fig. 2B). Our previously identified HLA-A*0201-restricted SARS-CoV peptides Md3 and Md3-C9 have an A62T substitution between SARS-CoV-2 and SARS-CoV [80]. However, both A and T can act as the P2 or P3 anchor for HLA-A*0201 peptides. Another peptide, Mn2, has only the V96I mutation in SARS-CoV-2. The amino acids valine (V) and isoleucine (I) are hydrophobic residues, which are the preferred residues of HLA-A*0201 binding to the PΩ of peptides. Likewise, peptide N1 has two mutations, Q346N and N350Q, in SARS-CoV-2, and the volume and chemical properties of the two amino acids are similar. Thus, mutations in these peptides may not affect the presentation by MHC and the recognition by TCRs. Given this, these conserved peptides may act as a target for cross-reactive T cells for both SARS-CoV-2 and SARS-CoV. The certain degree of cross-T cell immunity between different coronavirus strains will shed light on a universal vaccine [82].
Summary of published pMHC structures of SARS-CoV-derived peptides in the PDB. (A) The pMHC structure of SARS-CoV-derived peptides in the PDB, including three HLA-A*0201 types and one each of the HLA-A*1101, HLA-B*1501, HLA-B*2401, and HLA-B*4001 types. (B) Analysis of whether the SARS-CoV-derived peptides contained in the PDB are consistent with the amino acids at the same positions in SARS-CoV-2.
The pMHC structures guide the potential utilization of T cell epitopes in vaccines
The general strategy for vaccine development is to induce the largest possible T cell response. It has been proven that immunogenicity can be improved by modifying the T cell epitope, and the modified epitope can also induce an immune response to kill target cells [83, 84]. Although not all functional T cell epitopes have a high binding affinity to MHC molecules, most immunodominant T cell epitopes have typical MHC anchor residues and show tight binding to MHCs [85-87]. Structures of pMHC complexes with or without a TCR solved by X-ray diffraction provide a chance to visualize the peptide presentation by certain MHC molecules and the recognition by the TCR.
First, we can use the structures for peptide modification to generate more and stronger immunodominant peptides for T cell tests or vaccine development [61]. For example, in the study of Borbulevych et al., the modified peptide produced by substituting Thr to Met at the P2 position of peptide gp100209-217 has nine-fold higher binding affinity for HLA-A2, a seven-fold slower dissociation rate, and has more immunogenicity in vitro and in vivo [88]. Similarly, N159-167 displays weak binding to HLA-A*0201 due to the subdominant anchor P2-Q. "This residue can be upgraded to an HLA-A*0201-binding peptide with a preferred P2 anchor (i.e., L, V, or I), which may increase the immunogenicity of peptide N159-167.
Second, the structures can be used to understand the mechanism of VOCs escape. The K417N mutation of the KIA_S epitope results in the disruption of the interaction between the peptide P1-Lys and the W167 of HLA-A*0201, leading to disrupted peptide binding. Although the L452R mutation of the NYN_S epitope does not prevent the binding of the peptide to HLA-A*2402, the solvent-exposed mutation may alter the recognition by the TCR [65].
Third, these structures can be employed in T cell receptor (TCR) evolution for developing therapeutic TCR. The current structure of the public TCR for SARS-CoV-2 S269-277 suggests that the affinity of the TCR may be optimized by mutating Cα residues 157-165 within the TCR sequence [62].
Conclusion and perspective
Although the COVID-19 pandemic is still wreaking havoc around the world, we can take an optimistic view that it will die down with the continuous implementation of non-pharmacological interventions and the application of the vaccines and boosters [89]. However, the potential flare-ups caused by emerging VOCs and the unpredictable nature of SARS-CoV-2 will pose a consistent risk to humans [90]. Thus, a universal vaccine based on the more conserved T cell antigenic peptides may provide a ray of light for future efforts focused on emerging human-infective sarbecoviruses and variants. As for peptide-based vaccine development, the intrinsic challenges for these vaccines should be considered, i.e., the HLA restriction of the T cell epitopes, the selection of immunodominant epitopes, and the weak immunogenicity of the synthesized peptides. For the HLA restriction, it may be a term of settlement to screen and identify T cell epitopes with features of cross-HLA alleles and even cross-HLA supertypes presentation and reactivity [59, 91, 92]. For the selection of immunodominant epitopes, the systematic and comprehensive evaluation of the immunogenicity profile of the proteome of SARS-CoV-2 and other coronaviruses is needed. Both the Structural and nonstructural proteins of SARS-CoV-2 can trigger CD4+ and CD8+ T cell immune responses. However, the structural proteins S, M, and N are heavily expressed in SARS-CoV-2-infected cells and are the most immunodominant targets of human CD4+ and CD8+ T cell responses to the virus [19]. Thus, it may be an advantageous strategy to first assess the ability of these proteins to elicit an immune response and incorporate them into vaccine development. Further, the polymorphism and coverage of HLA alleles should also be considered. An effective vaccine should provide coverage for a large and diverse portion of the population. Although the HLA varies greatly between individuals, studies have demonstrated that selecting for only 18 HLA class I (HLA-A*0101, HLA-A*0201, HLA-A*0301, HLA-A*2402, HLA-HLA-B*0702, HLA-B*0801, HLA-B*1402, HLA-B*1501, HLA-B*2705, HLA-B*3501, HLA-B*3901, HLA-B*4001, HLA-B*4402, HLA-B*5201, HLA-B*5701, HLA-B*5801, HLA-B*8101, and HLA-Cw*0701) alleles can provide protection to over 99% of the global population [93-95]. It may still be a challenging task to cover all these 18 HLA alleles with dominant epitopes in vaccine design. Hence, a viable suggestion for vaccine design could be initially target the epitopes of the high-frequency HLA alleles belonging to various HLA supertypes, such as the dominant epitopes present in supertypes A02, A03, and B07. It is a massive and complicated body of work to investigate the antigenic spectrum of the sarbecoviruses, especially considering the large genomes of coronaviruses, the multiformity of the coronaviridae, and the diversity of HLA alleles among different ethnic groups. However, these knots will be untied with the development of cellular immunity-related techniques, structure-based tools, and currently ascendant artificial intelligence technology. Finally, proper carriers and adjuvants will help to intensify the immunogenicity of synthesized peptides. In addition, appropriate longer peptides may cover more T cell epitopes with different HLA alleles and some B cell epitopes folded in specific natural secondary structures. If we work together, there will be a happy ending in this game of catch me if you can.
The current COVID-19 pandemic raises public concerns about human immunity against viruses, from herd immunity by natural infection to the duration of immunity from vaccination. The emerging SARS-CoV-2 variants also pose a challenge to the long-term cross-protection to the VOCs of the current vaccine. T cell immunity provides long-term immune memory against SARS-CoV-2 and broad cross-reactivity to viral variants. Herein, the currently determined pMHC structures involving SARS-CoV-2-derived T cell epitopes and several T cell receptor complexes in the PDB were summarized. Most of the T cell epitope peptides show good conservation among the SARS-CoV-2 variants, implying potential cross-T cell immunity. Previously determined pMHC structures with SARS-CoV-derived T cell epitopes also indicate a potential cross-immunity to SARS-CoV-2. The structural investigations of the MHC presentation and TCR recognition of SARS-CoV-2-derived epitopes provide insight into human immunity to newly emerging viruses and may shed light on the development of a universal vaccine against sarbecoviruses.
Acknowledgements
Funding
The work was supported by the National Key Research and Development Program of China (2022YFC2604100) and the National Natural Science Foundation of China (grants 92269203). Kefang Liu was supported by Young Elite Scientists Sponsorship Program by CAST (2021QNRC001).
Author contributions
CY, PYW, KFL, and WJL wrote the manuscript. CY and JMT analyzed and organized the data. WJL, KFL, and GFG conceived and designed this subject. WJL revised the manuscript. All authors contributed to the article and approved the submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lineburg K, Grant E, Swaminathan S, Chatzileontiadou D, Szeto C, Sloane H. et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. 2021;54:1055-65
2. Tian F, Chen Z, Feng Q. Nirmatrelvir-ritonavir compared with other antiviral drugs for the treatment of COVID-19 patients: A systematic review and meta-analysis. J Med Virol. 2023;95(4):e28732
3. Magel T, Meagher E, Boulter T, Albert A, Tsai M, Muñoz C. et al. Fatigue presentation, severity, and related outcomes 12 weeks post-COVID-19 hospitalization. FrontMed. 2023;10:1179783
4. de Candia P, Prattichizzo F, Garavelli S, Matarese G. T cells: warriors of SARS-CoV-2 infection. Trends Immunol. 2021;42:18-30
5. Shang Z, Chan S, Liu W, Li P, Huang W. Recent insights into emerging coronavirus: SARS-CoV-2. ACS Infect Dis. 2021;7:1369-88
6. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin J, Olsson A. et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158-68
7. Geers D, Shamier MC, Bogers S, den Hartog G, Gommers L, Nieuwkoop NN. et al. SARS-CoV-2 variants of concern partially escape humoral but not T cell responses in COVID-19 convalescent donors and vaccinees. Sci Immunol. 2021 6
8. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457-62
9. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D. et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336-45
10. Agerer B, Koblischke M, Gudipati V, Montaño-Gutierrez LF, Smyth M, Popa A. et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8(+) T cell responses. Sci Immunol. 2021 6
11. Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M. et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490-500
12. Tan A, Linster M, Tan C, Le Bert N, Chia W, Kunasegaran K. et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728
13. Hu D, Li L, Shi W, Zhang L. Less expression of CD4+ and CD8+ T cells might reflect the severity of infection and predict worse prognosis in patients with COVID-19: Evidence from a pooled analysis. Clin Chim Acta. 2020;510:1-4
14. Gallais F, Velay A, Nazon C, Wendling M, Partisani M, Sibilia J. et al. Intrafamilial exposure to SARS-CoV-2 associated with cellular immune response without seroconversion, France. Emerg Infect Dis. 2021;27:113-21
15. Zhang J, Lu D, Li M, Liu M, Yao S, Zhan J. et al. A COVID-19 T cell response detection method based on a newly identified human cd8(+) T cell epitope from SARS-CoV-2 - Hubei Province, China, 2021. China CDC Wkly. 2022;4:83-7
16. Wang Z, Yang X, Zhong J, Zhou Y, Tang Z, Zhou H. et al. Exposure to SARS-CoV-2 generates T cell memory in the absence of a detectable viral infection. Nat Commun. 2021;12:1724 -
17. Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol. 2022;23:186-93
18. Zhang J, Lin H, Ye B, Zhao M, Zhan J, Dong S. et al. One-year sustained cellular and humoral immunities of COVID-19 convalescents. Clin Infect Dis. 2021
19. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR. et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489-501.e15
20. Szeto C, Lobos CA, Nguyen AT, Gras S. TCR recognition of peptide-MHC-I: Rule makers and breakers. Int J Mol Sci. 2020 22
21. Liu J, Gao G. Major histocompatibility complex: interaction with peptides. eLS. 2011
22. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H. et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Investig. 2020;130:2620-9
23. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D. et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533-5
24. Wen XS, Jiang D, Gao L, Zhou JZ, Xiao J, Cheng XC. et al. Clinical characteristics and predictive value of lower CD4(+)T cell level in patients with moderate and severe COVID-19: a multicenter retrospective study. BMC Infect Dis. 2021;21:57
25. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin J-B, Olsson A. et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158-68 e14
26. Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861-80
27. Dai L, Gao G. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73-82
28. Xu K, Dai L, Gao G. Humoral and cellular immunity and the safety of COVID-19 vaccines: a summary of data published by 21 May 2021. Int Immunol. 2021;33:529-40
29. Bueno S, Abarca K, González P, Gálvez N, Soto J, Duarte L. et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine in a subgroup of healthy adults in Chile. Clin Infect Dis. 2021
30. Wang H, Zhang Y, Huang B, Deng W, Quan Y, Wang W. et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713-21
31. Kauffman K, Webber M, Anderson D. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J Control Release. 2016;240:227-34
32. Walsh E, Frenck R Jr, Falsey A, Kitchin N, Absalon J, Gurtman A. et al. Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. N Engl J Med. 2020;383:2439-50
33. Fu Y, Chen F, Cui L, Zhao Y, Zhang H, Fu S. et al. Immunological analysis of people in northeast china after SARS-CoV-2 inactivated vaccine injection. Vaccines (Basel). 2021 9
34. Robbiani D, Gaebler C, Muecksch F, Lorenzi J, Wang Z, Cho A. et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437-42
35. Vabret N. Antibody responses to SARS-CoV-2 short-lived. Nat Rev Immunol. 2020;20:519
36. Röltgen K, Wirz O, Stevens B, Powell A, Hogan C, Najeeb J. et al. SARS-CoV-2 antibody responses correlate with resolution of RNAemia but are short-lived in patients with mild illness. medRxiv. 2020
37. Carreño JM, Alshammary H, Tcheou J, Singh G, Raskin AJ, Kawabata H. et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682-8
38. Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L. et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat Med. 2022;28:481-5
39. Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML. et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457-66.e4
40. Sapkal G, Kant R, Dwivedi G, Sahay RR, Yadav PD, Deshpande GR. et al. Immune responses against different variants of SARS-CoV-2 including Omicron following 6 months of administration of heterologous prime-boost COVID-19 vaccine. J Travel Med. 2022 29
41. Galvan V, Quarleri J. Editorial: Comparison of antibody and T cell responses elicited by BBIBP-CorV (Sinopharm) and BNT162b2 (Pfizer-BioNTech) vaccines against SARS-CoV-2 in healthy adult humans. Geroscience. 2022;44(1):57-61
42. Piccoli L, Park Y, Tortorici M, Czudnochowski N, Walls A, Beltramello M. et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024-42
43. Choi J, Smith D. SARS-CoV-2 variants of concern. Yonsei Med J. 2021;62:961-68
44. Sun C XC, Bu GL, Zhong LY, Zeng MS. et al. Molecular characteristics, immune evasion, and impact of SARS-CoV-2 variants. Signal Transduct Target Ther. 2022;7:202
45. Andrews N, Stowe J, Kirsebom F, Toffa S, Rickeard T, Gallagher E. et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N Engl J Med. 2022;386:1532-46
46. Chen X, Chen Z, Azman AS, Sun R, Lu W, Zheng N. et al. Neutralizing antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants induced by natural infection or vaccination: a systematic review and pooled analysis. Clin Infect Dis. 2021;74:734-42
47. Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H. et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654-6
48. Soresina A, Moratto D, Chiarini M, Paolillo C, Baresi G, Focà E. et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol. 2020;31:565-9
49. Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F. et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270-4
50. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin JB, Olsson A. et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158-68.e14
51. Naranbhai V, Nathan A, Kaseke C, Berrios C, Khatri A, Choi S. et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell. 2022;185:1041-51 e6
52. Bellamkonda N, Lambe UP, Sawant S, Nandi SS, Chakraborty C, Shukla D. Immune Response to SARS-CoV-2 Vaccines. Biomedicines. 2022;10:1464
53. Lim JME, Hang SK, Hariharaputran S, Chia A, Tan N, Lee ES. et al. A comparative characterization of SARS-CoV-2-specific T cells induced by mRNA or inactive virus COVID-19 vaccines. Cell Rep Med. 2022;3:100793
54. Heitmann J, Bilich T, Tandler C, Nelde A, Maringer Y, Marconato M. et al. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature. 2021
55. Liu J, Wu B, Zhang S, Tan S, Sun Y, Chen Z. et al. Conserved epitopes dominate cross-CD8+ T cell responses against influenza A H1N1 virus among Asian populations. Eur J Immunol. 2013;43:2055-69
56. Szeto C, Chatzileontiadou DS, Nguyen AT, Sloane H, Lobos CA, Jayasinghe D. et al. The presentation of SARS-CoV-2 peptides by the common HLA-A∗ 02: 01 molecule. iScience. 2021;24:102096
57. Nguyen AT, Szeto C, Jayasinghe D, Lobos CA, Halim H, Chatzileontiadou DSM. et al. SARS-CoV-2 spike-derived peptides presented by HLA molecules. Biophysica. 2021;1:194-203
58. Sun Y, Liu J, Yang M, Gao F, Zhou J, Kitamura Y. et al. Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses. J Gen Virol. 2010;91:919-30
59. Liu J, Zhang S, Tan S, Yi Y, Wu B, Cao B. et al. Cross-allele cytotoxic T lymphocyte responses against 2009 pandemic H1N1 influenza A virus among HLA-A24 and HLA-A3 supertype-positive individuals. J Virol. 2012;86:13281-94
60. Liu J, Wu P, Gao F, Qi J, Kawana-Tachikawa A, Xie J. et al. Novel immunodominant peptide presentation strategy: a featured HLA-A*2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J Virol. 2010;84:11849-57
61. Liu WJ, Lan J, Liu K, Deng Y, Yao Y, Wu S. et al. Protective T cell responses featured by concordant recognition of middle east respiratory syndrome coronavirus-derived CD8+ T Cell epitopes and host MHC. J Immunol. 2017;198:873-82
62. Wu D, Kolesnikov A, Yin R, Guest JD, Gowthaman R, Shmelev A. et al. Structural basis for recognition of two HLA-A2-restricted SARS-CoV-2 spike epitopes by public and private T cell receptors. Nat Commun. 2022;13(1):19
63. Dolton G, Rius C, Hasan MS, Wall A, Szomolay B, Behiry E. et al. Emergence of immune escape at dominant SARS-CoV-2 killer T cell epitope. Cell. 2022;185(16):2936-295164
64. Nguyen TH, Rowntree LC, Petersen J, Chua BY, Hensen L, Kedzierski L. et al. CD8+ T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity. 2021;54:1066-82 e5
65. Zhang H, Deng S, Ren L, Zheng P, Hu X, Jin T. et al. Profiling CD8(+) T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants. Cell Rep. 2021;36:109708
66. Szeto C, Nguyen AT, Lobos CA, Chatzileontiadou DSM, Jayasinghe D, Grant EJ. et al. Molecular basis of a dominant sars-cov-2 spike-derived epitope presented by HLA-A*02:01 recognised by a public TCR. Cells. 2021 10
67. Shomuradova AS, Vagida MS, Sheetikov SA, Zornikova KV, Kiryukhin D, Titov A. et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53:1245-57 e5
68. Kared H, Redd AD, Bloch EM, Bonny TS, Sumatoh H, Kairi F. et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J Clin Investig. 2021 131
69. Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and-B polymorphism. Immunogenetics. 1999;50:201-12
70. Ellis JM, Henson V, Slack R, Ng J, Hartzman RJ, Hurley CK. Frequencies of HLA-A2 alleles in five US population groups: Predominance of A∗ 02011 and identification of HLA-A∗ 0231. Hum Immunol. 2000;61:334-40
71. Islam F, Dhawan M, Nafady MH, Emran TB, Mitra S, Choudhary OP. et al. Understanding the omicron variant (B.1.1.529) of SARS-CoV-2: Mutational impacts, concerns, and the possible solutions. Ann Med Surg. 2022;78:103737
72. Schulien I, Kemming J, Oberhardt V, Wild K, Seidel LM, Killmer S. et al. characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat Med. 2021;27:78-85
73. Ferretti AP, Kula T, Wang Y, Nguyen DMV, Weinheimer A, Dunlap GS. et al. Unbiased screens show CD8(+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53:1095-107.e3
74. Habel JR, Nguyen THO, van de Sandt CE, Juno JA, Chaurasia P, Wragg K. et al. Suboptimal SARS-CoV-2-specific CD8(+) T cell response associated with the prominent HLA-A*02:01 phenotype. Proc Natl Acad Sci U S A. 2020;117:24384-91
75. Chaurasia P, Nguyen THO, Rowntree LC, Juno JA, Wheatley AK, Kent SJ. et al. Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein. J Biol Chem. 2021;297:101065 -
76. Liu WJ, Tan S, Zhao M, Quan C, Bi Y, Wu Y. et al. Cross-immunity against avian influenza A(H7N9) virus in the healthy population is affected by antigenicity-dependent substitutions. J Infect Dis. 2016;214:1937-46
77. Røder G, Kristensen O, Kastrup JS, Buus S, Gajhede M. Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:459-62
78. Ji W, Niu L, Peng W, Zhang Y, Cheng H, Gao F. et al. Salt bridge-forming residues positioned over viral peptides presented by MHC class I impacts T cell recognition in a binding-dependent manner. Mol Immunol. 2019;112:274-82
79. Blicher T, Kastrup JS, Buus S, Gajhede M. High-resolution structure of HLA-A*1101 in complex with SARS nucleocapsid peptide. Acta Crystallogr D Biol Crystallogr. 2005;61:1031-40
80. Liu J, Sun Y, Qi J, Chu F, Wu H, Gao F. et al. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J Infect Dis. 2010;202:1171-80
81. Liu J. et al. Functional and structural definition of a clustering region of hla-a2-restricted cytotoxic T lymphocyte epitopes. Sci Technol Soc. 2011;29(27):19-26
82. Zhao M, Liu K, Luo J, Tan S, Quan C, Zhang S. et al. Heterosubtypic protections against human-infecting avian influenza viruses correlate to biased cross-T cell responses. mBio. 2018 9
83. Ekeruche-Makinde J, Clement M, Cole DK, Edwards ESJ, Ladell K, Miles JJ. et al. T cell receptor-optimized peptide skewing of the T cell repertoire can enhance antigen targeting*. J Biol Chem. 2012;287:37269-81
84. Zaremba S, Barzaga E, Zhu M, Soares N, Tsang K-Y, Schlom J. Identification of an enhancer agonist cytotoxic T lymphocyte peptide from human carcinoembryonic antigen. Cancer Res. 1997;57:4570-7
85. Liu J, Zhang S, Tan S, Zheng B, Gao G. Revival of the identification of cytotoxic T-lymphocyte epitopes for immunological diagnosis, therapy and vaccine development. Exp Biol Med. (Maywood, NJ). 2011;236:253-67
86. Liu J, Dai L, Qi J, Gao F, Feng Y, Liu W. et al. Diverse peptide presentation of rhesus macaque major histocompatibility complex class I Mamu-A 02 revealed by two peptide complex structures and insights into immune escape of simian immunodeficiency virus. J Virol. 2011;85:7372-83
87. Yue C, Xiang W, Huang X, Sun Y, Xiao J, Liu K. et al. Mooring stone-like Arg(114) pulls diverse bulged peptides: first insight into African swine fever virus-derived T cell epitopes presented by swine MHC class I. J Virol. 2021: Jvi0137821.
88. Borbulevych OY, Baxter TK, Yu Z, Restifo NP, Baker BM. Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: implications for vaccine design. J Immunol (Baltimore, Md: 1950). 2005;174:4812-20
89. Gao GF, Liu WJ. Let's get vaccinated for both flu and COVID-19: on the world flu day 2021. China CDC Wkly. 2021;3:915-7
90. Liu WJ, Liu S. A tale of two cities: from influenza HxNy to SARS-CoV-z. China CDC Wkly. 2021;3:1052-6
91. Zhu S, Liu K, Chai Y, Wu Y, Lu D, Xiao W. et al. Divergent peptide presentations of HLA-A(*)30 alleles revealed by structures with pathogen peptides. Front Immunol. 2019;10:1709
92. Zhang S, Liu J, Cheng H, Tan S, Qi J, Yan J. et al. Structural basis of cross-allele presentation by HLA-A*0301 and HLA-A*1101 revealed by two HIV-derived peptide complexes. Mol Immunol. 2011;49:395-401
93. Nathan A, Rossin EJ, Kaseke C, Park RJ, Khatri A, Koundakjian D. et al. Structure-guided T cell vaccine design for SARS-CoV-2 variants and sarbecoviruses. Cell. 2021;184:4401-13.e10
94. Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201-12
95. Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1
Author contact
![]() Corresponding authors: William J. Liu, liujunchinacdc.cn; Kefang Liu, liukfac.cn.
Corresponding authors: William J. Liu, liujunchinacdc.cn; Kefang Liu, liukfac.cn.
Received 2022-11-3
Accepted 2023-7-7
Published 2023-8-6
