ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2024; 20(2):751-764. doi:10.7150/ijbs.83205 This issue Cite
Review
Molecular Modulators and Receptors of Selective Autophagy: Disease Implication and Identification Strategies
State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macau, China.
*These authors contributed equally to this paper.
Abstract
Autophagy is a highly conserved physiological process that maintains cellular homeostasis by recycling cellular contents. Selective autophagy is based on the specificity of cargo recognition and has been implicated in various human diseases, including neurodegenerative diseases and cancer. Selective autophagy receptors and modulators play key roles in this process. Identifying these receptors and modulators and their roles is critical for understanding the machinery and physiological function of selective autophagy and providing therapeutic value for diseases. Using modern researching tools and novel screening technologies, an increasing number of selective autophagy receptors and modulators have been identified. A variety of Strategies and approaches, including protein-protein interactions (PPIs)-based identification and genome-wide screening, have been used to identify selective autophagy receptors and modulators. Understanding the strengths and challenges of these approaches not only promotes the discovery of even more such receptors and modulators but also provides a useful reference for the identification of regulatory proteins or genes involved in other cellular mechanisms. In this review, we summarize the functions, disease association, and identification strategies of selective autophagy receptors and modulators.
Keywords: selective autophagy, autophagy, screening technology, protein-protein interaction, genome-wide screening
Introduction
Macroautophagy (hereafter autophagy) is a conserved degradation pathway in eukaryotes to help recycle cytoplasmic components to maintain cellular homeostasis [1]. The initiation of autophagy is usually subject to the mammalian target of rapamycin (mTOR) inhibition and AMP-activating protein kinase (AMPK) activation, which stimulate the Unc-51 like autophagy activating kinase 1 (ULK1) complex, including autophagy-related protein 13 (ATG13), focal adhesion kinase family interacting protein of 200 kD (FIP200) and autophagy-related protein 101 (ATG101), and facilitate the formation of phagophore fragments [2]. ULK1 then transduces autophagic signaling to activate the PI3KC3 complex (BECN1, class III PI 3-kinase (VPS34), PI 3-kinase regulatory subunit 4 (VPS15), autophagy-related protein 14, and nuclear receptor binding factor 2 (NRBF2)) for autophagosome nucleation [3]. The production of PI3P and the engagement of WD-repeat protein interacting with phosphoinositides (WIPI) induce the autophagosome expansion. Two ubiquitin-like conjugation modules were required to promote autophagosome formation. First, autophagy-related protein 7 (ATG7) and autophagy-related protein 10 (ATG10) are involved in the conjugation of autophagy-related protein 5 (ATG5), autophagy-related protein 12 (ATG12) and autophagy-related protein 16 (ATG16), which acts as E3 enzyme to stimulate conjugation of phosphatidylethanolamine (PE) to autophagy-related protein 8 (ATG8)-family. ATG7, autophagy-related protein 4 (ATG4) and autophagy-related protein 3 (ATG3) are then responsible for the cleavage of the ATG8-family membranes [4]. Lipidated ATG8 ultimately interacts with LC3-interacting region (LIR)-containing proteins, such as receptors and substrates, and proceeds to autophagosome maturation. Following phagophore closure, the autophagosomes fuse with the lysosomes, resulting in the degradation of autophagic substrates by acidic lysosomal hydrolases.
Selective autophagy is the selective form of autophagy, achieved through the specificity of cargo recognition [5]. There are various terms to describe diverse selective autophagy according to different autophagic substrates: mitophagy [6], ER-phagy [7], aggrephagy [8], lipophagy [9], xenophagy [10], nucleophagy [11], pexophagy [12], ferritinophagy [13], lysophagy [14] and fluidophagy [15]. Studies have revealed that selective autophagy is implicated in various human diseases such as cancers, neurodegenerative diseases, and infectious diseases [16-22]. For example, impaired mitophagy has been observed in Alzheimer's disease (AD) models with the downregulation of BCL2 interacting protein 3 (BNIP3), fun14 domain containing 1 (FUNDC1), and optineurin (OPTN) [16, 17]. Family with sequence 134, member B (FAM134B), an ER-phagy receptor, plays a tumor-suppressing or tumor-promoting role in different types of cancers, such as colon cancer, esophageal squamous cell carcinoma and pancreatic cancer [18-20]. Many selective autophagy stimulators exhibit potential therapeutic effects on diseases [23, 24]. Therefore, identification of novel receptors and modulators of selective autophagy is not only necessary for mechanistic studies of the selective autophagy process, but also important for understanding the pathogenesis of many diseases.
In selective autophagy, selective autophagy receptors (SARs) recognize specific cargoes and tether them to the autophagosomes by interacting with the ATG8/LC3 family [25]. Typically, an LC3 interacting region (LIR) and a cargo-binding site, such as Ub-binding domains (UBDs), are two necessary components in SARs. FAM134B, NIX, OPTN, TAX1BP1, CCT2, etc., are all well-known selective autophagy receptors with typical or atypical LIRs [26]. SARs directly affect the recognition and binding of cargos and autophagosomes. Unlike SARs, selective autophagy modulators usually do not directly recruit the autophagosomes but regulate the process of selective autophagy, such as WIPI2, PTEN-L and Parkin [27, 28]. Some of the soluble autophagy modulators tether to the substrates indirectly by recognizing polyubiquitin on the substrates, while parts of autophagy modulators bind to the substrate directly, accompanied with post-translational modifications (PTMs) [4, 29-32]. Selective autophagy modulators can also utilize other motifs, for example, ubiquitin-interacting motifs (UIM) bound to Atg8/LC3 proteins and ubiquitin-associated (UBA) domains [33]. Taken together, selective autophagy is a comprehensive regulatory process and understanding the modules of selective autophagy is the essential parts for mechanistic exploration of selective autophagy. And due to the structural properties of SARs, new SARs can be found by protein-protein interaction-based screening strategies, while by genome-wide screening both SARs and selective autophagy modulators can be found. Different screening strategies determine whether it is possible to discover these proteins. Our review summarizes screening strategies and technologies used to identify selective autophagy receptors and modulators, and we discuss the strengths and challenges of these strategies and technologies.
Selective autophagy modulators and receptors in human diseases
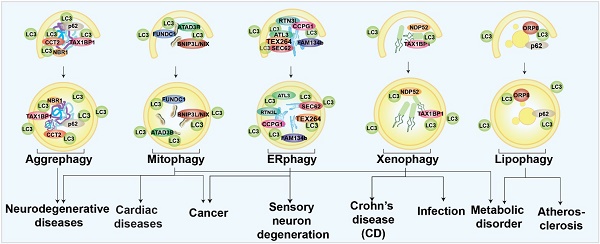
In selective autophagy, certain autophagy receptors target specific autophagic cargoes [34]. These receptors bind autophagy cargoes and ATG8s on the inner membrane surface of phagophore and drive the core autophagy process. Autophagy receptors can be ubiquitin-dependent or not. Ubiquitin-dependent selective receptors, such as sequestosome 1 (p62), Neighbor of BRCA1 gene 1 (NBR1), NDP52/CALCOCO2, Tax1 binding protein 1 (TAX1BP1), and OPTN, are the most well-studied [32, 35]. These receptors contain oligomerization domains, ATG8-binding LIR domains, and ubiquitin-binding domains that facilitate receptor recognition of substrates [36-39]. Among the ubiquitin-independent receptors [29], the glycophagy receptor starch binding domain 1 (STBD1) has a carbohydrate-binding domain for cargo identification. In contrast, the ferritinophagy receptor nuclear receptor coactivator 4 (NCOA4) binds directly to ferritin [38-40]. Understanding the roles of selective autophagy in human health and diseases can be inspired by the studies of its receptors and modulators. The different types of selective autophagy involved in different human diseases are briefly summarized in Fig. 1. Several forms of selective autophagy, such as lysophagy, pexophagy, fluidophagy, etc., are not mentioned here, due to the limited evidence to support the disease associations.
Schematic diagram of selective autophagy modulation in human diseases.
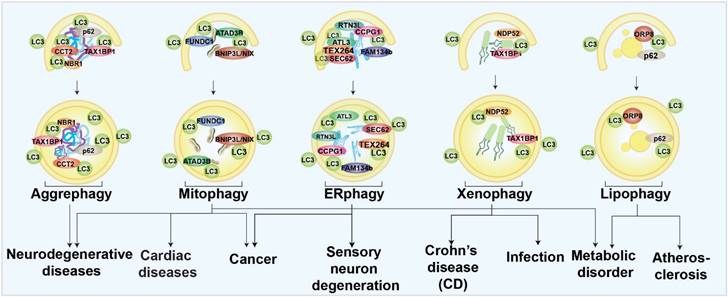
Aggrephagy
Aggrephagy is characterized by specific autophagic degradation of protein aggregates or inclusion bodies within the cell [41]. Studies found that targeting certain aggrephagy receptors was a potential treatment for neurodegenerative diseases due to the wide-spread recognition of misfolded protein aggregates as pathogenic factors [42]. For example, the lysosomal protein, TECPR1, has been found to facilitate aggrephagy and degrade chemical-induced or overexpressed huntingtin (HTT) protein aggregates neural cells [43]. Recently, chaperonin-containing TCP-1 subunit 2 (CCT2) has been identified as novel receptor for solid protein aggrephagy through a LC3-labeled HTT inclusion bodies purification strategy followed by proteomic determination [44]. Some potent pharmacological aggrephagy inducers have also been revealed and show neuroprotective capabilities in neurodegenerative diseases, such as PD180970 and XCT 790 [45-48].
Mitophagy
Mitochondria are specific organelles with their own genomes; they govern cellular energy supply [49]. Once mitochondria are damaged, mitophagy will be invoked and damaged mitochondria will be specifically eliminated to maintain cellular homeostasis. Mitophagy has been mostly studied especially in neurodegenerative diseases [50]. Mice lacking Parkin- and Pink-dependent mitophagy are found to have an accumulation of mutant mtDNAs, which may be a pathogenic factor for PD [51]. Mitophagy is also related to AD. In a recent work, our group used machine learning to identify two novel small molecules as mitophagy enhancers that have demonstrated the capacity to improve AD-associated pathologies [52]. Mitophagy has also been discussed extensively in cancer therapy due to the accumulation of reactive oxygen species (ROS) and mtDNAs caused by mitochondrial dysfunction. The elevated toxins, such as ROS, mtDNA, ATP, and N-formyl peptides from damaged mitochondria have been shown to increase genomic instability and accelerate the development of cancer [53]. Some mitophagy-related genes, such as BNIP3 and PARKIN, act as tumor suppressors [53, 54]. Mitophagy inhibition can be effective in cancer treatment if mitophagy is considered as an adaptive process. Mitophagy inhibitors, such as liensinine and MDIVI-1, have been found to enhance the sensitivity of tumor cells to chemotherapy [55, 56]. In the aging heart, mitophagy has been regarded as the center of rejuvenation [57]. Mitophagy defects exacerbated cardiac injury and reduced survival following myocardial infarction in one in vivo study [58]. Meanwhile, mitophagy-associated proteins, such as FUNDC1 and RAB9, have been proved to be protective in heart ischemia models [59, 60]. Mitophagy can also trigger muscle-adipose crosstalk to alleviate dietary obesity [61]. Mitochondria are tightly coupled with cellular metabolism, and mitophagy has emerged as a player in metabolic diseases. Studies have found that mitophagy in adipose tissue increases cellular lipid metabolic processing capacity. The diabetes susceptibility gene CLEC16A is involved in mitophagy regulation in β cells [62-64]. In summary, regulation of mitophagy might be a promising strategy for many diseases.
Endoplasmic reticulum (ER)-phagy
Endoplasmic reticulum (ER)-phagy is an autophagic process to degrade specific parts of the ER for ER quality control and function maintenance [65]. By analyzing ER-phagy-related proteins, researchers determined that ER-phagy is closely associated with a series of neurological disorders and cancers. In mammalian cells, FAM134B, Reticulon-3 (RTN3L), SEC62, cell cycle progression 1 (CCPG1), atlastin GTPases 1 (ATL1), and testes expressed gene 264 (TEX264) are well-known ER-phagy receptors [65]. Most of the ER-phagy-related genes were found to function in the neurological system. Mutations of ATL1 have been found to be a cause of hereditary sensory neuropathy type 1 [66]. FAM134B is necessary for long-term survival of nociceptive and autonomic ganglion neurons [67]. RTN3 is involved in the etiology of AD [68]. In summary, ER-phagy appears to be closely linked to neurological function, especially regulation of sensory neurons. Dexmedetomidine, a sedative, has been proved to induce ER-phagy thereby alleviating ER stress in a spinal cord neuropathic pain model [24]. Cancers are also linked to ER-phagy. Studies have found that Sec62 is a potential oncogene in non-small cell lung cancer [69], and is proposed as a prognostic marker in advanced head and neck squamous cell carcinoma [70]. Moreover, novel FAM134B (JK1) mutations have been identified in oesophageal squamous cell carcinoma [19]. Although many ER-phagy regulators have been found to be involved in several diseases, exactly how ER-phagy is related to these diseases, in terms of mechanism, is not understood.
Xenophagy
Xenophagy, which targets bacterial pathogens for lysosomal degradation, has been mostly linked to infectious diseases as a host defensive mechanism [10, 71]. Xenophagy is closely related with Crohn's disease (CD), a kind of inflammatory bowel disease type. Numerous proteins, including ATG16L1, immunity-related GTPase M (IRGM), and nucleotide-binding oligomerization domain 2 (NOD2), have been linked to the development of CD [72-75]. Patients with severe Covid-19 have also shown signs of virus-induced xenophagy [76]. It has been demonstrated that increased xenophagy helps to protect the host against infection. For example, hypoxia-inducible factor (HIF-1) is a mediator to activate xenophagy and limit salmonella infection and spread [21]. Resveratrol also activates xenophagy and promotes intracellular bacteria clearance in intestinal epithelial cells and macrophages [77].
Lipophagy
The selective autophagy that targets lipid droplets is called lipophagy [9]. Since metabolic disorders frequently involve lipid droplet build-up, controlling droplet degradation has been proposed to alleviate lipid stresses [78-81]. Lipophagy is thought to regulate the breakdown of lipid droplets in the liver, and much research have examined the functions of lipophagy in regulating the progression of non-alcoholic fatty liver disease (NAFLD) [78, 82, 83]. Elevating lipophagy alleviates NAFLD. For example, iridoids and phillygenin have been identified as lipophagy enhancers and have been found to ameliorate NAFLD and hepatic steatosis [82, 83]. However, lack of hepatic autophagy in mice was resistant to physiological steatosis due to activation of nuclear factor erythroid 2-related factor 2 (NRF2) and maintenance of nuclear receptor corepressor 1 (NCoR1) [84-86], indicating that compensatory regulations may be upregulated in the organistic system, especially inhibition of fasting-induced de novo lipogenesis in the liver. Lipophagy is also critical to maintaining the lipid homeostasis of vascular endothelial cells, vascular smooth muscle cells, and macrophages. As lipid accumulation in these cells is closely related to atherosclerosis, induction of lipophagy may have therapeutic value for atherosclerosis [80]. Multiple sclerosis and diabetic nephropathy are also linked to lipophagy. Thus, targeting lipophagy shows extensive potential to regulate wide range of serious diseases [79, 87].
Screening for selective autophagy receptors and modulators
As targeting selective autophagy is promising as a strategy to treat diseases, it is important to understand disease pathogenesis in terms of autophagy receptors and modulators. Screening autophagy receptors and modulators using various modern technologies is an efficient methodology in this regard. We will briefly summarize and discuss emerging screening methods in sections below.
In selective autophagy receptor and modulator identification, the first step is usually establishment of robust reporters to monitor selective autophagy. For example, GFP-mcherry-LC3, mito-keima, and mcherry-eGFP-RAMP4 are all reporter systems for autophagy and selective autophagy determination, which have been successfully used for modulator screening [88-90]. Other fluorescent reporters which have not applied in autophagy regulator screening are also expected to be used, such as Ribo-Keima reporter [91]. The second step is to establish specific selective autophagy induction conditions [92, 93]. The third, last step is to use target-based, or phenotype-based approaches [38, 94, 95]. Selective autophagy receptors and modulators can also be identified by isolating subcellular organelles or components from cells with different levels of autophagy, such as lipid droplets (lipophagy) [94] and inclusion bodies (aggrephagy) [44].
Schematic representation of strategy for screening selective autophagy receptors and modulators.
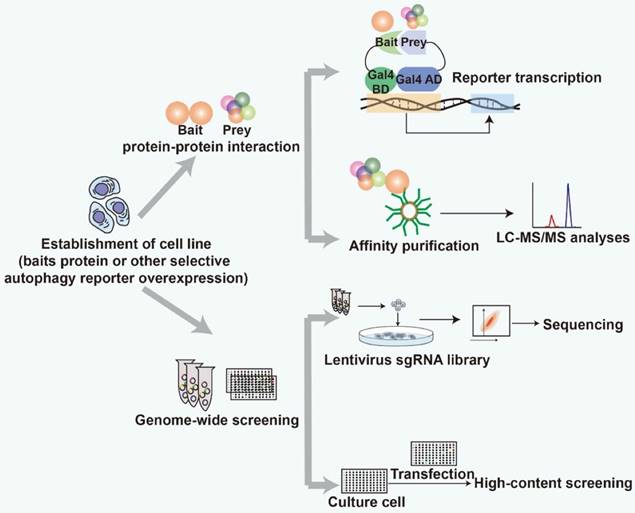
Strategies based on protein-protein interactions (PPIs) and genome-wide screening have been used to find specific selective autophagy receptors and modulators (Fig. 2). Both strategies require robust detection methods to screen for the receptors and modulators. The identified proteins can be defined as selective autophagy receptors and modulators after further experimental validation, such as co-immunoprecipitation, fluorescence co-localization and functional characterization. Transmission election microscopy (TEM) is a gold standard to assess the status of autophagy. Recently, it has been successfully applied to analyze peripheral autophagy markers associated with PD [96]. TEM is recommended to be used to evaluate the efficacy of the screened receptors and modulators and investigate their functional implications for autophagy.
Approaches based on protein-protein interactions (PPIs)
Autophagic related proteins form functional complexes to achieve the fine regulation of the autophagy process. For example, ULK1-FIP200-ATG13, ATG14L-BECN1-VPS34-VPS15, and ATG5-ATG12-ATG16 are three protein complexes involved in autophagy initiation, autophagosome elongation, and maturation, respectively. In selective autophagy, receptor-cargo recognition is a critical step to obtain specificity [25]. Based on the recognition modules of selective autophagy receptors, an efficient strategy for objectively identifying novel selective autophagy receptors, is bait-based identification of interacting proteins. Atg8/LC3 family proteins are widely utilized as bait [43, 44, 93, 97-104]. The bait-interacting proteins or proteins with LIR motifs are candidate receptors for further validation [105]. In recent years, strategies based on PPIs are widely used to identify selective autophagy regulators [94, 101, 105-107]. Three strategies will be described below: computational LIR prediction, yeast two-hybrid (Y2H) screening, and mass spectrometry-based proteomic analysis.
Computational LIR prediction
Atg8-family proteins play a central role in mediating selective autophagy [105]. Some of the well-known selective autophagy receptors, such as p62, NBR1 and BNIP3, achieve autophagosome targeting by binding with LC3 at its interacting region (LIR) [32]. Jacomin, et al., have developed a free database to predict new LIR-containing proteins (LIRCPs) using a computational approach [105, 108]. Many regulators have been successfully identified using this method. In 2019, Yifan Zhang, et al., revealed that Listeria monocytogenes can induce mitophagy independent of known mitophagy receptors (Nix, BNIP3 and FUNDC1) [109]. Applying an iLIR web server, they successfully screened and identified a novel mitophagy regulator, NLR family member X1 (NLRX1), which contains an LIR motif in the pattern-recognition receptors (PPRs). Computational prediction is an easy approach to screening for receptors that contain the LIR motif. However, it's not able to predict the noncanonical LIR motif-containing receptor, such as CALCOCO2/NDP52 [110] and additional procedures required to be done to get convincing results.
LIR is a short linear motif (SLiM) present in many proteins, including those that do not interact with LC3 [111]. To eliminate some spurious LC3-binding proteins, intrinsically disordered regions are required to be identified. The Disprot-Database of Protein Disorder (http://www.disprot.org/pondr-fit.php) is a free website for confirming disordered regions. It's better to exclude disordered regions on Disprot before prediction. In the cases without typical LIR, and updated prediction based on the artificial intelligence (AI) system, named AlphaFold2-multimer, can be used [112]. This newly established AI system has successfully predicted some atypical LIR motifs, such as ILVV in NDP52, MLVV in TAX1BP1, and YDFM in ATG40. Post-translational modification (PTM) is an important manner to regulate LIR-binding efficiency. Phosphorylating of S177 adjacent to the LIR of OPTN and S332 in the LIR motif of p62 may regulate the LC3 binding efficiency [113, 114]. In addition, acetylation of Lys49 on the LIR has been found to disrupt the LC3 binding [115]. Therefore, the prediction of the PTM domain in LIR could be an easy way to find out the potential PTM-related regulatory pathway. There are several online databases and related tools for PTM prediction. For example, DAPPLE (http://saphire.usask.ca/saphire/dapple2) focuses on phosphorylation modification, dbPTM (http://dbPTM.mbc.nctu.edu.tw/) can predict the modifications of phosphorylation, glycosylation and sulfation, and PTM-ssMP (http://bioinformatics.ustc.edu.cn/PTM-ssMP/index/) that can predict different types of modifications. All these prediction servers can be useful tools for screening and greatly assist in the identification of PTM-associated regulators in selective autophagy.
Yeast two-hybrid (Y2H) screening
The yeast two-hybrid (Y2H) system is a well-established genetics-based system to screen for bait-binding proteins from a large-scale cDNA library [116]. In selective autophagy study, bait proteins are fused with the DNA-binding domain (BD) of Gal4. The Gal4 activation domain is fused to prey. Reporter gene expression indicates successful interaction of bait proteins and prey proteins. Atg8 family proteins mostly used bait [117-120]. A successful example was the identification of ER-phagy receptor, FAM134[98]. In this case, LC3B and GATE16/GABARAPL2 are bait. After screening, FAM134 family members were identified [98]. FKBP prolyl isomerase 8 (FKBP8) was also identified as a mitophagy regulator in a human thymus cDNA library screening using LC3B as bait [100]. Proteins other than those in the LC3 family were also designed as bait for screening selective autophagy regulators [121-123]. ZZ and LB domains of p62 were used as bait in yeast two-hybrid screening. The prefoldin-like chaperone ubiquitously expressed transcript protein (UXT) was identified as an aggrephagy adaptor that binds to the LB domain of p62 [123]. In a xenophagy study, NLR family pyrin domain containing 4 (NLRP4) was used as bait, and ARHGDIA was finally identified as a regulator protein during Group A Streptococcus (GAS) infection [124]. Compared to the in vitro approaches based on bacterial expression, Y2H is a more advanced in vivo technique using yeast as a host cell [125]. However, not all modification-driving interactions can be observed by using this method. The construction of cDNA libraries is challenging, and the screening process is time-consuming and labor-intensive.
Mass spectrometry (MS)-based proteomic analysis
MS-based proteomic analysis is a powerful approach that has been widely used in the identification of receptors and modulators that bind with bait proteins. Proteomics analysis unbiasedly provides plenty of protein information by its large-scale protein screening capacity and flexible application with different labeling methods [25]. Another advantage is that proteomics can read the protein composition comprehensively without utilizing conventional fluorescence labelling-based autophagy assays [126].
Affinity purification coupled with MS-proteomics
Affinity purification is widely used for selective autophagy study. In selective autophagy study, Atg8 families (LC3s), selective autophagy receptors, or selective autophagy substrates are usually chosen as bait proteins for affinity purification [93, 106, 127]. Positive hits are analyzed and subjected to secondary screening to sort out those potentially involved in selective autophagy.
In most cases, ATG8 families are widely used as bait proteins for screening selective autophagy regulators. By expressing YFP-FLAG-His6 (YFH)-tagged ATG8 in Schizosaccharomyces pombe, two research groups performed affinity-purification coupled with mass spectrometry (AP-MS) analysis and then identified an ER-phagy receptor, Mug185 [93, 128]. Using ATG8 as a bait in Saccharomyces cerevisiae, autophagy-related protein 39 (ATG39) and autophagy-related protein 40 (ATG40) were identified as receptors for nucleophagy and ER-phagy, respectively, after mass spectrometry analysis of ATG8 immunoprecipitates [106]. In plants, researchers have used tunicamycin to trigger stress-induced-ER-phagy and immunoprecipitation-MS analysis to obtain the GFP-ATG8A binding proteins. In this work, cytosolic protein C53 was identified as an ER-phagy regulator [99]. To identify PolyQ-huntingtin (HTT) inclusion bodies (IBs)-associated aggrephagy regulator, IBs with high (H) and low (L) LC3 recruitment were isolated by a fluorescence-activated particle sorting (FAPS) system, which can enhance the accuracy of screening. Then by mass spectrometry, CCT2 was identified as a novel aggrephagy regulator that interacted with ATG8s through a VLIR motif [44].
Besides the Atg8 family, autophagy receptors and substrates have also been applied for IP-MS analysis. To identify novel regulators associated with p62 or autophagy-linked FYVE protein (ALFY), one study compared the immunoprecipitated proteins from ALFY-/- vs WT cells, or from EGFP-p62 over-expressing cells vs EGFP over-expressing cells. The study finally identified two proteins, nipsnap homolog 1 (NIPSNAP1) and nipsnap homolog 2 (NIPSNAP2, that selectively interacted with ALFY and p62 and were further confirmed as mitophagy regulators [129]. Another study performed co-immunoprecipitation (Co-IP) to obtain the zinc finger FYVE-type containing 1 (ZFYVE1) or WIPI1-associated proteins after inducing mitophagy. In this study, breast carcinoma amplified sequence 3 (BCAS3) and chromosome 16 open reading frame 70 (C16orf70) were identified as selective and non-selective autophagy regulators, respectively [127].
More specifically, mutation in the binding domain of LC3B has been established for identification of interacting proteins. One study used K51A mutant LC3B to screen for novel LC3B-interacting autophagy regulators [101]. Researchers identified TEX264 as having a high binding score by analyzing the IP-MS results using LC3B WT and the LC3B K51A mutant proteins as bait. Subsequently, TEX264 was determined as being an ER-phagy receptor by its cellular localization and by other confirmation experiments.
The Immunoprecipitation-Mass Spectrometry (IP-MS) may also identify unexpected receptors and modulators. For example, to figure out the new function of mitochondrial fission protein 1 (Fis1), flag-tagged Fis1-expressing Hela cells were used to identify proteins that it interacted with [90]. Syntaxin 17 (STX17), a soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) protein, was identified as a novel regulator interacting with Fis1. Functional study showed that Fis1 restrained the localization of STX17 onto mitochondria and ER-mitochondria contact sites to prevent STX17-mediated mitophagy. Ribophagy receptor, nuclear fragile X mental retardation-interacting protein 1 (NUPFIP1), was identified after analyzing the mTORC1-regulated lysosomal proteome [130]. Transmembrane protein 192 (TMEM192) is a lysosomal/endosomal protein, which has been used as a marker to target and pull-down lysosomal proteins in this study. Researchers utilized 3-HA-taged TMEM192 or 2xFLAG-tagged to perform IP after triggering autophagy by starving or treating with Torin 1, then, from the proteomic results, defined the mTORC1 dependent-lysosomal proteins. At last, they confirmed nuclear fragile X mental retardation-interacting protein 1 (NUFIP1) as a ribophagy receptor that binds to LC3B to regulate ribosomal degradation.
MS-proteomic analysis with advanced labeling methods
Despite being a powerful tool in biological study, conventional proteomic analysis is not highly quantitative [131]. Additionally, weak, or temporary binding partners are difficult to detect based on original MS conditions. To solve these problems, stable isotopes are incorporated into proteins or peptides to achieve accurate and sensitive quantification. By labeling proteins or peptides in a specific group, the stable isotope labeling by amino acids in cell culture (SILAC) approach allows researchers to perform highly quantitative analysis of proteomic changes in two or more different groups. Moreover, the novel proximity labeling-MS method can label “prey” proteins near the bait and detect transient and weak binding partners [132].
SILAC is a commonly used approach for quantitative analysis [133]. Metabolic labeling allows analyzation of cell mixtures from two different groups as a single sample, which greatly reduces the variation of quantitative results due to subsequent experimental procedures and can be used to detect low-level protein alteration. This method can provide condition-dependent quantitative information. A classic example was the identification of NCOA4 as the cargo receptor mediating ferritinophagy [38]. In this study, PANC-1 and PA-TU-8988T cell lines and MCF7 cell line with different autophagy levels were studied. Autophagosomes were purified by density gradient separation and subjected to MS analysis. NCOA4 was identified as ferritinophagy receptor based on candidate overlap. Interestingly, later analysis of MS data from this research identified CALCOCO1 as an ER-phagy receptor [134]. To identify autophagy dependent ATG9A-interacted proteins, stable isotope labeling by amino acids (SILAC) was applied to nutrient-rich and amino acid-starved ATG9A-overexpressing HEK293 cells. MS-based proteomic analysis of immuno-isolated ATG9A-positive membrane proteins revealed that BAR domain-containing proteins were associated with ATG9A vesicles [135].
Proximity labeling- MS is an effective approach to identify regulatory proteins with weak transient interactions that can be used to bait proteins. Diverse enzymes, such as biotin ligases, PTM ligases, and peroxidases, are utilized for labeling neighboring proteins [136, 137]. Peroxidases label proteins by converting biotin phenol to a biotin-phenoxyl radical in presence of H2O2 [25]. MS coupled with proximity-dependent biotin identification (BioID) has been used to study known and novel protein-protein high-confidence proximity interactions in the autophagy pathway. For example, a tandem MS was performed among BioID bait proteins within the autophagy pathway before streptavidin purification. Subsequently, spermine binding protein-like (SBPL) family proteins were identified as Atg8-family interacting proteins in selective autophagy [138]. In a study, co-expressing HA-tagged hATG8s and BioID-tectonin beta-propeller repeat containing 1 (TECPR1) has been used to analyze biotinylation of hATG8s in cell lysates. Finally, researchers found that LC3C was strongly biotinylated and TECPR1 selectively interacted with LC3C to facilitate autophagosome-lysosome fusion to promote aggrephagy [43]. Proximity labeling-mass spectrometry provides an abundance of protein interaction information. In Wei, L. et al.'s study, APEX2 was used as a bait to identify potential cargos of SQSTM1-like receptors (SLR) in autophagosomes. In this study, a panel of Hela cell lines overexpressing NBR1, NDP52, OPTN, p62, TAX1BP1, and toll interacting protein (TOLLIP) that fused with apurinic endodeoxyribonuclease 2 (APEX2) was established. These APEX2 overexpressing cells were treated with BafA1 prior to H2O2. TMEM106B is an endosome/lysosome marker, and the authors first established a TMEM106B-APEX2 over-expressing cell line to optimize the vesicle content profiling and then used APEX2-LC3B-expressing cells as an autophagy control. Above 200 proteins were filtered and identified as proximity partners of SLRs and LC3B [139].
SILAC and proximity labeling has been utilized together to distinguish enrichment proteins in specific conditions. Using SILAC-labeled AP2-hATG8s-overexpressing cells in the absence and presence of bafilomycin A1 (BafA1) prior to addition of H2O2 can enrich the biotinylated proteins in autophagosomes. The mitochondrial protein metaxin 1 (MTX1) has been identified as an autophagosome cargo in mitophagy [97]. MS is not only widely used in identifying protein-protein interactions, but also applicable for cell proteomics, organelle proteomics and post-translational modifications detection to achieve unbiased screening of selective autophagy regulators. S. Robichaud et al. isolated lipid droplets (LDs) from macrophage foam cells under basal conditions or upon chloroquine treatment and then performed MS. By comparing the proteomes of chloroquine-treated LDs and untreated LDs, they identified several lipophagy regulators, most of which contain LIR motifs [94]. To isolate mono-Ub modified proteins in the identification of ubiquitylation-modified regulators upon dengue infection, J. S. Zhang et al. utilized purified HA-Ub-L73P∗. Among 40 candidates, a lipid droplet protein, ancient ubiquitous protein 1 (AUP1), was identified as a lipophagy regulator [140].
PPIs provide important information for understanding selective autophagy machinery. The general approach is to design autophagy or selective autophagy-related protein bait, and then to screen and identify bait-associated proteins in different conditions. MS-based proteomic analysis is the most widely used approach for protein identification. Using tag-associated “bait” protein overexpressing cells can enrich “prey” proteins for further MS-based protein identification. The approach can also be combined with other advanced labeling techniques for more informative analysis. However, how to obtain high-confident candidates from large amounts of data is a challenge. In addition, MS is a necessary technique for PPIs, but not all proteins can be covered by MS, which can be a major limitation in application.
Genome-wide selective autophagy screening
Another effective and objective method for screening selective autophagy receptors and modulators is the functional genomics screening approach, which is an unbiased strategy to identify genes or genetic elements related to phenotype of interest [141, 142]. High-throughput loss-of-function screening and identification of selective autophagy receptors and modulators based on cellular phenotypes has been well established in recent years. Genome-wide RNAi and yeast library screening are two methods dependent on a large library data pool. In recent years, CRISPR-Cas9-based screening has rapidly developed, and this method has been widely used in selective autophagy modulator screening [22, 103, 143, 144].
RNAi-based screening
RNAi-based screening is usually performed in an arrayed or pooled format. By combining high-content screening or high through-put screening platforms, it can efficiently identify regulator proteins [102, 145-147]. Mandell et al. transfected mRFP-GFP-LC3B Hela cells with SMARTpool siRNAs to screen functional regulators of autophagy according to the RFP and GFP puncta. TRIM5α was identified as a selective autophagy receptor of HIV-1 capsid in this study [146]. TRIM has also been identified as a selective autophagy receptor for inflammatory proteins in another similar study [102]. In this study, TRIM family was silenced using SMARTpool siRNA in THP-1 cells. Image-based high-content analysis of LC3 puncta revealed TRIM family proteins were involved in IFN-γ induced-autophagy. They found that tripartite motif-containing (TRIM) family proteins can selectively target key components for autophagic degradation in inflammasome and type I interferon responses [102]. A directed siRNA screening of 67 lipid kinase and phosphatase genes was performed to identify the lipid kinases and phosphatases regulating xenophagy. By evaluating the changes of host bacterial defense and further experiments, SAC1 was identified as a xenophagy regulator [148]. However, RNAi-based screening is limited by the relative high cost of siRNA library, and the excessive amounts of time and labor require.
CRISPR-Cas9-based screening
As CRISPR-Cas9-based genome editing methods have developed, the CRISPR-Cas 9 system has been quickly adapted for functional genomic screening. Compared with RNAi pool screening, the CRISPR-Cas9 technique is more efficient for genome screening, and it can deliver quantitative results [149]. Liang JR et al. have established an ER-phagy tandem reporter (EATR) system, and performed genome-wide flow cytometry-based CRISPRi screening for ER-phagy regulatory genes [144]. The screening successfully identified mitochondrial metabolism and ER-resident UFMylation related genes involved in ER-phagy [89]. This approach can analyze the ER-phagy-regulatory pathway and modifications systemically and without bias.
By screening IFN-β-EGFP reporter cell lines with CRISPRi, coiled-coil domain-containing protein 50 (CCDC50) was identified as a negative regulator for IFN-β expression [22]. Further evidence proved that CCDC50 was a selective autophagy receptor participating in retinoic acid-inducible gene 1 (RIG-I)/MDA5 degradation during viral infection. To identify xenophagy regulatory factors, Yue Xu et al. performed CRISPRi screening of FIP200-/- GFP-LC3 Hela cells expressing LAMP1-mCherry [103]. FIP200 is a canonical autophagic protein but is dispensable in bacteria-induced autophagy. FIP200 deficiency cell type is a good sorting cell type to avoid basal autophagy flux interference, but not bother xenophagy formation. The xenophagy-positive cells were sorted after BFP-labeled bacterial infection, and V-ATPase was identified as a xenophagy regulator.
Genome-wide yeast library screening
The genome-wide yeast library has also been used for screening selective autophagy regulators. To investigate ER-phagy regulators, yeasts were transformed with Sec61-mCherry or Rtn1-GFP plasmids and positive clones were identified based on imaging analysis when the fusion of mCherry and vacuoles was impaired after rapamycin stimulation. GFP-Atg8 was used as a control to assess whether these mutants lead to defects in bulk autophagy. This study identified Vps13 as one of the regulators of ER-phagy. Further mechanistic studies showed that Vps13 mutant cells exhibited a 70% reduction in the packaging of the ER into autophagosomes but didn't alter the number of autophagosomes [104]. In another study, authors used ER-shaping mutant yeasts to identify whether ER-shaping proteins play a role in ER-phagy. ER-phagy receptor Sec61-GFP was utilized. And after treating cells with rapamycin (mutant clones failed to increase GFP puncta), Lnp1 was finally identified based on imaging analysis. Further evidence proved that Lnp1 was required to stabilize the actin-dependent ER remodeling in ER-phagy [150]. Using ATG gene knock-out yeast library, autophagy-related protein 24 (ATG24)/SNX4 was identified to be required for selective autophagic degradation of proteasomes [151].
Other forward genetic screening studies have been used for selective autophagy regulator screening and identification [152-154]. Screening of a Drosophila eye lethal mutant library revealed that Tre-2/Bub2/Cdc16 (TBC) protein dTBC1D22 modulated Rab40 to regulate lipophagy [81].
Comparison of different screening approaches.
| Forms of selective autophagy | Methods description | Hits | Verified receptors and modulators | Reference | |
|---|---|---|---|---|---|
| Bait/Target | Screening method | ||||
| ER-phagy | ATG8 | MS-proteomic | >400 | Epr1 | [93] |
| Sec61, Rtn1 and ATG8 | Genetic Screening | 21 | Vps13 | [104] | |
| Sec61 | Genetic Screening | / | Lnp1 | [150] | |
| LC3B and GABARAPL2 | Y2H Screening | / | FAM134 | [98] | |
| ATG8 | MS-proteomic | / | C53 | [99] | |
| RAMP4 and LC3B | CRISPRi Screening | 200 | RPN1, etc. | [89] | |
| LC3B | MS-proteomic | 87 | TEX264 | [101] | |
| ALFY and p62 | MS-proteomic | >11 | NIPSNAP1 and NIPSNAP2 | [129] | |
| Mitophagy | Fis1 | MS-proteomic | / | STX17 and Fis1 | [90] |
| / | LIR prediction | / | NLRX1 | [109] | |
| ATG8 | Y2H Screening | / | FKBP8 | [100] | |
| PRKN, ZFYVE1 and WIPI1 | MS-proteomic | 35 | BCAS3 and C16orf70 | [127] | |
| Om45 | Genetic Screening | / | YIL146C, ECM37, and ATG32 | [155] | |
| Clec16a | MS-proteomic | 800 | Nrdp1 | [64] | |
| ATG8s | MS-proteomic | 1147 | MTX1, LC3C | [97] | |
| OPTN | CRISPRi Screening | 273 | VDAC2, TOMM20, etc. | [156] | |
| PARK2 | CRISPRi Screening | >50 | PARK2, PINK1, aTG14, etc. | [156] | |
| Aggrephagy | NBR1, NDP52, OPTN, p62, TAX1BP1, and TOLLIP | MS-proteomic | 279 | autophagy substrates | [139] |
| p62 | Y2H Screen | 12 | UXT | [123] | |
| LC3 | MS-proteomic | >30 | CCT2 | [44] | |
| Xenophagy | LAMP1 and LC3 | Genetic Screening | 1 | SopF, etc. | [103] |
| Lipophagy | / | MS-proteomic | 91 | More than 10 | [94] |
| / | MS-proteomic | 40 | AUP1 | [140] | |
| Ribophagy | mTORC1 | MS-proteomic | 343 | NUFIP1 | [130] |
| IFN-γ induced autophagy | LC3 | RNAi Screening | 24 | 24 different TRIMs | [102] |
| Selective autophagy degradation for RIG-I/MD54 | IFN-β | CRISPRi Screening | 1000 | CCDC50 | [22] |
The RNAi technique and yeast mutant library which are usually handled as pre-setting array formats limit researchers to choosing detection methods, such as high-content imaging analysis with pooled population [102, 104, 150]. In comparison with these two approaches, the CRISPR-Cas9-based technique is more flexible in that cell phenotype detection methods can be chosen without format limitation. However, dependence on flow cytometry-based fluorescence detection restricts the selection of models [22, 103, 144].
Genome-wide screening is undoubtedly a powerful tool to screen for selective autophagy-related genes. It can screen out genes that are associated with changes in selective autophagy. However, the candidate genes may not encode receptors and modulators in selective autophagy, and further biological confirmation is required for refinement. PPI may be a better way to identify specific selective autophagy receptors or modulators, but low abundance proteins may be missed due to the limitation of the technique. Combination of multiple screening strategies may enhance the target identification efficiency and facilitate off-targets deconvolution process.
Summary and future perspectives
Identification of selective autophagy receptors and modulators is critical for understanding the selective autophagy machinery and regulatory network. Our review summarizes current screening strategies and technologies applied in the discovery of selective autophagy receptors and modulators and discusses the advantages and disadvantages of these technologies. It is a resource for the selection of strategy and technology to be used in future studies. However, most of the screening models used to identify interaction partners involve over-expression of selective autophagy markers. We foresee that, in the future, label-free technologies, like cellular thermal shift assay (CESTA), will be developed [157, 158].
An increasing number of tools to monitor selective autophagy have been developed. Before applying these new tools in screening, it is important to confirm the reliability and sensitivity of the tools. For example, ER-phagy probe GFP-mCherry-RAMP4 indicates a higher level of ER-phagy when there are more red-only puncta. However, flow cytometry results indicate that the ratio of red to green puncta must be used, rather than the number of red puncta. Fluorescent images provide more dimensional information regarding the distribution pattern of interested probes, but efficient quantification from a powerful analytic system is required for screening. Fortunately, the latest high-content imaging systems are equipped with powerful image analysis tools that can perform quantification analysis. Recently, artificial intelligence (AI) technology has also been successfully applied in image analysis, allowing more efficient and accurate image quantification [159, 160]. Although an increasing number of selective autophagy small molecule compounds have been identified, most of them fail to precisely regulate a specific form of selective autophagy without affecting other forms of autophagy. For example, carbonyl cyanide m-chlorophenyl-hydrazone (CCCP), the most commonly used mitophagy inducer, induces both mitophagy and bulk autophagy [161]. The limitation in selectivity results in misleading data that confuses the screening analysis. In future studies, more effort focused on the identification of highly selective and druggable receptors and modulators, including kinases and GPCRs, for selective autophagy regulation will enable us to achieve the goal of efficient modulation of selective autophagy under physiological and pathological conditions.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 82271455), Science and Technology Development Fund, Macau SAR (No. 0025/2022/A1, 005/2023/SKL, 0128/2019/A3), Shenzhen Fundamental Research Program (No. SGDX20210823103804030), Guangdong Basic and Applied Basic Research Foundation (No. 2022A1515012416) and the University of Macau grants (No. MYRG2022-00094-ICMS) awarded to Jia-Hong Lu. We thank Martha Dahlen for her English editing of this manuscript.
Author Contributions
Conceptualization, J.L. and M.W.; writing the original draft preparation, visualization, M.W. and Z.L.; writing the review and editing, funding acquisition, supervision, J.L.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Choi AMK, Ryter SW, Levine B. Autophagy in Human Health and Disease. New Engl J Med. 2013;368:1845-6
2. Nishimura T, Mizushima N. The ULK complex initiates autophagosome formation at phosphatidylinositol synthase-enriched ER subdomains. Autophagy. 2017;13:1795-6
3. Nishimura T, Tooze SA. Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 2020;6:32
4. Lamark T, Johansen T. Mechanisms of Selective Autophagy. Annu Rev Cell Dev Biol. 2021;37:143-69
5. Jin MY, Liu X, Klionsky DJ. SnapShot: Selective Autophagy. Cell. 2013;152:368-U400
6. Lemasters JJ. Perspective - Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuv Res. 2005;8:3-5
7. Bernales S, Schuck S, Walter P. Selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285-7
8. Overbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007;3:300-22
9. Singh R, Kaushik S, Wang YJ, Xiang YQ, Novak I, Komatsu M. et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131-U64
10. Levine B. Eating oneself and uninvited guests: Autophagy-related pathways in cellular defense. Cell. 2005;120:159-62
11. Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonakal I. et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5:795-804
12. Dunn WA, Cregg JM, Kiel JAKW, van der Klei IJ, Oku M, Sakai Y. et al. Pexophagy - The selective autophagy of peroxisomes. Autophagy. 2005;1:75-83
13. Young ARJ, Chan EYW, Hu XW, Koch R, Crawshaw SG, High S. et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888-900
14. Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun. 2013;4:2111
15. Agudo-Canalejo J, Schultz SW, Chino H, Migliano SM, Saito C, Koyama-Honda I. et al. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature. 2021;591:142-6
16. Martin-Maestro P, Gargini R, Garcia E, Perry G, Avila J, Garcia-Escudero V. Slower Dynamics and Aged Mitochondria in Sporadic Alzheimer's Disease. Oxid Med Cell Longev. 2017. 2017
17. Roca-Agujetas V, Barbero-Camps E, de Dios C, Podlesniy P, Abadin X, Morales A. et al. Cholesterol alters mitophagy by impairing optineurin recruitment and lysosomal clearance in Alzheimer's disease. Molecular Neurodegeneration. 2021 16
18. Kasem K, Sullivan E, Gopalan V, Salajegheh A, Smith RA, Lam AKY. JK1 (FAM134B) represses cell migration in colon cancer: a functional study of a novel gene. Exp Mol Pathol. 2014;97:99-104
19. Haque MH, Gopalan V, Chan KW, Shiddiky MJA, Smith RA, Lam AKY. Identification of Novel FAM134B (JK1) Mutations in Oesophageal Squamous Cell Carcinoma. Sci Rep-Uk. 2016 6
20. Zhang ZQ, Chen J, Huang WQ, Ning D, Liu QM, Wang C. et al. FAM134B induces tumorigenesis and epithelial-to-mesenchymal transition via Akt signaling in hepatocellular carcinoma. Mol Oncol. 2019;13:792-810
21. Dowdell AS, Cartwright IM, Kitzenberg DA, Kostelecky RE, Mahjoob O, Saeedi BJ. et al. Essential role for epithelial HIF-mediated xenophagy in control of Salmonella infection and dissemination. Cell Rep. 2022 40
22. Hou PP, Yang KX, Jia PH, Liu L, Lin YX, Li ZB. et al. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res. 2021;31:62-79
23. Wang WW, Han RY, He HJ, Wang Z, Luan XQ, Li J. et al. Delineating the Role of Mitophagy Inducers for Alzheimer Disease Patients. Aging and Disease. 2021;12:852-67
24. Liu YD, Wang S, Wang ZB, Ding MM, Li XY, Guo J. et al. Dexmedetomidine Alleviated Endoplasmic Reticulum Stress via Inducing ER-phagy in the Spinal Cord of Neuropathic Pain Model. Frontiers in Neuroscience. 2020 14
25. Sankar DS, Dengjel J. Protein complexes and neighborhoods driving autophagy. Autophagy. 2021;17:2689-705
26. Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167-78
27. Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ. et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506-22
28. Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA. et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211-21
29. Khaminets A, Beh C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6-16
30. Gubas A, Dikic I. A Guide To horizontal ellipsis The regulation of selective autophagy receptors. Febs J. 2022;289:75-89
31. Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233-42
32. Johansen T, Lamark T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J Mol Biol. 2020;432:80-103
33. Marshall RS, Hua ZH, Mali S, McLoughlin F, Vierstra RD. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell. 2022;185:1101-2
34. Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279-96
35. Kirkin V, Rogov VV. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol Cell. 2019;76:268-85
36. Mandell MA, Jain A, Kumar S, Castleman MJ, Anwar T, Eskelinen EL. et al. TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. J Cell Sci. 2017;130:1194
37. Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T. et al. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev Cell. 2016;39:13-27
38. Mancias JD, Wang XX, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105-109
39. Dowdle WE, Nyfeler B, Murphy L. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Mol. Biol. Cell. 2014 25
40. Jiang SX, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. 2011;413:420-5
41. Hyttinen JMT, Amadio M, Viiri J, Pascale A, Salminen A, Kaarniranta K. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res Rev. 2014;18:16-28
42. Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med. 2015;47:e147
43. Wetzel L, Blanchard S, Rama S, Beier V, Kaufmann A, Wollert T. TECPR1 promotes aggrephagy by direct recruitment of LC3C autophagosomes to lysosomes. Nat Commun. 2020 11
44. Ma XY, Lu CJ, Chen YT, Li SL, Ma NJ, Tao X. et al. CCT2 is an aggrephagy receptor for clearance of solid protein aggregates. Cell. 2022;185:1325-1345
45. Lyu L, Chen Z, McCarty N. TRIM44 links the UPS to SQSTM1/p62-dependent aggrephagy and removing misfolded proteins. Autophagy. 2022;18:783-98
46. Ikari S, Yang Q, Lu SL, Liu Y, Hao F, Tong G. et al. Quercetin in Tartary Buckwheat Induces Autophagy against Protein Aggregations. Antioxidants (Basel). 2021 10
47. Suresh SN, Pandurangi J, Murumalla R, Vidyadhara DJ, Garimella L, Acharya A. et al. Small molecule modulator of aggrephagy regulates neuroinflammation to curb pathogenesis of neurodegeneration. Ebiomedicine. 2019;50:260-273
48. Suresh SN, Rao MJ, Manjithaya R. XCT 790 is a pharmacological aggrephagy inducer in a yeast model of proteotoxicity. Cell Biol Int. 2021;45:654-61
49. Annesley SJ, Fisher PR. Mitochondria in Health and Disease. Cells. 2019 8
50. Hsieh CH, Shaltouki A, Gonzalez AE, da Cruz AB, Burbulla LF, St Lawrence E. et al. Functional Impairment in Miro Degradation and Mitophagy Is a Shared Feature in Familial and Sporadic Parkinson's Disease. Cell Stem Cell. 2016;19:709-724
51. Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD. et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258-262
52. Xie CL, Zhuang XX, Niu ZM, Ai RX, Lautrup S, Zheng SJ. et al. Amelioration of Alzheimer's disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nat. Biomed. Eng. 2022;6:76-93
53. Chourasia AH, Macleod KF. Tumor suppressor functions of BNIP3 and mitophagy. Autophagy. 2015;11:1937-1938
54. Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y. et al. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002-6011
55. Qian W, Wang J, Roginskaya V, McDermott LA, Edwards RP, Stolz DB. et al. Novel combination of mitochondrial division inhibitor 1 (mdivi-1) and platinum agents produces synergistic pro-apoptotic effect in drug resistant tumor cells. Oncotarget. 2014;5:4180-4194
56. Zhou J, Li GB, Zheng Y, Shen HM, Hu X, Ming QL. et al. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy. 2015;11:1259-1279
57. Liang WJJ, Gustafsson AB. The Aging Heart: Mitophagy at the Center of Rejuvenation. Front. Cardiovasc. Med. 2020 7
58. Kubli DA, Zhang XX, Lee Y, Hanna RA, Quinsay MN, Nguyen CK. et al. Parkin Protein Deficiency Exacerbates Cardiac Injury and Reduces Survival following Myocardial Infarction. J Biol Chem. 2013;288:915-926
59. Zhang WL, Siraj S, Zhang R, Chen Q. Mitophagy receptor FUNDC1 regulates mitochondrial homeostasis and protects the heart from I/R injury. Autophagy. 2017;13:1080-1081
60. Saito T, Nah J, Oka S, Mukai R, Monden Y, Maejima Y. et al. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J Clin Investig. 2019;129:802-819
61. Fu TT, Xu ZS, Liu L, Guo QQ, Wu H, Liang XJ. et al. Mitophagy Directs Muscle-Adipose Crosstalk to Alleviate Dietary Obesity. Cell Rep. 2018;23:1357-1372
62. Goldman SJ, Zhang Y, Jin S. Autophagic degradation of mitochondria in white adipose tissue differentiation. Antioxid Redox Signal. 2011;14:1971-1978
63. Barcena C, Mayoral P, Quiros PM. Mitohormesis, an Antiaging Paradigm. Int Rev Cell Mol Biol. 2018;340:35-77
64. Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J. et al. The Diabetes Susceptibility Gene Clec16a Regulates Mitophagy. Cell. 2014;157:1577-1590
65. Ferro-Novick S, Reggiori F, Brodsky JL. ER-Phagy, ER Homeostasis, and ER Quality Control: Implications for Disease. Trends Biochem. 2021;46:630-639
66. Guelly C, Zhu PP, Leonardis L, Papic L, Zidar J, Schabhuttl M. et al. Targeted High-Throughput Sequencing Identifies Mutations in atlastin-1 as a Cause of Hereditary Sensory Neuropathy Type I. Am. J. Hum. Genet. 2011;88:99-105
67. Murphy SM, Davidson GL, Brandner S, Houlden H, Reilly MM. Mutation in FAM134B causing severe hereditary sensory neuropathy. J. Neurol. Neurosurg. Psychiatry. 2012;83:119-120
68. Zou YY, He WX, Wang KL, Han HL, Xiao TT, Chen XM. et al. Identification of rare RTN3 variants in Alzheimer's disease in Han Chinese. Human Genetics. 2018;137:141-150
69. Linxweiler M, Linxweiler J, Barth M, Benedix J, Jung V, Kim YJ. et al. Sec62 Bridges the Gap from 3q Amplification to Molecular Cell Biology in Non-Small Cell Lung Cancer. Am. J. Pathol. 2012;180:473-483
70. Wemmert S, Lindner Y, Linxweiler J, Wagenpfeil S, Bohle R, Niewald M. et al. Initial evidence for Sec62 as a prognostic marker in advanced head and neck squamous cell carcinoma. Oncol. Lett. 2016;11:1661-1670
71. Campoy E, Colombo MI. Autophagy in intracellular bacterial infection. Biochim Biophys Acta. 2009;1793:1465-1477
72. Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V. et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242-245
73. Brest P, Lapaquette P, Mograbi B, Darfeuille-Michaud A, Hofman P. Risk predisposition for Crohn disease: a "menage a trois" combining IRGM allele, miRNA and xenophagy. Autophagy. 2011;7:786-787
74. Alsaadi RM, Losier TT, Tian WS, Jackson A, Guo ZH, Rubinsztein DC. et al. ULK1-mediated phosphorylation of ATG16L1 promotes xenophagy, but destabilizes the ATG16L1 Crohn's mutant. Embo Rep. 2019 20
75. Parkes M. Evidence from Genetics for a Role of Autophagy and Innate Immunity in IBD Pathogenesis. Dig Dis. 2012;30:330-333
76. Garcia C, Duong JA, Poeette M, Ribes A, Payre B, Memier V. et al. Platelet activation and partial desensitization are associated with viral xenophagy in patients with severe COVID-19. Blood Adv. 2022;6:3884-3898
77. Al Azzaz J, Rieu A, Aires V, Delmas D, Chluba J, Winckler P. et al. Resveratrol-Induced Xenophagy Promotes Intracellular Bacteria Clearance in Intestinal Epithelial Cells and Macrophages. Front. Immunol. 2019 9
78. Grefhorst A, van de Peppel IP, Larsen LE, Jonker JW, Holleboom AG. The Role of Lipophagy in the Development and Treatment of Non-Alcoholic Fatty Liver Disease. Front. Endocrinol. 2021;11:1099
79. Han YC, Xiong S, Zhao H, Yang SK, Yang M, Zhu XJ. et al. Lipophagy deficiency exacerbates ectopic lipid accumulation and tubular cells injury in diabetic nephropathy. Cell Death Dis. 2021;12:1031
80. Liu Q, Wang YM, Gu HF. Lipophagy in atherosclerosis. Clinica Chimica Acta. 2020;511:208-214
81. Duan XY, Xu LN, Li YW, Jia LJ, Liu W, Shao WX. et al. Regulation of lipid homeostasis by the TBC protein dTBC1D22 via modulation of the small GTPase Rab40 to facilitate lipophagy. Cell Rep. 2021;36:109541
82. Zhou WL, Yan X, Zhai YY, Liu H, Guan LL, Qiao Y. et al. Phillygenin ameliorates nonalcoholic fatty liver disease via TFEB-mediated lysosome biogenesis and lipophagy. Phytomedicine. 2022;103:154235
83. Lee DH, Park SH, Huh YH, Kim MJ, Seo HD, Ha TY. et al. Iridoids of Valeriana fauriei contribute to alleviating hepatic steatosis in obese mice by lipophagy. Biomed Pharmacother. 2020;125:109950
84. Ding WX, Ni HM, Waguri S, Komatsu M. Lack of hepatic autophagy promotes severity of liver injury but not steatosis. J Hepatol. 2022;77:1458-1459
85. Li Y, Chao X, Yang L, Lu Q, Li T, Ding WX. et al. Impaired Fasting-Induced Adaptive Lipid Droplet Biogenesis in Liver-Specific Atg5-Deficient Mouse Liver Is Mediated by Persistent Nuclear Factor-Like 2 Activation. Am J Pathol. 2018;188:1833-1846
86. Takahashi SS, Sou YS, Saito T, Kuma A, Yabe T, Sugiura Y. et al. Loss of autophagy impairs physiological steatosis by accumulation of NCoR1. Life Sci Alliance. 2020;3:e201900513
87. Haidar M, Loix M, Vanherle S, Dierckx T, Vangansewinkel T, Gervois P. et al. Targeting lipophagy in macrophages improves repair in multiple sclerosis. Autophagy. 2022;18:2697-2710
88. Wen W, Li XM, Yin MG, Wang HY, Qin LX, Li H. et al. Selective autophagy receptor SQSTM1/p62 inhibits Seneca Valley virus replication by targeting viral VP1 and VP3. Autophagy. 2021;17:3763-3775
89. Liang JR, Lingeman E, Luong T, Ahmed S, Muhar M, Nguyen T. et al. A Genome-wide ER-phagy Screen Highlights Key Roles of Mitochondrial Metabolism and ER-Resident UFMylation. Cell. 2020;180:1160-1177
90. Xian HX, Yang QY, Xiao L, Shen HM, Liou YC. STX17 dynamically regulated by Fis1 induces mitophagy via hierarchical macroautophagic mechanism. Nat Commun. 2019;10:2059
91. An H, Harper JW. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol. 2018;20:135-143
92. Kumar R, Rahman MA, Nazarko TY. Nitrogen Starvation and Stationary Phase Lipophagy Have Distinct Molecular Mechanisms. Int J Mol Sci. 2020;21:9094
93. Zhao D, Zou CX, Liu XM, Jiang ZD, Yu ZQ, Suo F. et al. A UPR-Induced Soluble ER-Phagy Receptor Acts with VAPs to Confer ER Stress Resistance. Mol Cell. 2020;79:963-977.e3
94. Robichaud S, Fairman G, Vijithakumar V, Mak E, Cook DP, Pelletier AR. et al. Identification of novel lipid droplet factors that regulate lipophagy and cholesterol efflux in macrophage foam cells. Autophagy. 2021;17:3671-3689
95. An H, Ordureau A, Paulo JA, Shoemaker CJ, Denic V, Harper JW. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol Cell. 2019;74:891-908.e10
96. Biagioni F, Ferese R, Giorgi FS, Modugno N, Olivola E, Lenzi P. et al. An attempt to dissect a peripheral marker based on cell pathology in Parkinson's disease. J. Neural Transm. 2021;128:1599-1610
97. Le Guerroue F, Eck F, Jung J, Starzetz T, Mittelbronn M, Kaulich M. et al. Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway. Mol Cell. 2017;68:786-796.e6
98. Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M. et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354-358
99. Stephani M, Picchianti L, Gajic A, Beveridge R, Skarwan E, Hernandez VSD. et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife. 2020;9:e58396
100. Bhujabal Z, Birgisdottir AB, Sjottem E, Brenne HB, Overvatn A, Habisov S. et al. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. Embo Rep. 2017;18:947-961
101. Chino H, Hatta T, Natsume T, Mizushima N. Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol Cell. 2019;74:909-921.e6
102. Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T. et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015;210:973-989
103. Xu Y, Zhou P, Cheng S, Lu QH, Nowak K, Hopp AK. et al. A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy. Cell. 2019;178:552-566.e20
104. Chen SL, Mari M, Parashar S, Liu DM, Cui YX, Reggiori F. et al. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. P Natl Acad Sci USA. 2020;117:18530-18539
105. Jacomin AC, Samavedam S, Promponas V, Nezis IP. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy. 2016;12:1945-1953
106. Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y. et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359-362
107. Chang J, Hwang HJ, Kim B, Choi YG, Park J, Park Y. et al. TRIM28 functions as a negative regulator of aggresome formation. Autophagy. 2021;17:4231-4248
108. Kalvari I, Tsompanis S, Mulakkal NC, Osgood R, Johansen T, Nezis IP. et al. iLIR A web resource for prediction of Atg8-family interacting proteins. Autophagy. 2014;10:913-925
109. Zhang YF, Yao YK, Qiu XX, Wang GD, Hu Z, Chen SY. et al. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat Immunol. 2019;20:433-446
110. von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ. et al. An essential role for the ATG8 ortholog LC3C in antibacterial autophagy. Autophagy. 2013;9:784-786
111. Popelka H, Klionsky DJ. Analysis of the native conformation of the LIR/AIM motif in the Atg8/LC3/GABARAP-binding proteins. Autophagy. 2015;11:2153-2159
112. Ibrahim T, Khandare V, Mirkin FG, Tumtas Y, Bubeck D, Bozkurt TO. AlphaFold2-multimer guided high-accuracy prediction of typical and atypical ATG8-binding motifs. PLoS Biol. 2023;21:e3001962
113. Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279-289
114. Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228-233
115. Fan S, Yue L, Wan W, Zhang Y, Zhang B, Otomo C. et al. Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein. Angew Chem Int Ed Engl. 2021;60:26105-26114
116. Yu HY, Tardivo L, Tam S, Weiner E, Gebreab F, Fan CY. et al. Next-generation sequencing to generate interactome datasets. Nat Methods. 2011;8:478-480
117. Johansen T, Birgisdottir AB, Huber J, Kniss A, Dotsch V, Kirkin V. et al. Methods for Studying Interactions Between Atg8/LC3/GABARAP and LIR-Containing Proteins. Method Enzymol. 2017;587:143-169
118. Tsapras P, Nezis IP. A yeast two-hybrid screening identifies novel Atg8a interactors in Drosophila. Autophagy. 2022;18:1211-1212
119. Zhou J, Wang Z, Wang XT, Li XF, Zhang ZC, Fan BF. et al. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy. 2018;14:487-504
120. Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA. et al. A Role for NBR1 in Autophagosomal Degradation of Ubiquitinated Substrates. Mol Cell. 2009;33:505-516
121. Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG. et al. The Cargo Receptor NDP52 Initiates Selective Autophagy by Recruiting the ULK Complex to Cytosol-Invading Bacteria. Mol Cell. 2019;74:320-326.e6
122. Sakowski ET, Koster S, Celhay CP, Park HS, Shrestha E, Hetzenecker SE. et al. Ubiquilin 1 Promotes IFN-gamma- Induced Xenophagy of Mycobacterium tuberculosis. Plos Pathog. 2015;11:e1005076
123. Yoon MJ, Choi B, Kim EJ, Ohk J, Yang C, Choi YG. et al. UXT chaperone prevents proteotoxicity by acting as an autophagy adaptor for p62-dependent aggrephagy. Nat Commun. 2021;12:1955
124. Nozawa T, Aikawa C, Minowa-Nozawa A, Nakagawa I. The intracellular microbial sensor NLRP4 directs Rho-actin signaling to facilitate Group A Streptococcus-containing autophagosome-like vacuole formation. Autophagy. 2017;13:1841-1854
125. Van Criekinge W, Beyaert R. Yeast Two-Hybrid: State of the Art. Biol Proced Online. 1999;2:1-38
126. Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy. 2021;17:1-382
127. Kojima W, Yamano K, Kosako H, Imai K, Kikuchi R, Tanaka K. et al. Mammalian BCAS3 and C16orf70 associate with the phagophore assembly site in response to selective and non-selective autophagy. Autophagy. 2021;17:2011-2036
128. Liu XM, Yamasaki A, Du XM, Coffman VC, Ohsumi Y, Nakatogawa H. et al. Lipidation-independent vacuolar functions of Atg8 rely on its noncanonical interaction with a vacuole membrane protein. Elife. 2018;7:e41237
129. Abudu YP, Pankiv S, Mathai BJ, Lystad AH, Bindesboll C, Brenne HB. et al. NIPSNAP1 and NIPSNAP2 Act as "Eat Me" Signals for Mitophagy. Dev Cell. 2019;49:509-525
130. Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA. et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science. 2018;360:751-758
131. Chen XL, Wei SS, Ji YL, Guo XJ, Yang FQ. Quantitative proteomics using SILAC: Principles, applications, and developments. Proteomics. 2015;15:3175-3192
132. Kustatscher G, Collins T, Gingras AC, Guo TN, Hermjakob H, Ideker T. et al. Understudied proteins: opportunities and challenges for functional proteomics. Nat Methods. 2022;19:774-779
133. Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376-386
134. Nthiga TM, Shrestha BK, Sjottem E, Bruun JA, Larsen KB, Bhujabal Z. et al. CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. Embo Journal. 2020;39:e103649
135. Judith D, Jefferies HBJ, Boeing S, Frith D, Snijders AP, Tooze SA. ATG9A shapes the forming autophagosome through Arfaptin 2 and phosphatidylinositol 4-kinase III beta. J Cell Biol. 2019;218:1634-1652
136. Gingras AC, Abe KT, Raught B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr Opin Chem Biol. 2019;48:44-54
137. Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801-810
138. Tu YX, Sydor AM, Coyaud E, Laurent EMN, Dyer D, Mellouk N. et al. Global Proximity Interactome of the Human Macroautophagy Pathway. Autophagy. 2022;18:1174-1186
139. Zellner S, Schifferer M, Behrends C. Systematically defining selective autophagy receptor-specific cargo using autophagosome content profiling. Mol Cell. 2021;81:1337-1354.e8
140. Zhang JS, Lan Y, Li MY, Lamers MM, Fusade-Boyer M, Klemm E. et al. Flaviviruses Exploit the Lipid Droplet Protein AUP1 to Trigger Lipophagy and Drive Virus Production. Cell Host Microbe. 2018;23:819-831.e5
141. Cui Y, Cheng X, Chen Q, Song B, Chiu A, Gao Y. et al. CRISP-view: a database of functional genetic screens spanning multiple phenotypes. Nucleic Acids Res. 2021;49:D848-D854
142. Mohr SE, Perrimon N. RNAi screening: new approaches, understandings, and organisms. Wiley Interdiscip Rev RNA. 2012;3:145-158
143. Wei L, Lee D, Law CT, Zhang MS, Shen JL, Chin DWC. et al. Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat Commun. 2019;10:13 4681
144. Liang JR, Lingeman E, Ahmed S, Corn JE. Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol. 2018;217:3354-3367
145. Heath RJ, Goel G, Baxt LA, Rush JS, Mohanan V, Paulus GLC. et al. RNF166 Determines Recruitment of Adaptor Proteins during Antibacterial Autophagy. Cell Rep. 2016;17:2183-2194
146. Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C. et al. TRIM Proteins Regulate Autophagy and Can Target Autophagic Substrates by Direct Recognition. Dev Cell. 2014;30:394-409
147. Hale CM, Cheng Q, Ortuno D, Huang M, Nojima D, Kassner PD. et al. Identification of modulators of autophagic flux in an image-based high content siRNA screen. Autophagy. 2016;12:713-726
148. Liu K, Kong LJ, Graham DB, Carey KL, Xavier RJ. SAC1 regulates autophagosomal phosphatidylinositol-4-phosphate for xenophagy-directed bacterial clearance. Cell Rep. 2021;36:109434
149. Housden BE, Perrimon N. Comparing CRISPR and RNAi-based screening technologies. Nat Biotechnol. 2016;34:621-623
150. Chen SL, Cui YX, Parashar S, Novick PJ, Ferro-Novick S. ER-phagy requires Lnp1, a protein that stabilizes rearrangements of the ER network. P Natl Acad Sci USA. 2018;115:E6237-E6244
151. Howell LA, Nemec AA, Murray MA, Tomko RJ. Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. Faseb J. 2018;32:21466-21480
152. Liang QQ, Yang PG, Tian E, Han JH, Zhang H. The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy. 2012;8:1426-1433
153. Zhang GM, Lin L, Qi D, Zhang H. The composition of a protein aggregate modulates the specificity and efficiency of its autophagic degradation. Autophagy. 2017;13:1487-1495
154. Lu Q, Yang PG, Huang XX, Hu WQ, Guo B, Wu F. et al. The WD40 Repeat PtdIns(3)P-Binding Protein EPG-6 Regulates Progression of Omegasomes to Autophagosomes. Dev Cell. 2011;21:343-357
155. Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 Is a Mitochondrial Protein that Confers Selectivity during Mitophagy. Dev Cell. 2009;17:98-109
156. Heo JM, Harper NJ, Paulo JA, Li MM, Xu QK, Coughlin M. et al. Integrated proteogenetic analysis reveals the landscape of a mitochondrial-autophagosome synapse during PARK2-dependent mitophagy. Sci Adv. 2019;5:eaay6424
157. Martinez Molina D, Nordlund P. The Cellular Thermal Shift Assay: A Novel Biophysical Assay for In Situ Drug Target Engagement and Mechanistic Biomarker Studies. Annu Rev Pharmacol Toxicol. 2016;56:141-161
158. Song Y, Zhao M, Wu Y, Yu B, Liu HM. A multifunctional cross-validation high-throughput screening protocol enabling the discovery of new SHP2 inhibitors. Acta Pharm Sin B. 2021;11:750-762
159. Lahiri V, Klionsky DJ. ATG4-family proteins drive phagophore growth independently of the LC3/GABARAP lipidation system. Autophagy. 2021;17:1293-1795
160. Kanfer G, Sarraf SA, Maman Y, Baldwin H, Dominguez-Martin E, Johnson KR. et al. Image-based pooled whole-genome CRISPRi screening for subcellular phenotypes. J Cell Biol. 2021;220:e202006180
161. Kane MS, Paris A, Codron P, Cassereau J, Procaccio V, Lenaers G. et al. Current mechanistic insights into the CCCP-induced cell survival response. Biochem. Pharmacol. 2018;148:100-110
Author contact
![]() Corresponding author: Jia-Hong Lu, University of Macau, Macau, China. E-mail: jiahongluedu.mo; Tel.: +853-8822-4508.
Corresponding author: Jia-Hong Lu, University of Macau, Macau, China. E-mail: jiahongluedu.mo; Tel.: +853-8822-4508.
Received 2023-2-4
Accepted 2023-8-31
Published 2024-1-1
