ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2023; 19(15):4778-4792. doi:10.7150/ijbs.83098 This issue Cite
Review
Novel insights from spatial transcriptome analysis in solid tumors
1. Department of Hematology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, 160 Pujiang Road, Shanghai, 200127, China.
2. School of Medicine, Shanghai Jiao Tong University, Shanghai, 200025, China.
3. Ganzhou Key Laboratory of Hematology, Department of Hematology, The First Affiliated Hospital of Gannan Medical University, Ganzhou, Jiangxi, 341000, China.
4. Ministry of Education Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China.
# These authors contributed equally to this work.
* These authors contributed equally to this work.
Abstract
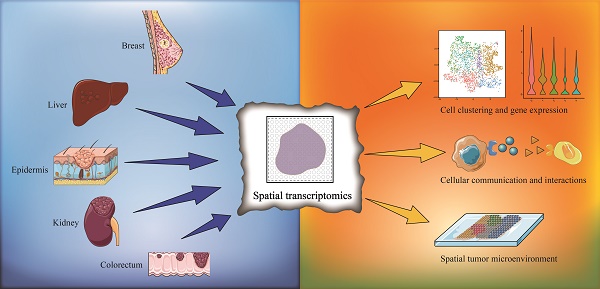
Since its first application in 2016, spatial transcriptomics has become a rapidly evolving technology in recent years. Spatial transcriptomics enables transcriptomic data to be acquired from intact tissue sections and provides spatial distribution information and remedies the disadvantage of single-cell RNA sequencing (scRNA-seq), whose data lack spatially resolved information. Presently, spatial transcriptomics has been widely applied to various tissue types, especially for the study of tumor heterogeneity. In this review, we provide a summary of the research progress in utilizing spatial transcriptomics to investigate tumor heterogeneity and the microenvironment with a focus on solid tumors. We summarize the research breakthroughs in various fields and perspectives due to the application of spatial transcriptomics, including cell clustering and interaction, cellular metabolism, gene expression, immune cell programs and combination with other techniques. As a combination of multiple transcriptomics, single-cell multiomics shows its superiority and validity in single-cell analysis. We also discuss the application prospect of single-cell multiomics, and we believe that with the progress of data integration from various transcriptomics, a multilayered subcellular landscape will be revealed.
Keywords: spatial transcriptomics, solid tumor, tumor heterogeneity, tumor microenvironment
