10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(7):715-732. doi:10.7150/ijbs.9126 This issue Cite
Research Paper
Type-IV Antifreeze Proteins are Essential for Epiboly and Convergence in Gastrulation of Zebrafish Embryos
State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, University of the Chinese Academy of Sciences, Wuhan 430072, China.
*These authors contributed equally to this work.
Received 2014-3-16; Accepted 2014-5-18; Published 2014-6-24
Abstract
Many organisms in extremely cold environments such as the Antarctic Pole have evolved antifreeze molecules to prevent ice formation. There are four types of antifreeze proteins (AFPs). Type-IV antifreeze proteins (AFP4s) are present also in certain temperate and even tropical fish, which has raised a question as to whether these AFP4s have important functions in addition to antifreeze activity. Here we report the identification and functional analyses of AFP4s in cyprinid fish. Two genes, namely afp4a and afp4b coding for AFP4s, were identified in gibel carp (Carassius auratus gibelio) and zebrafish (Danio rerio). In both species, afp4a and afp4b display a head-to-tail tandem arrangement and share a common 4-exonic gene structure. In zebrafish, both afp4a and afp4b were found to express specifically in the yolk syncytial layer (YSL). Interestingly, afp4a expression continues in YSL and digestive system from early embryos to adults, whereas afp4b expression is restricted to embryogenesis. Importantly, we have shown by using afp4a-specific and afp4b-specifc morpholino knockdown and cell lineage tracing approaches that AFP4a participates in epiboly progression by stabilizing yolk cytoplasmic layer microtubules, and AFP4b is primarily related to convergence movement. Therefore, both AFP4 proteins are essential for gastrulation of zebrafish embryos. Our current results provide first evidence that AFP such as AFP4 has important roles in regulating developmental processes besides its well-known function as antifreeze factors.
Keywords: antifreeze protein, yolk syncytial layer, epiboly, convergence, morphogenesis.
Introduction
During vertebrate embryogenesis, embryonic cell movements are very critical for morphogenesis and establishment of normal embryo architecture [1]. Through this process, the blastoderm transforms into three germ layers of endoderm, mesoderm and ectoderm as well as two major body axes of anterior-posterior axis and ventral-dorsal axis, and these changes mainly result from the harmonized morphogenetic movements, including epiboly, involution, convergence and extension (CE) [2, 3]. Numerous studies have shed light on some signaling molecules [3, 4], and some key factors that are related to cell adhesion, cytoskeletal rearrangement and cell interactions between the enveloping layer (EVL) and the yolk syncytial layer (YSL) have been suggested to play significant roles in gastrulation cell movements and embryonic development [5]. For example, the lipid pregnenolone, produced from cholesterol by Cyp11a1 enzyme, was shown to promote zebrafish (Danio rerio, Dr) embryonic cell movements by increasing yolk cytoplasmic layer (YCL) microtubule abundance [6]. In addition, Npc1 (Niemann-Pick disease, type C1), a 13 transmembrane-spanning protein containing a sterol-sensing domain, was also demonstrated to contribute early morphogenetic movements including epiboly and CE movements [7]. Nevertheless, most of the significant regulators remain to be further identified because embryonic cell movements have been believed to be driven by a variety of cellular behaviors and other molecules [8].
Following our systematic studies on maternal factor screening and gene function identification [9-13] in polyploid gibel carp (Carassius auratus gibelio, Cag) with multiple modes of unisexual and sexual reproduction [14-19], we have also identified and characterized a type-IV antifreeze protein gene (afp4) [20]. Antifreeze proteins (AFPs) have been found in fungi, bacteria, plants and animals, and they can bind to ice crystals to inhibit growth and recrystallization of ice [21]. According to their molecular structures, four types of AFPs, type I-IV AFPs, have been characterized in teleost fishes [22]. The type-IV antifreeze protein (AFP4) was firstly discovered from longhorn sculpin (Myoxocephalus octodecimspinosis) in 1997 [23], and its homologues had been identified in other teleost fishes, ranging from polar to tropic region and from seawater to freshwater [20, 24, 25]. As a new type of AFP, their molecular structures have been characterized by the conserved four-helix bundle [24-26], and the antifreeze activity has been demonstrated in several fishes [23-26]. Significantly, along with wide discoveries of afp4 homologues, their abundant expression has been detected recently in oocytes [27, 28] and in embryos [20] from some teleost fishes. So far, AFPs have been studied for more than 30 years, but almost all the previous studies have focused on the structural and biochemical properties in freezing avoidance [29-31], and little is known about the biological functions in embryonic development. The finding about abundant expression of afp4 during embryogenesis in gibel carp provides a good chance for us to explore the biological roles. In this study, we firstly identify two tandem duplicated afp4 genes from gibel carp and zebrafish, analyze their genomic organization, and characterize their expression pattern. Then, we use zebrafish as a model to reveal their biological functions as two key regulators in early morphogenetic movements of zebrafish embryogenesis.
Materials and Methods
Full-length cDNA cloning
A positive BAC clone of afp4 was isolated from gibel carp BAC library [32] by PCR screening based on the cDNA sequence of afp4 in gibel carp (Cagafp4, GenBank accession No. AY365004) and it was sequenced as described previously [10]. Zebrafish database search was performed on the web server of NCBI by BLAST using the cDNA of Cagafp4 as a query of nucleotide collection (nr/nt) database.
To achieve full-length cDNA sequence of the other Cagafp4 (Cagafp4a) and reexamine those of afp4s in zebrafish (Drafp4s), total RNAs of gibel carp or zebrafish embryos were purified with TRIzol Reagent (Ambion, USA) as described previously [33], and then SMART cDNA libraries were constructed according to the SMARTer™ PCR cDNA Synthesis Kit User Manual (Clontech, USA). The Cagafp4a and Drafp4s cDNAs were completed by 3' and 5' rapid amplification of cDNA ends (RACE), with primers (Table 1) based on the obtained Cagafp4s or Drafp4s cDNA sequences. The complete cDNA sequences of Cagafp4a and Drafp4a were deposited in GenBank (accession No. KJ183062 and KJ183061, respectively).
Sequence and phylogenetic analyses
The genomic structures of Cagafp4s and Drafp4s were achieved by comparative analyses between cDNA sequences and corresponding genomic sequences.
The complete sequences of other functionally characterized AFP4s in Myoxocephalus octodecemspinosus (Mo) [23, 26], Pleuragramma antarcticum (Pa), Notothenia coriiceps (Nc) [25] and Gadus morhua (Gm) [27, 28] were downloaded from GenBank non-redundant protein database. An unrooted maximum likelihood (ML) phylogenetic tree of AFP4s was constructed by MEGA 6 [34] with 1000 bootstrap replications, in which their evolutionary distances were computed using the JTT+F model, which was selected by ProtTest 2.4 [35] based on the corrected Akaike information criterion. Multiple amino acid sequence alignment was performed by Clustalx program (EMBL-EBI, UK). Signal peptides were predicted by a web tool SignalP 3.0 Server (CBS, Denmark). Identities and similarities between DrAFP4a and the other seven AFP4s were acquired by pairwise alignment using a web tool EMBOSS Needle (EMBL-EBI, UK). Sequence alignment of Drafp4a and Drafp4b cDNA was performed by DNAMAN 6.0 software (Lynnon Biosoft, Canada).
Zebrafish manipulation
Wild type (WT) zebrafish were maintained as described by Westerfield, and all the zebrafish embryos were obtained from natural spawning, and rinsed with embryos medium [36]. Embryos were incubated at 28.5°C and staged according to Kimmel et al [2]. The animal protocol for this research was approved by the Institute of Hydrobiology Institutional Animal Care and Use Committee (Approval ID: keshuizhuan 0829).
Primers used in this study.
| Name | Sequence (5'→3') | Usage |
|---|---|---|
| Cagafp4a-5'Race | AACCAGCATACATACGAAGAG | RACE |
| Cagafp4a-3'Race | TTCCTCATCGCTGTCCTTGTTAC | |
| Drafp4a-5'Race | CAGAGGCTTGATCTGCTCT | |
| Drafp4a-3'Race | CAATCTCTGGACGCAAGG | |
| Drafp4b-5'Race | GGCAATAGGTTTAATCTGGTCC | |
| Drafp4b-3'Race | TATCGTCACATTGACACAAGG | |
| 5' PCR Primer | AAGCAGTGGTATCAACGCAGAGTAC | |
| 3' PCR Primer | AAGCAGTGGTATCAACGCAGAGTACTTTTT | |
| Drβ-actin-F | AGCACGGTATTGTGACTAACTG | qPCR and semi-quantitative RT-PCR |
| Drβ-actin-R | TCGAACATGATCTGTGTCATC | |
| Drafp4a-RT-F | CAATCTCTGGACGCAAGG | |
| Drafp4a-RT-R | CAGAGGCTTGATCTGCTCT | |
| Drafp4b-RT-F | TATCGTCACATTGACACAAGG | |
| Drafp4b-RT-R | GGCAATAGGTTTAATCTGGTCC | |
| Drafp4a-WISH-F | TCTGGACGCAAGGCAACT | WISH |
| Drafp4a-WISH-R | TCTAATACGACTCACTATAGGGAAACAAAACCATCAGCATAC | |
| Drafp4b-WISH-F | AGAGCCCAGTTTCAGCCCAT | |
| Drafp4b-WISH-R | TAATACGACTCACTATAGGGACAATCAACTCCAAAATATCAGG | |
| Drafp4a-EGFP-F | GGATCCTCTGGACGCAAGGCAACT | Synthesizing afp4:EGFP mRNA |
| Drafp4a-EGFP-R | AAGCTTTGGCCTGGTCAGCAACAAAT | |
| Drafp4b-EGFP-F | GGATCCGACACACCATCACACGACA | |
| Drafp4b-EGFP-R | AAGCTTCTCTGCTCTCCTCCAGGTAAG | |
| EGFP-F | AAGCTTATGGTGAGCAAGGGCGA | |
| EGFP-R | CTCGAG TTACTTGTACAGCTCGTCCAT | |
| Drafp4a-F | TGGATCCATGAAATTCTCCCTCATCG | Synthesizing afp4 mRNA |
| Drafp4a-R | CGCTCGAGAAACAAAACCATCAGCATAC | |
| Drafp4b-F | TGGATCCATGAAACTCTCCCTCATC | |
| Drafp4b-R | CGCTCGAGGGATACTCATATCTGGAG |
RNA isolation, real-time PCR and semi-quantitative RT-PCR
Total RNAs were isolated from zebrafish embryos or tissues of adults using TRIzol Reagent as described previously [33], and cDNAs were then generated using M-MLV Reverse Transcriptase (Promega, USA) as described previously [37]. Real-time PCR (qPCR) and semi-quantitative RT-PCR analyses were performed with specific primers for afp4a, afp4b and β-actin (Table 1) as described previously [37, 38].
Western blot analyses
Western blot analyses were performed as reported previously using the anti-CagAfp4 antibody (1:200) [20]. Immunoreactive bands were visualized by using BCIP/NBT staining.
Whole-mount in situ hybridization
Whole-mount in situ hybridization (WISH) was carried out as previously described [39]. For antisense probe synthesis, T7 RNA polymerase promoter was added to the 5' end of reverse primers and a DIG RNA labeling kit (Roche, Germany) was used. In brief, DNA templates of Drafp4a/b were amplified by RT-PCR from zebrafish embryos cDNA with the primers Drafp4a/b-WISH-F and Drafp4a/b-WISH-R (Table 1). The riboprobe produced for Drafp4a was targeted to nucleotides 20-525 (accession No. KJ183061) and for Drafp4b to nucleotides 292-844 (accession No. BC153962). In addition, antisense probes of the following mRNAs were synthesized and used: foxA3, gsc, ntl, myoD, hgg1, dxl3b, pax2.1, sox17, eve1. For sectioning, 15 μm sections of the stained shield to bud stage embryos were made on a CM 1850 frozen microtomy (Leica, Germany) according to the previous report [40]. Images were acquired by Leica S8APO dissecting microscope (Leica, Germany). To measure the extent of epiboly or mediolateral length of somites, the pictures of embryos stained by ntl probe or myoD probe were measured by ImageJ software 1.47v (National Institutes of Health, USA) and analyzed as described previously [6, 41].
Morpholinos, RNAs and microinjection
Morpholinos (MOs, Gene Tools, LLC, USA) were designed to target the 5' untranslated region (tb-MO) or the intron 3/ exon 4 boundary (sb-MO) of Drafp4a or Drafp4b. Sequences were as following: afp4a-tb-MO: 5'-AAGAGTTGCCTTGCGTCCAGAGATT-3'; afp4a-sb-MO: 5'-CACTGTGCCATGAACAAACAAAAAC-3'; afp4b-tb-MO: 5'-ATGATTGTGGGATGAGCCAGGGTTG-3'; afp4b-sb-MO: 5'-AAGCACTGCATCACAAAGACAATAC-3'. In addition, several control MOs, such as standard control MO (Ctrl-MO, 5′-CCTCCTACCTCAGTTACAATTTATA-3′), afp4a-sb-miMO (afp4a-sb-MO with five-nucleotide mismatches indicated in lowercase, 5'-CACTcTcCCATGAAgAAtCtAAAAC-3'), and afp4b-tb-miMO (afp4b-tb-MO with five-nucleotide mismatches indicated in lowercase, 5'-ATcATTGTcGGATcAGCCAcGcTTG-3′) were also synthesized.
For rescue experiments, afp4a and afp4b cDNAs were amplified with primers containing BamHI and XhoI restriction sites from full-length cDNA without 5' untranslated region (UTR) and cloned into pCS2+. To test the efficiency and specificity of tb-MOs, 5' UTR and part of the N-terminal open reading frame (ORF) of afp4a or afp4b were fused in frame with the EGFP ORF, and cloned into pCS2+ (primers are listed in Table 1). Plasmid for in vitro transcription of Kaede was generous gift from Dr. Brian Ciruna. Capped RNAs were prepared with the mMESSAGE mMACHINE kit (Ambion, USA) as previously described [12].
MOs or mRNAs were injected at the one-cell stage. The amount of MO or mRNA injected for each embryo was as below: afp4a-sb-MO or afp4a-sb-miMO, 4 ng; afp4b-tb-MO or afp4b-tb-miMO, 2.5 ng; afp4a-tb-MO or afp4b-sb-MO, 8 ng; afp4-MOs, 4 ng afp4a-sb-MO and 2.5 ng afp4b-tb-MO; Ctrl-MO was at the same amount with afp4-MO using in the same experiment; to detect the translation blocking efficiency, about 100 pg afp4a:EGFP or afp4b:EGFP mRNA was injected into a random subset of afp4-MO-injected embryos (co-injection); for rescue experiment, about 150 pg afp4a or afp4b mRNA was co-injected with afp4-MO.
Reagent treatments
For nocodazole treatment, the dechorionated embryos were incubated in 1 μg/mL nocodazole (Sigma, USA), which was diluted in 1 x Danieau buffer as previously described [6]. For pregnenolone treatment, the dechorionated embryos were incubated with 0.1, 1, 10 or 20 μM pregnenolone (Sigma, USA) from 1k-cell stage until the late epiboly stage.
Microtubule and F-actin staining
Embryos with chorion removed were fixed in microtubule-stabilizing buffer (MSB) and the whole-mount microtubule staining with primary antibody mouse anti-β-tubulin (1:500 in blocking buffer, Sigma, USA) and secondary antibody rhodamine-conjugated goat anti-mouse IgG (1:200 in blocking buffer, Thermo, USA) was performed according to previous study [42].
Images were acquired with a Leica TCS-SP2 confocal microscope (Leica, Germany), and confocal Z-series image stacks collected at 2 μm intervals were assembled by LSM510 basic software. Microtubule area percentage (%) was calculated as following steps. Firstly, several 100 x 100 pixel (about 72.2 x 72.2 μm) images of the YCL microtubules were randomly intercepted from the blastoderm margin of each embryo picture, as indicated by a white square in the corresponding picture. Then, these images were changed to gray-scale images by Adobe Photoshop CS (Adobe, USA). Moreover, the gray-scale images were converted to white (microtubules) and black (background) by Make Binary of ImageJ. Finally, the pixel number of white (n) in each image was measured by Histogram of ImageJ, and the proportion in the 100 x 100 pixel (n/10000) was computed as microtubule area percentage (%) in each image. For this calculation, more than 20 embryos were used, and about five images for each embryo were computed.
F-actin staining was carried out with 1 μg/mL rhodamine-phalloidin (Sigma, USA) as previously described [43]. Rhodamine-phalloidin was prepared as stock solutions of 1 mg/mL in methanol, and then diluted in PBTD/BSA just before use. Images were acquired with a Leica TCS-SP2 confocal microscope and confocal Z-series image stacks collected at 2 μm intervals were assembled by LSM510 basic software. Phalloidin signal intensity, which indicated F-actin intensity, was analyzed using ImageJ software as previously described [44].
Cell lineage tracing
Cell lineage tracing was performed according to previous reports [41, 45]. Briefly, 100 pg Kaede mRNA was injected with afp4b-tb-miMO or afp4b-tb-MO to embryos at the one-cell stage. The injected embryos were kept in the dark until the shield stage. The Kaede fluorophore was converted from green to red by focusing a 40-sec pulse of ultraviolet (UV) light, specifically on a group of cells in dorsal or lateral blastoderm margin, using the pinhole of a Leica LCS SP2 confocal microscope. Photos were acquired at the indicated developmental stages by a Leica MZ16 FA stereomicroscope (Leica, Germany). Figures were constructed using Adobe Photoshop CS. The angle and length were measured by utilizing ImageJ software.
Statistical analyses
For statistical analyses, means ± standard deviation (SD) were acquired by Microsoft Excel 2003 (Microsoft, USA), and one-way analyses of variance (ANOVA) and cross-table analyses were performed with SPSS 13.0 software (SPSS, USA).
Results
Identification and molecular characterization of two tandem afp4s in gibel carp and zebrafish
Previous report has characterized a full-length cDNA of afp4 in gibel carp (GenBank accession No. AY365004) [20]. To characterize its genomic organization, we obtained a positive BAC by PCR screening from gibel carp BAC library [32] as described previously [10]. Sequencing the BAC clone revealed the two tandem duplicated gene sequences, and full-length cDNA of the other afp4 was achieved by RACE (GenBank accession No. KJ183062). Zebrafish database searches also found a similar genomic organization of the two tandem duplicated genes on the chromosome 16 (Supplementary material Fig. S1), and revealed two afp4 homologues (GenBank accession No. BC133822 and BC153962). Then, full-length cDNAs of the two afp4s was reexamined and acquired by RACE (GenBank accession No. KJ183061 and BC153962). Genomic organization comparison between gibel carp and zebrafish revealed almost identical exons in size and similar exon/intron boundaries (Supplementary material Table S1).
In order to clarify evolutionary relationship between the four AFPs and other functionally characterized AFP4s with antifreeze activity, we constructed a phylogenetic tree. As shown in Fig. 1A, although the classification position of these species belongs to different orders including Ostariophysi, Paracanthopterygii and Acanthopterygii, the eight known AFP4s are gathered into two main clades. Clade A contains DrAFP4s and CagAFP4s, and clade B includes the four functionally characterized AFP4s. Multiple amino acid sequence alignment of DrAFP4s and other AFP4s also revealed their evolutionary conservation. Significantly, the newly characterized AFPs in zebrafish and gibel carp shared common properties of the functionally characterized AFP4s, including the four α-helical regions, signal peptide, unrelated coding region 1 and 2 (UCR1 and UCR2), the 33-codon block, and 11-mer repeats [23-26]. Moreover, the amino acid identities between DrAFP4a and other AFP4 ranged from 31.1% to 83.8%, and their signal peptide and α-helical region showed higher consensus than the complete amino acid sequences (Fig. 1B). Therefore, the newly identified and characterized genes should belong to homologs of afp4. Drafp4a and Drafp4b were abbreviated to afp4a and afp4b for the common use.
To further investigate the biological functions of afp4s in zebrafish, we analyzed the full-length cDNA sequences of zebrafish afp4a and afp4b (GenBank accession No. KJ183061 and BC153962, respectively). As shown in Fig. 1C, their nucleotide sequence alignment shows 88.8% of high identity in the ORFs, but much lower identity exists in the 5' and 3' UTR regions, and the 3' UTR of afp4b is 275 nt longer than that of afp4a. Moreover, significant sequence differences were found in their predicted promoter regions in 5' upstream sequences (data not shown). All the sequence differences imply the existence of their divergence in expression pattern and biological function.
Divergent and dynamic temporal expression patterns between afp4a and afp4b
Subsequently, we examined expression patterns of afp4a and afp4b in zebrafish by qPCR and Western blot analyses. Firstly, two sets of specific primers were designed according to their most divergent sequences to detect and distinguish afp4a transcript and afp4b transcript, respectively (Fig. 1C), and the transcript specificity was verified by sequencing the amplified products of afp4a and afp4b. Significantly, the qPCR revealed differential expression pattern and dynamic changes between afp4a and afp4b during embryogenesis and early larval development. As shown in Fig. 2A, afp4a initially transcribes from 4 hours postfertilization (hpf) at which the embryos develop to sphere stage, whereas afp4b transcribes from 6 hpf when the embryos are at shield stage. As development proceeds, afp4a transcript increases progressively and reaches to the highest level at 12 hpf when the embryos develop to 6-somite stage, whereas the amount of afp4b transcript exceeds that of afp4a at 12 hpf, and peaks at 16 hpf. At the afp4b peak duration, afp4a transcript gradually decreases from 12 hpf to 20 hpf. Then, when the afp4b transcript reduces gradually from 20 hpf to 28 hpf, afp4a transcript rises again from 24 hpf to 28 hpf. Around 32 hpf, afp4b increases again, and quickly declines to the bottom from 32 hpf to 48 hpf. After 48 hpf, almost no afp4b transcript is detected in the later embryos and larvae. However, the afp4a transcript decreases again from 32 hpf to 72 hpf, but its transcript gains once again in 96 hpf and 120 hpf larvae. The AFP4s protein expression pattern during embryogenesis was also detected by Western blot assay using the antibody against both AFP4s. As shown in Fig. 2B, the specific AFP4s protein band appears from 6 hpf at shield stage, reaches to the peak level at 16 hpf, and then reduces gradually to a certain level from 24 hpf to 96 hpf during later embryogenesis.
Phylogenetic relationship and molecular characterization of DrAFP4s and other functionally characterized AFP4s. (A) An unrooted ML phylogenetic tree of the known AFPs from Danio rerio (DrAFP4a, NP_001038953; DrAFP4b, XP_697091), Carassius auratus gibelio (CagAFP4a, AHZ08737; CagAFP4b, AAR12991), Gadus morhua (GmAFP4, Q56TU0), Myoxocephalus octodecemspinosus (MoAFP4, ABA41379), Notothenia coriiceps (NcAFP4, ADU02181) and Pleuragramma antarcticum (PaAFP4, ADU02180). Their classification positions, such as Ostariophysi, Paracanthopterygii and Acanthopterygii, are indicated on the right. (B) Multiple amino acid sequence alignment of DrAFP4s and other AFP4s. Amino acids consensus is shown below the alignment by three kinds of consensus symbols. An asterisk (*) means a position where has a single conserved residue; a colon (:) means a position where process high similarity (scoring > 0.5); a period (.) means a position where hold low similarity (scoring =< 0.5). Asterisks (*) above protein sequences show the predicate α-helical regions. Three exons (E2, E3 and E4) and boundary of eight parts of proteins [signal peptide, unrelated coding region 1 and 2 (UCR1 and UCR2), the 33-codon block, and 11-mer repeats (4 to 9)] are indicated according to a previous report [25]. Identities and similarities between DrAFP4a with other AFP4s are shown behind the alignment. (C) Nucleotide sequence alignment of Drafp4a and Drafp4b cDNAs. Identical nucleotides are indicated by the black background; the start code (ATG) and stop code (TAA) are lined by black lines; putative polyadenylation signals are boxed; the targets of the antisense probe to afp4a or afp4b used in whole-mount in situ hybridization are lined by red line above or blue line below sequences, respectively; two sets of specific primers used in RT-PCR to detect afp4a or afp4b transcript are respectively indicated by red or blue boxes; targets of tb-MOs are pointed out by brackets.
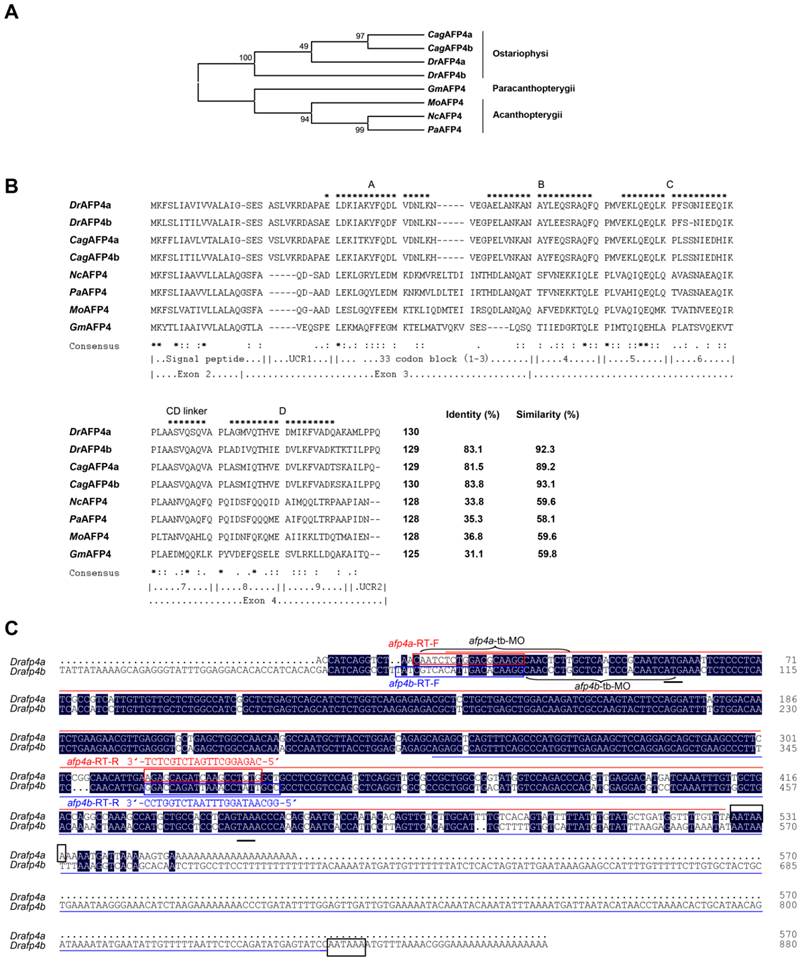
Temporal and spatial expression pattern of afp4s. (A) Expression of afp4a and afp4b in embryonic development stages and early larvae evaluated by qPCR and normalized to β-actin. The histograms stand for the relative transcript level of afp4a (red) or afp4b (blue), and the relative transcript level of afp4a at the sphere stage is set to 1. Data shown are means ± SD of three independent assays. (B) Western blot assay from sphere stage to 96 hpf. β-actin protein is internal control. (C, D) In situ hybridized WT embryos for afp4a/afp4b (C) or afp4b (D). Stages of embryos are shown in the bottom right corner of each panel. Black arrows indicate the liver, and white arrows indicate the intestine. Embryos from sphere stage to 6-somite stage are oriented animal pole up and dorsal right, and embryos from 24 hpf to 120 hpf are heading to the left and dorsal up. (E, F) Expression of afp4a and afp4b in adult tissues evaluated by qPCR (E) and semi-quantitative RT-PCR (F), and normalized to β-actin. (E) The histograms stand for the relative transcript level of afp4a (red) or afp4b (blue), and the relative transcript level of afp4a in the hindgut is set to 1. Data shown are means ± SD of three independent assays.
Yolk syncytial layer and digestive system-specific expression of afp4s in embryos, early larvae, and adults
Moreover, the expression patterns of afp4a and afp4b were investigated in embryos and larvae by WISH using two different antisense probes. The first probe, targeted to a 5'-terminal 506 nt sequence of afp4a transcript (Fig. 1C), can recognize both transcripts of afp4a and afp4b because of their high sequence similarity. The second probe, which is mainly against to extra 3'-terminal of afp4b with 553 nt (Fig. 1C), is only able to examine the afp4b transcript. These WISH data further revealed a differential and dynamic expression fashion between afp4a and afp4b. As shown in Fig. 2C, the positive signal hybridized with the first probe initiates to appear in YSL from 30% epiboly stage at about 4.7 hpf. Then, the signal quickly enhances from shield stage at about 6 hpf, and reaches to the strongest from 8 hpf to 12 hpf during which 75% epiboly, tail-bud formation and somitogenesis (6-somite) have progressed. The sectioned observations of the stained embryos from shield to bud stages showed restricted expression in YSL (Supplementary material Fig. S2A-C). Along with the embryonic development, the specific expression domain in YSL becomes clearer, especially in 24 hpf embryos. After yolk absorption, the signal gradually becomes weak from the posterior to anterior YSL in the hatched larvae, and finally restricts to the formed digestive organs, including intestine and liver, in 96 hpf and 120 hpf larvae. In comparison with the above data, the afp4b-specific transcript expresses later, and also appears within YSL from the shield stage embryos at about 6 hpf. Then, its expression strengthens rapidly, and reaches the strongest at about 12 hpf when the embryos develop to 6-somite stage (Fig. 2D). The longitudinal section of the stained embryos also showed specific expression in YSL (Supplementary material Fig. S2D-F). After 24 hpf, the afp4b-specific transcript reduces gradually, and not any afp4b-specific transcript is observed in the corresponding expression position after 72 hpf (Fig. 2D).
Furthermore, tissue distribution of afp4a and afp4b transcripts was examined in adult zebrafish by qPCR and semi-quantitative RT-PCR. As shown in Fig. 2E and 2F, the afp4a transcript is abundant in liver and foregut, slight in hindgut, and no signal exists in other tissues, including heart, spleen, kidney, brain, skin, muscle, ovary, testis and swimming bladder, whereas afp4b mRNA is absent in all the examined tissues. All the data indicate that both transcripts of afp4a and afp4b express specifically in YSL, but the later expressed afp4b appears only in embryogenesis, whereas afp4a expresses continually in YSL and digestive system from early embryos to adults, suggesting that afp4a and afp4b might play similar but different biological roles during zebrafish embryogenesis and early larval development.
AFP4a and AFP4b contribute to early morphogenetic movements during zebrafish embryogenesis
To explore their respective biological functions, we designed two kinds of antisense MOs specific to afp4a or afp4b. One is translation-blocking MO (tb-MO), which is directed to the 5′ UTR of afp4a (afp4a-tb-MO) or afp4b (afp4b-tb-MO) (Fig. 1C). The efficiency and specificity of each tb-MO were confirmed by co-injecting with afp4a:EGFP or afp4b:EGFP mRNA (100 pg/embryo) into one-cell stage zebrafish embryos. The test showed that about 8 ng afp4a-tb-MO for each embryo could largely deplete the EGFP fluorescence produced by afp4a:EGFP mRNA, but did not inhibit that expressed by afp4b:EGFP mRNA. On the other hand, lower dose afp4b-tb-MO (2.5 ng/embryo) could completely deplete the EGFP fluorescence produced by afp4b:EGFP mRNA, but did not reduce that produced by afp4a:EGFP mRNA. However, if the dose of afp4b-tb-MO was increased to 5 ng, the proportion of afp4a:EGFP-injected embryos with EGFP fluorescence was significantly reduced to 62% (Supplementary material Fig. S3A). These data indicate that afp4a-tb-MO can specifically knockdown AFP4a expression, whereas afp4b-tb-MO can specifically deplete AFP4b expression only at a lower dose, and a higher dose can interfere with the specificity and lead to cross-reaction with afp4a.
The other kind of MO is splice-blocking morpholino (sb-MO), which is targeted to the intron 3/exon 4 boundary of afp4a (afp4a-sb-MO) or afp4b (afp4b-sb-MO), since significant nucleotide difference only exists in the boundary sequence between afp4a and afp4b premature mRNAs (Supplementary material Table S2). The sb-MO resulted in a transcript with intron 3 insertion, and introduced a premature stop codon right after the exon 3/intron 3 junction. This early stop codon led to a truncated protein (58 amino acid residues), which lost the entire product of exon 4, including the characteristic 11-mer conserved repeats in all AFP4s (Fig. 1B, Supplementary material Fig. S3B and Table S2). Semi-quantitative RT-PCR results showed that 4 ng afp4a-sb-MO could specifically change the splice of all the endogenous afp4a mRNA, while 8 ng afp4b-sb-MO could only specifically change the splice of about half the endogenous afp4b mRNA (Supplementary material Fig. S3B).
Then, 4 MOs were respectively injected with appropriate concentration described above. When Ctrl-MO (8 ng/embryo) embryos developed to bud stage, both afp4a-tb-MO (8 ng/embryo) morphants and afp4a-sb-MO (4 ng/embryo) morphants displayed an epibolic delay; nevertheless, a more serious defect in epiboly was observed in afp4a-sb-MO morphants. At the same time, both afp4b-tb-MO (2.5 ng/embryo) morphants and afp4b-sb-MO (8 ng/embryo) morphants showed a longer anterior-posterior axis, and afp4b-sb-MO led to a similar but milder defect (Supplementary material Fig. S3C). The two distinct MOs targeted to the same gene caused the similar defect, confirming the phenotypes-specific to afp4a or afp4b. Moreover, afp4a-sb-MO and afp4b-tb-MO could produce the more obvious defect with lower individual MO dose than the other MO targeted to the same gene, and the lower dose was likely to cause fewer non-specific defects. Therefore, afp4a-sb-MO and afp4b-tb-MO were used in subsequent experiments.
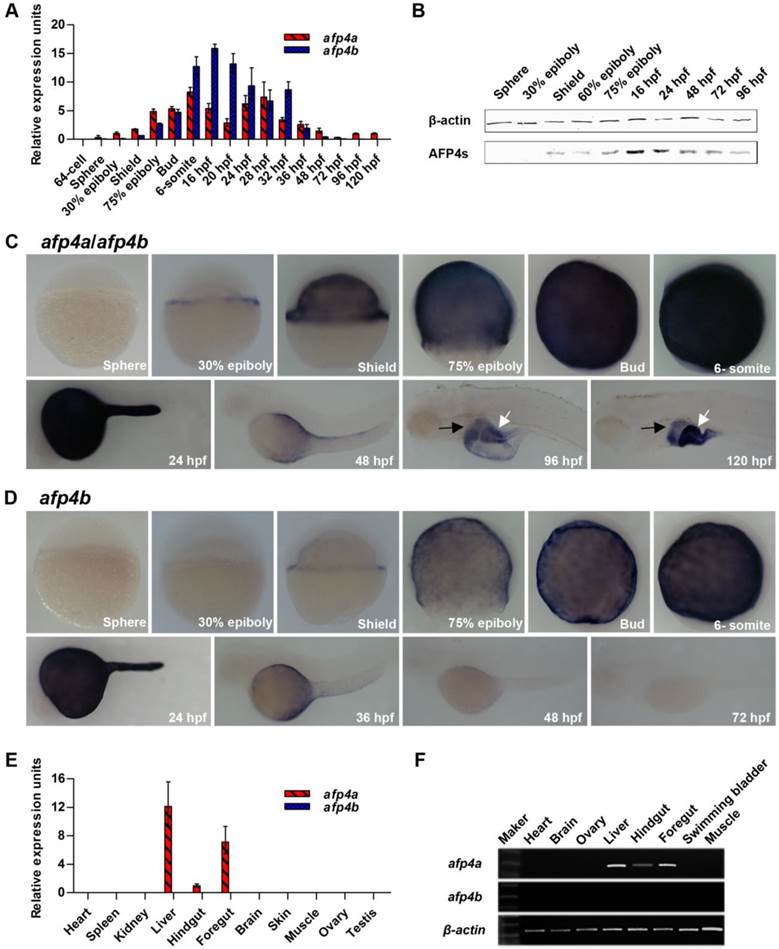
Subsequently, afp4a-sb-MO (4 ng/embryo) and afp4b-tb-MO (2.5 ng/embryo) were respectively or simultaneously (afp4-MOs) injected into zebrafish embryos at the one-cell stage. Western blot detection further confirmed the specificity and effectiveness. A significant decrease of AFP4s level was observed from shield stage (about 92% reduction) to 6-somite stage (about 64% reduction) in the afp4a-sb-MO morphants, whereas a contrary change was detected from shield stage (about 6% reduction) to 6-somite stage (about 38% reduction) in the afp4b-tb-MO morphants (Fig. 3A and 3B). These are consistent with the higher transcript level of afp4a than that of afp4b before 6-somite stage. Significantly, all the AFP4s were completely depleted from shield stage to 6-somite stage in the afp4a-sb-MO and afp4b-tb-MO co-injected (afp4-MOs) morphants.
Moreover, morphologic changes of embryos in each group were closely monitored. When Ctrl-MO embryos developed to shield stage, at which time their blastoderm covered 50% of the yolk and formed shield, afp4a-sb-MO morphants also formed shield, but their blastoderm could not cover half of the yolk. While Ctrl-MO embryos reached 75% epiboly, afp4a-sb-MO morphants displayed an obvious delay in epiboly. At the bud stage, when epiboly was completed in Ctrl-MO embryos, about 20% yolk could not be covered by blastomeres in afp4a-sb-MO morphants (Fig. 3C, D). Significantly, the afp4b-tb-MO injected embryos did not show any obvious defects until 75% epiboly, but the morphants began to display a longer anterior-posterior axis from bud stage. Moreover, delayed developments were observed during somitogenesis, in which their somites became wider in the mediolateral axis and the boundary of somites was faint. At 24 hpf, the afp4b-tb-MO injected embryos displayed shorter anterior-posterior body axis without distinct v-shape somites, and approximately 49.3% of the morphants died (Fig. 3E, G). In contrast, the afp4a-sb-MO injected embryos showed a more normal phenotype than the afp4b-tb-MO morphants after bud stage. At 6-somite stage, about 85.4% morphants, which had closed their blastopores, showed a slight increased linear distance between the head and tail, and about 8.0% morphants still could not finish epiboly (data not shown). However, some obvious defects, such as curving trunk, reduction of yolk extension and diminution of head growth, were observed at 24 hpf. Meanwhile, the 8.0% afp4a-sb-MO morphants, which failed to finish epiboly, displayed a curly tail (Fig. 3D). In comparison with the afp4a-sb-MO (Fig. 3D) or afp4b-tb-MO (Fig.3E) morphants, the morphogenetic defects during gastrulation (from shield stage to bud stage) in the afp4-MOs co-injected morphants (Fig. 3F) were basically similar to that in the embryos merely injected with afp4a-sb-MO (Fig. 3D). However, the proportion of the dead afp4-MOs embryos was higher (about 33.3%) than that of the embryos only injected with afp4a-sb-MO (about 2.4%) or afp4b-tb-MO (about 2.5%) at bud stage (Fig. 3G). Two hours later, when embryos developed to 6-somite stage, the survived afp4-MOs morphants showed the delayed development of head, elongated anterior-posterior axis and faint somite boundary, or even crooked notochord. Furthermore, the proportion of dead afp4-MOs-injected embryos increased substantially to approximately 83.1%, whereas those of single MO injected embryos were still very low (about 3.5% for afp4a-sb-MO, and 3.9% for afp4b-tb-MO). At 24 hpf, the survived afp4-MOs morphants showed similar phenotypes with afp4b-tb-MO morphants, and the proportion of dead embryos increased to about 86.8%. (Fig. 3F, G)
To confirm the specificity for above data, we synthesized mRNA of afp4a and afp4b lacking 5' UTR, where was the target of tb-MO for rescue examination. Remarkably, the morphogenetic defects caused by the afp4-MOs could be rescued by the synthesized mRNA of afp4a and afp4b, because significant death decrease and significant wild type increase appeared in the co-injected embryos of each or mixture of afp4a-mRNA and afp4b-mRNA (P < 0.001). However, phenotype and death were still at high levels. (Fig. 3G) This observation could because that the RNA amount injected to embryos was not most suitable for rescue. These data suggest that AFP4a and AFP4b might contribute to early morphogenetic movements during zebrafish embryogenesis.
Reduced AFP4a and/or AFP4b expression causes serious morphogenetic defects during early embryonic development. (A) Test of decrease of AFP4s by Western blotting at shield stage, 75% epiboly stage, bud stage and 6-somite stage after injection of afp4a-sb-MO and/or afp4b-tb-MO. afp4-MOs means afp4a-sb-MO and afp4b-tb-MO co-injection. β-actin protein is internal control. (B) Statistical data of the relative AFP4s expression level in afp4a-sb-MO and/or afp4b-tb-MO morphants from shield stage to 6-somite stage. Normalized to the expression of β-actin protein and the corresponding relative expression level of AFP4s in WT embryos is set to 1. Results are presented as means ± SD of three independent experiments. (C-F) Morphology of embryos from shield stage to 24 hpf after injecting Ctrl-MO (C), afp4a-sb-MO (D), afp4b-tb-MO (E), or afp4-MOs (F). Lateral views of embryos from shield to bud stage, dorsal to the right, anterior at the top; lateral or dorsal views of embryos at 6-somite stage with their animal pole facing upward; lateral views of embryos at 24 hpf, anterior towards the left and dorsal to the top. The arrows in shield stage embryos indicate the shield; the arrow in 24 hpf afp4a-sb-MO morphants indicates the unclosed blastopore; the lines in lateral-viewed 6-somite stage embryos indicate head-to-tail distance; the lines in dorsal-viewed embryos indicate somites width; the dotted lines show the boundary of a curve notochord. Morphology was scored as mild (white) or severely defective (black) in afp4a-sb-MO or afp4b-tb-MO injected embryos at 24 hpf. (G) Percentage of embryos at bud stage, 6-somite stage and 24 hpf in each class after injecting Ctrl-MO, afp4a-sb-MO, afp4b-tb-MO, or afp4-MOs (the left columns indicate by a grey box. Results represent means ± SD of three independent experiments, N=76 to 116 embryos per condition); or percentage of afp4-MOs-injected embryos in each class at 6-somite stage after injecting afp4a-mRNA, afp4b-mRNA, or afp4a-mRNA together with afp4b-mRNA (afp4-mRNAs) (the right columns indicate by a black box. Results represent means ± SD of three independent experiments, N=55 to 89 embryos per condition. * means P < 0.001, when compared to afp4-MOs).
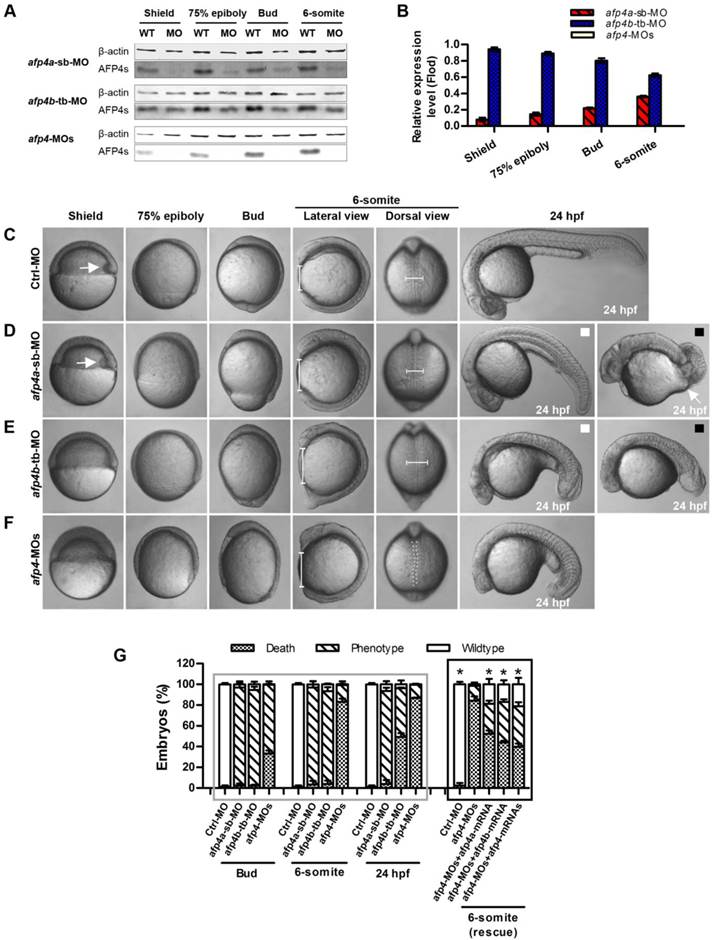
The reduced AFP4a expression leads to epibolic delay. (A) ntl expression in 8 hpf groups injected with afp4a-sb-miMO, afp4a-sb-MO, afp4a-sb-MO plus afp4a-mRNA, or afp4a-sb-MO plus afp4b-mRNA. Lateral views of embryos, dorsal to the right, anterior to the top. The calculating method for the epiboly percentage is shown. (B) Histogram represents mean ± SD of the epiboly percentage in each group. N, the number of embryos that were analyzed from three independent experiments. *P < 0.01.
AFP4a is required for epiboly progression
To confirm whether the morphogenetic defect in the afp4a-sb-MO morphants during gastrulation was resulted from AFP4a depletion, we further synthesized a control MO containing 5-base mismatches relative to afp4a-sb-MO (afp4a-sb-miMO). Identically to the above Ctrl-MO, the embryos injected with 4 ng of afp4a-sb-miMO were normal, and thus the afp4a-sb-miMO was utilized as a control in following studies on the role of AFP4a. In addition, the synthesized afp4a or afp4b mRNA was co-injected with afp4a-sb-MO for rescue examination, and the mesodermal marker ntl was used to visualize the blastoderm margin through WISH (Fig. 4A) and thereby to quantify the epiboly percentage (Fig. 4B). In comparison with normal epiboly progression (80.2% ± 3.7%, N=213) at 8 hpf in the afp4a-sb-miMO embryos, the afp4a-sb-MO embryos displayed significant epiboly delay (56.5% ± 5.0%, N=156), and the delay could be rescued by co-injection with 150 pg afp4a-mRNA (73.1% ± 9.3%, N=155), but could not be restored by the same amount of afp4b-mRNA (58.3% ± 6.1%, N=170). These data indicate that AFP4a depletion leads to epiboly defect.
Since the afp4a-expressed YSL had been revealed to play significant roles in germ layer differentiation and early morphogenetic movements [46], we further tested whether the AFP4a depletion affected other early embryonic development except for epiboly progression. Firstly, we detected germ layer differentiation by endodermal marker foxA3, mesendodermal marker gsc and mesodermal marker ntl in shield stage embryos. In comparison with the afp4a-sb-miMO control embryos, their expression domains at the gastrulation onset stage were only located slightly towards animal-pole in the afp4a-sb-MO morphants (Supplementary material Fig. S4A-C). These changes should be caused by epiboly delay, and germ layer differentiation is little affected in the AFP4a-knockdown embryos. At the same stage, AFP4a-depleted embryos also showed the prominent embryonic shield similar to that in control embryos, even though the blastoderms in these embryos covered less than 50% of the yolk cell (Fig. 3D). This phenotype implies that involution movement is normal in the morphants.
Moreover, localization of ntl in axial chordal mesoderm and myoD in adaxial cells at bud stage confirmed the presence of a slight wider and shorter axis with the unclosed blastopore in afp4a-sb-MO morphants (Supplementary material Fig. S4D and E), but the expression domains of myoD in adaxial cells and somites were comparable between afp4a-sb-MO morphants and afp4a-sb-miMO morphants at 8-somite stage when most afp4a-sb-MO morphants finished epiboly (Supplementary material Fig. S4F). These data indicate that mediolateral convergence seems a little change because of the unclosed blastopore. On the other hand, the normality in anterior part of ntl expression domain was observed (Supplementary material Fig. S4G). Moreover, the expression of hgg1, which marked the prechordal plate, and of dlx3b, which expressed in the anterior edge of the neural plate, were almost the same as afp4a-sb-miMO embryos at the bud stage (Supplementary material Fig. S4H). In addition, at the 8-somite stage, the expression domains of pax2.1 in the optic stalk, the midbrain hindbrain boundary, and the otic vesicles were comparable between afp4a-sb-MO morphants and afp4a-sb-miMO morphants (Supplementary material Fig. S4I). These data indicate that the anterior extension of the axial mesendoderm is not affected, and the convergence movement is slightly affected by AFP4a deficiency.
YCL microtubule arrays of afp4a-sb-MO morphants are less stable than that of afp4a-sb-miMO control embryos. (A-D) YCL microtubules are thinner in afp4a-sb-MO embryos (B, D) compared to those in afp4a-sb-miMO embryos (A, C), at 50% epiboly after nocodazole treatment. (C or D) is the enlarged image of the areas in the square in (A or B). Embryos are oriented with anterior up. Scale bars, 100 µm. (E) Average percentage of the microtubule area in YCL alone EVL margin in afp4a-sb-miMO embryos and afp4a-sb-MO morphants at about 50% epiboly after nocodazole treatment. (F) Epibolic defect in AFP4a-deficient embryos can be rescued by 1 and 10 µM pregnenolone (P5). (E and F) Data are represented as means ± SD. N, the number of embryos that were analyzed from three independent experiments. *P < 0.01.
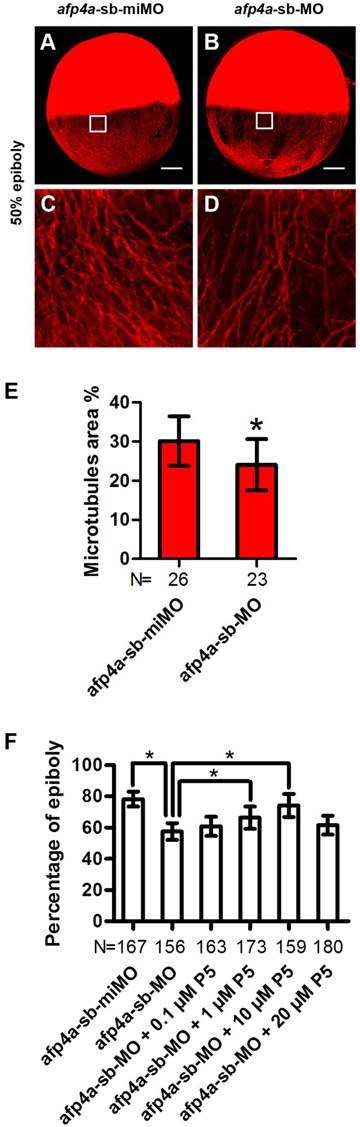
AFP4a participates in epiboly progression by stabilizing YCL microtubules
Above observed phenotypes in the afp4a-sb-MO morphants, such as obviously delayed epiboly, mildly affected CE, and normal germ layer induction, involution and anterior axial formation, are highly similar to those of YCL microtubule disrupted embryos [6, 47]. This strong resemblance promoted us to visualize microtubule array in YCL by staining them with antibody against β-tubulin. Firstly, we compared the afp4a-sb-miMO and afp4a-sb-MO embryos fixed at 50% epiboly stage, and no any changes were observed (data not shown). Then, we treated the afp4a-sb-miMO and afp4a-sb-MO embryos at 50% epiboly stage with 1 μg/ml nocodazole for 20 min at 28.5℃, and examined the microtubules again in the fixed embryos. Obviously, the YCL microtubules in the afp4a-sb-MO embryos were thinner than those in the afp4a-sb-miMO embryos (Fig. 5A-D). To quantify the data, the percentages of microtubule area in 100 x 100 pixel images were obtained, and a statistical analysis was performed. The average percentage of microtubule area was 30.1% (SD=6.3%, N=26) in the afp4a-sb-miMO embryos, whereas it was only 24.1% (SD=6.6%, N=23) in the afp4a-sb-MO morphants (Fig. 5E), indicating that the stability of microtubules is reduced in the afp4a-sb-MO morphants.
To confirm that the epiboly delay resulted from the microtubule disruption, we used pregnenolone (P5, 0.1, 1, 10 and 20 µM) to incubate the afp4a-sb-MO morphants, since pregnenolone was demonstrated to promote epiboly by stabilizing microtubules [6]. Significantly, when afp4a-sb-miMO embryos reached to 78.2% epiboly (SD=4.8%, N=167), the blastoderms only covered 57.4% yolk cells (SD=5.4%, N=156) in the afp4a-sb-MO morphants. In addition, the epibolic defect of afp4a-sb-MO morphants could be partially rescued by the pregnenolone treatment, in which the significant rescuing increases were observed from 1 to 10 μM pregnenolone treatment (66.3% ± 7.2%, N=173; 74.1% ± 7.3%, N=159). However, a rescuing decrease was observed in 20 μM treatment (61.4% ± 6.0%, N=180) (Fig. 5F). A similar epibolic defect was also observed in embryos treated by high dose of microtubule stabilizing agent taxol [48]. In this respect, this decrease could be interpreted as a result of too stable microtubule arrays. These results indicate that YCL microtubule cytoskeleton is disturbed in the afp4a morphants, suggesting that AFP4a might participate in epiboly progression by stabilizing YCL microtubules.
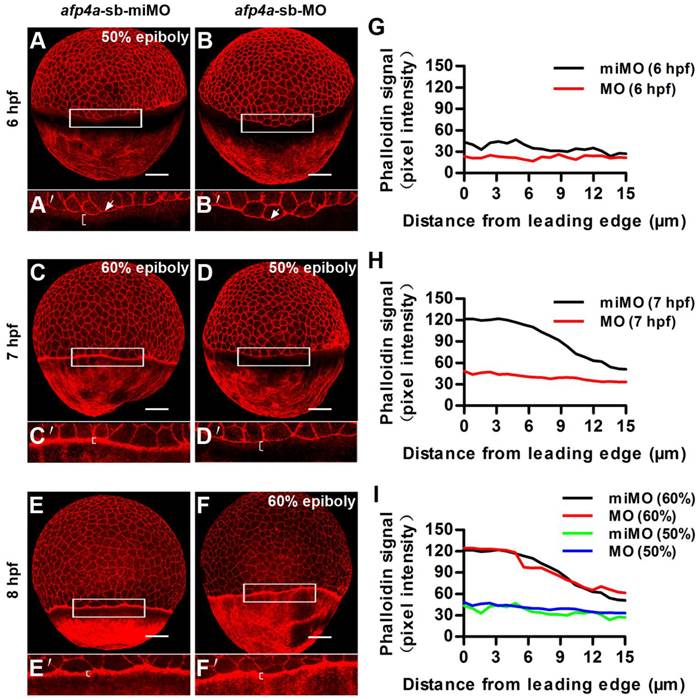
Another actin cytoskeleton structure, including F-actin rings at the vegetal margin of deep cells and EVL, and punctuate actin band in the external-YSL lying vegetal to the EVL leading margin, which form after 50% epiboly, had been revealed to be an important drive of the second half epiboly [43]. To reveal whether the actin cytoskeleton was also affected in the afp4a morphants, we performed F-actin staining with rhodamine-phalloidin. In afp4a-sb-miMO embryos, the punctuate actin band initiated to form at 6 hpf and became very clear at 7 hpf (Fig. 6A, C and E), whereas in the afp4a-sb-MO morphants, it began to appear at 7 hpf and displayed obviously at 8 hpf (Fig. 6B, D and F). This difference might be resulted from epiboly progression delay. Quantitative analyses indicated that there were obviously different phalloidin signals between the afp4a-sb-miMO embryos and the afp4a morphants at their corresponding 6 hpf and 7 hpf (Fig. 6G, H), but basic similar phalloidin intensity signals were observed from their same development stages, when 50% or 60% epiboly embryos were comparatively analyzed (Fig. 6I). The results suggest that the actin cytoskeleton should be not affected in the afp4a morphants, and AFP4a might specifically interact with YCL microtubules to participate in epiboly progression.
Formation of the punctate actin band in YSL seems to be timed by an epiboly-dependent clock in afp4a-sb-MO morphants. (A-F) afp4a-sb-miMO embryos (A, C, E) and the same-age afp4a-sb-MO embryos (B, D, F) at 6 hfp (A, B), 7 hpf (C, D) and 8 hpf (E, F) stained with rhodamine-phalloidin (F-actin). Among them, afp4a-sb-miMO morphants at 6 hpf (A) and afp4a-sb-MO morphants at 7 hpf (D) reach about 50% epiboly, and afp4a-sb-miMO morphants at 7 hpf (C) and afp4a-sb-MO morphants at 8 hpf (F) reach about 60% epiboly. (A'-F') are the enlarged images of the areas in the rectangle in corresponding (A-F). Embryos are oriented with their animal pole up; brackets in A'-F' indicate the punctuate actin bands; white arrows in A' and B' indicate leading edge of the EVL; scale bars, 100 µm. (G-I) Average intensity of phalloidin signal in the YSL along the EVL margin in afp4a-sb-miMO (miMO) embryos and afp4a-sb-MO (MO) morphants at 6 hpf (G), 7 hpf (H) and 50% and 60% epiboly (I). The intensity of the phalloidin signal was plotted along a line perpendicular to the EVL margin. Average plots of ten embryos for each group are shown.
AFP4b is required for CE movement without affecting dorsoventral axis patterning
To investigate the role of AFP4b in embryogenesis, we synthesized a 5-base mismatched control MO to afp4b-tb-MO (afp4b-tb-miMO), and observed a normal embryo phenotype similar to the Ctrl-MO injection at same concentration of 2.5 ng/embryo (data not shown). So, the afp4b-tb-miMO was served as a control of the afp4b-tb-MO in subsequent research about the function of AFP4b. In comparison with control afp4b-tb-miMO embryos, the somite width was significantly enhanced in the afp4b-tb-MO morphants at 8-somite stage, as revealed by myoD probe. This abnormity could be rescued only by co-injection of 150 pg afp4b-mRNA, but not by the same amount of afp4a-mRNA (Fig. 7A). A quantitative and statistical analysis showed a significant increase of average somite width (251.4 ± 45.2 μm, N=176, P < 0.01) in the afp4b-tb-MO morphants relative to that in the afp4b-tb-miMO embryos (192.0 ± 10.3 μm, N=160). Moreover, this increase could be effectively recovered by the afp4b-mRNA co-injection (195.6 ± 40.1 μm, N=149), but could not be rescued by the same amount of afp4a-mRNA (245.6 ± 37.6 μm, N=153) (Fig. 7B). These data indicate that the somite defect is resulted from specific reduction of AFP4b expression.
The above somite defect implied that AFP4b might be involved in CE movements, because it was very similar to the previous observation in has2 and rhoA morphants [49, 50]. For this reason, we firstly used dorsal (gsc) and ventral (eve1) makers to detect dorsoventral patterning change in the afp4b-tb-MO morphants. In comparison with afp4b-tb-miMO embryos, no any obvious change was observed in the afp4b-tb-MO morphants at 75% epiboly (Fig. 7C, D), indicating that AFP4b knockdown did not disrupt dorsoventral axis patterning and differentiation.
Reduced AFP4b expression affects CE movement. (A) myoD expression in 8-somite groups injected with afp4b-tb-miMO, afp4b-tb-MO, afp4b-tb-MO plus afp4b-mRNA, or afp4b-tb-MO plus afp4a-mRNA. Dorsal views of embryos, anterior to the top. (B) Average mediolateral length of somites in each group. Error bars show SD. N, the number of embryos that were analyzed from three independent experiments. *P < 0.01. (C-I) Dorsoventral pattern is normal, but CE is affected in afp4b-tb-MOs morphants showing by whole-mount in situ hybridization. In each section, expression of marker genes in afp4b-tb-miMO morphants (left) and afp4b-tb-MO morphants (right) are shown. (C) gsc, 75% epiboly stage. (D) eve1, 75% epiboly stage. (E) sox17, bud stage. (C-E) Lateral views, dorsal to the right. (F) hgg1 and dlx3b, bud stage, top view, ventral up. (G) ntl, bud stage, dorsal view, animal pole to the top. (H and I) pax2.1 and myoD, 9-somite stage. (H) Flat-mounted embryos, head to the left. os, optic stalk; m, the midbrain hindbrain boundary; o, otic vesicles; p, pronephros. (I) Lateral view, dorsal to the right.
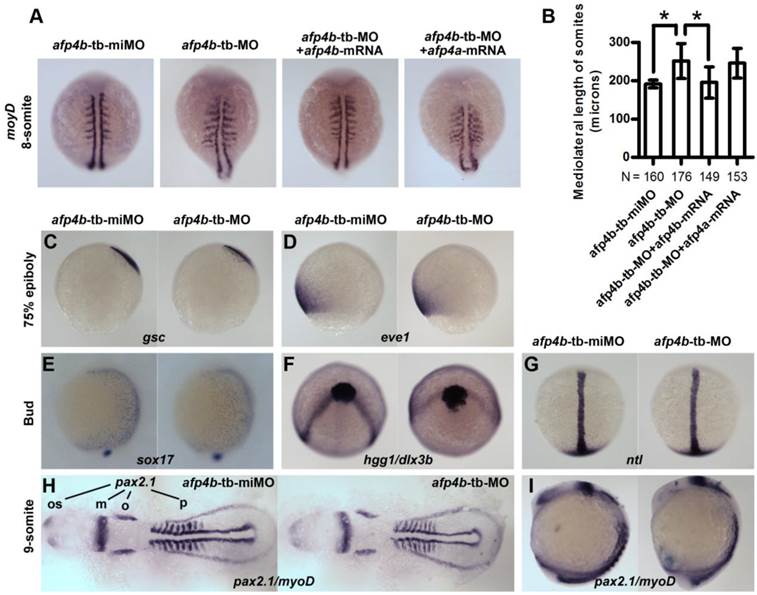
Moreover, we detected CE movements by marker genes specific to endoderm (sox17), axial and paraxial mesoderm (hgg1, ntl, myoD), and neuroectoderm (dlx3b and pax2.1). The sox17-labeled endoderm cells moved slower dorsally during gastrulation in afp4b-tb-MO morphants, as indicated by the earlier dorsoventral expression pattern of sox17 at the end of gastrulation (Fig. 7E). Thus, the CE defect in afp4b-tb-MO morphants might be associated with dorsal migration of ventrolateral endoderm cells. In addition, the neural plate was much wider in afp4b-tb-MO morphants, as reflected by the laterally expansion expression domain of dlx3b at bud stage (Fig. 7F) and pax2.1 (the midbrain hindbrain boundary, the otic vesicles) at 9-somite stage (Fig. 7H). Thus, the CE movement of neuroectoderm was delayed in afp4b-tb-MO morphants. Moreover, afp4b-tb-MO morphants showed indistinguishable notochord from control embryos, as revealed by the expression of ntl at bud stage (Fig. 7G). However, afp4b morphants displayed a broader somatic mesoderm and short head to tail distance during somitogenesis, as represented by the expression patterns of myoD and pax2.1 (the optic stalk to the posterior end of the pronephros) in Flat-mounted embryos at 9-somite stage (Fig. 7H). At the same time, afp4b-tb-MO morphants still showed a longer anterior-posterior axis (Fig. 7I). Similar observations were also reported in research of has2 and npc1 [7, 49]. Therefore, the CE movement of axial mesoderm was least affected, but that of paraxial mesoderm was significantly changed in afp4b-tb-MO morphants. The above data indicate that CE movement of ventrolateral cells is severely impaired, whereas CE movement of axial cells and dorsoventral axis patterning are normal after the AFP4b knockdown, implying that AFP4b is required for CE movement without affecting dorsoventral axis patterning.
AFP4b is primarily related to convergence
To directly clarify how CE movements are affected by AFP4b depletion during gastrulation, we respectively injected Kaede mRNA with afp4b-tb-miMO or afp4b-tb-MO into one-cell stage embryos. Then, Kaede protein was activated from green to a bright and stable red fluorescence by UV illumination at a specific time and in a specific cell group, and cell movements were traced as previously described [41]. Firstly, the lateral marginal cells, 90° from the dorsal shield, were activated at 6 hpf by a focused UV pulse (Fig. 8A and D), and the movement trajectories of the labeled red fluorescence cells were traced. In comparison with normal movement trajectories in afp4b-tb-miMO embryos (Fig. 8B and C), the labeled red fluorescence cells in the afp4b-tb-MO embryos underwent normal animal and vegetal migrations, but the movement towards the dorsal side was severely affected, and the labeled cells did not reach to the midline (Fig. 8E and F).
Cell tracing experiments. (A-F) Distribution of labeled lateral cells in afp4b-tb-miMO embryos (A-C) and afp4b-tb-MO embryos (D-F) at 6 hpf (A, D), 8 hpf (B, E) and 10 hpf (C, F). (G-L) Distribution of labeled dorsal cells in afp4b-tb-miMO embryos (G-I) and afp4b-tb-MO embryo (J-L) at 6 hpf (G, J), 8 hpf (H, K) and 10 hpf (I, L). (A-L) Lateral views, animal pole is up and dorsal is to the right. (M) Graph displaying the average degree from the dorsal axis to labeled cells (indicated by angle) at 6 hpf, 8 hpf and 10 hpf, **P < 0.001. (N) Graph displaying the average extension in labeled dorsal cells (indicated by arc) at 6 hpf, 8 hpf and 10 hpf, *P < 0.05. Ten embryos per treatment were evaluated, and error bars show SD.
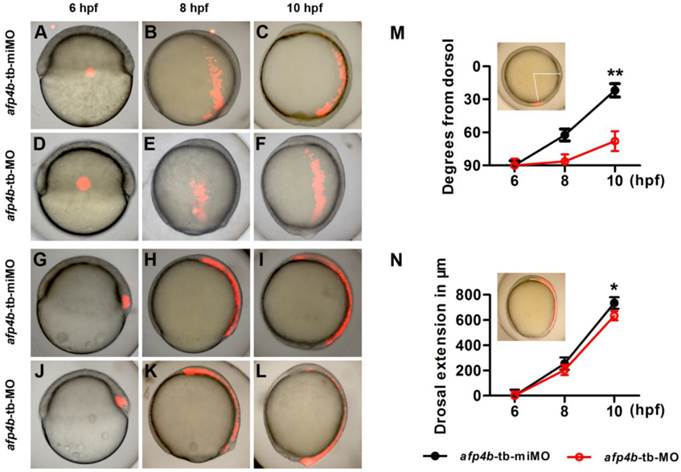
A quantitative and statistical analysis showed a significant difference of distance (P < 0.001) from dorsal axis between afp4b-tb-miMO (22 ± 6°) embryos and afp4b-tb-MO morphants (68 ± 9°) at 10 hpf (Fig. 8M), implicating that AFP4b might be related to convergent movement of lateral cells towards the dorsal side.
To measure the relationship between AFP4b and dorsal extension movement, a group of cells within the dorsal embryonic shield was marked at 6 hpf (Fig. 8G, J), and the movement of labeled cells was recorded at 8 hpf and 10 hpf. In both the afp4b-tb-miMO embryos and the afp4b-tb-MO morphants at 10 hpf, the marked cells were similarly distributed in the dorsal axial cells, and only a slight dorsal extension delay was observed in the afp4b-tb-MO morphants (Fig. 8G-L). A quantitative and statistical analysis also showed a slight reduction (P < 0.05) in the dorsal extension of the afp4b-tb-MO morphants (634 ± 36 μm) relative to the afp4b-tb-miMO embryos (735 ± 46 μm) (Fig. 8N). Together, these data indicate that AFP4b is primarily related to convergence movement.
Discussion
In this study, we have identified two head-to-tail tandem duplicated afp4 genes (afp4a and afp4b) from gibel carp and zebrafish, and found that they possess similar genomic structures and protein sequences. Comparison of cDNA sequences between afp4a and afp4b shows high similarity in their ORF sequences, whereas significant differences exist in their UTR sequences and promoter sequences. RT-PCR analysis and WISH have revealed differential expression patterns and dynamic changes between afp4a and afp4b during embryogenesis, early larval development and adults. Thereby, we have found for the first time that both afp4a and afp4b are specifically expressed in YSL, but the later expressed afp4b exists only in embryogenesis, whereas afp4a expresses continuously in YSL and digestive system from early embryos to adults. Subsequently, we have studied the roles of AFP4a and AFP4b by using afp4a-specific and afp4b-specifc morpholino knockdown approaches, and found that AFP4a and AFP4b contribute to early morphogenetic movements during zebrafish embryogenesis. Moreover, we have observed that the YCL microtubule cytoskeleton is disturbed in the afp4a morphants, while the actin cytoskeleton is not affected in the afp4a morphants, suggesting that AFP4a might participate in epiboly progression by stabilizing YCL microtubules. In addition, we have revealed that AFP4b is required for CE movement without affecting dorsoventral axis patterning, and demonstrated that AFP4b is primarily related to convergence movement. Therefore, this current study has confirmed that both AFP4a and AFP4b are key regulators during zebrafish embryogenesis, and contribute to epiboly progression and convergence movement, respectively.
AFP4 was firstly isolated from the serum of longhorn sculpin in 1997 [23], and genomic structure of afp4 had not been described until 2011 by Lee et al [25]. Previously, two afp4 homologs have been reported from databases of three-spined stickleback (Gasterosteus aculeatus) and Atlantic salmon (Salmo salar) [25], but their distributions in the chromosomes have not been clarified. Here, for the first time, we have shown the genomic organization and the head-to-tail tandem distribution of the two duplicated afp4s in gibel carp and zebrafish. Gene duplication has been believed to be a major evolutionary driving force for organism complexity, and expression divergence has been proposed as the first step of functional divergence between duplicate genes [51]. The afp4a and afp4b are highly similar in their ORFs, but significant differences are found in their UTR sequences and promoter sequences, and differential expression patterns between afp4a and afp4b are also observed during their embryogenesis and early larval development. These differences should be consistent with their functional divergence. Previous studies have demonstrated that the cis-regulatory elements in the promoters can result in expression divergence [52] and the divergent sequences in 3' UTR also affect the expression pattern of duplicate genes [53, 54].
Along with AFP4s have been identified in temperate, subtropical and tropical fishes that have no need to prevent freezing [20, 24, 25], their biological roles have been speculated to bind to lipid or ligand other than ice because of the helix bundle structure similarity to certain apolipoproteins (Apos) [24, 26, 28], but the exact physiological functions have been unknown up to the present. Our current study has revealed the biological roles of AFP4a and AFP4b in embryonic development, and has confirmed that AFP4a and AFP4b contribute to epiboly progression and convergence movement during early zebrafish embryogenesis. Recently, some Apos, such as Apo-14 [55, 56], ApoA-II [57], ApoB [58], ApoC1 [45], have been also reported to participate in embryonic morphogenesis and organogenesis in gibel carp and zebrafish. Significantly, homological searches also show about 20% identities between AFP4s and some Apos that include mammalian ApoA-II and fish Apo-14, and the conserved amphipathic alpha-helices within AFP4s are also a common property in Apos. Therefore, we propose that afp4s might share a common ancestral gene with some apos genes, and still maintain the original function in temperate, subtropical and tropical fishes. In addition, afp4a and afp4b show similar expression patterns with Apo-14 [55] and ApoC1 [45] during embryogenesis, and all of them are expressed in YSL. As it has been previously reported that some genes expressed in YSL function during early embryonic morphogenesis [45, 55, 57-60], AFP4a and AFP4b also play the related roles.
Another significant finding in this study is about the association between AFP4a and YCL microtubule cytoskeleton, because our results have revealed that the afp4a-MO morphants display an obvious defect in epiboly progression, and YCL microtubule arrays are less stable in afp4a-MO morphants. Previous studies have indicated that microtubule arrays in YCL are important drive of epiboly, and their homeostasis is crucial for normal epibolic movement [47, 48]. Two groups of related factors have been identified. The first group, which can stabilize YCL microtubules, includes Cyp11a1 [6] and calcium channel β4 subunits (CACNB4) [60]. The second group, which are involved in microtubules organization, includes pou domain, class 5, transcription factor 1 (Pou5f1) [61] and eomesodermin A (Eomesa) [62]. Of these epiboly mutants and morphants, afp4a-MO morphants most closely resemble cyp11a1-MO morphants. Firstly, afp4a and cyp11a1 have the common expression domain in YSL. Secondly, they both display serious epibolic defect and slight CE defect, but do not change germ layer patterning and involution. Thirdly, stability of YCL microtubules is lowered in both embryos. In addition, pregnenolone, the catalytic product of Cyp11a1, can partially rescue the epibolic defect of AFP4a-knockdown embryos. Our study indicates that AFP4a either directly or indirectly stabilizes the microtubules in YCL, which extends our knowledge of YCL microtubule regulation.
Supplementary Material
Figures S1 - S4, Tables S1 - S2.
Abbreviations
AFP: Antifreeze protein; AFP4: type-IV antifreeze proteins; afp4: type-IV antifreeze protein gene; Cag: Carassius auratus gibelio; Dr: Danio rerio; Cagafp4: afp4 in gibel carp; Drafp4s: afp4s in zebrafish; YSL: yolk syncytial layer; YCL: yolk cytoplasmic layer; CE: convergence and extension; RACE: rapid amplification of cDNA ends; WT: wild type; qPCR: Real-time PCR; ML: maximum likelihood; WISH: whole-mount in situ hybridization; SD: standard deviation; ANOVA: analysis of variance; UTR: untranslated region; ORF: open reading frame; UV: ultraviolet; hpf: hours postfertilization; EVL: enveloping layer; Apo: apolipoprotein.
Acknowledgements
This work was funded by the National Key Basic Research Program of China (2010CB126301), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA08030201), the earmarked fund for Modern Agro-industry Technology Research System (NYCYTX-49), the Innovation Project of Chinese Academy of Sciences (KSCX3-EW-N-04), and the Autonomous Project of the State Key Laboratory of Freshwater Ecology and Biotechnology (2011FBZ17).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Keller R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 2002;298:1950-4
2. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253-310
3. Solnica-Krezel L, Sepich DS. Gastrulation: making and shaping germ layers. Annu Rev Cell Dev Biol. 2012;28:687-717
4. Solnica-Krezel L. Gastrulation in zebrafish — all just about adhesion? Curr Opin Genet Dev. 2006;16:433-41
5. Rohde LA, Heisenberg CP. Zebrafish gastrulation: cell movements, signals, and mechanisms. Int Rev Cytol. 2007;261:159-92
6. Hsu HJ, Liang MR, Chen CT, Chung BC. Pregnenolone stabilizes microtubules and promotes zebrafish embryonic cell movement. Nature. 2006;439:480-3
7. Schwend T, Loucks EJ, Snyder D, Ahlgren SC. Requirement of Npc1 and availability of cholesterol for early embryonic cell movements in zebrafish. J Lipid Res. 2011;52:1328-44
8. Yin C, Ciruna B, Solnica-Krezel L. Convergence and extension movements during vertebrate gastrulation. Curr Top Dev Biol. 2009;89:163-92
9. Dong CH, Yang ST, Yang ZA, Zhang L, Gui JF. A C-type lectin associated and translocated with cortical granules during oocyte maturation and egg fertilization in fish. Dev Biol. 2004;265:341-54
10. Peng JX, Xie JL, Zhou L, Hong YH, Gui JF. Evolutionary conservation of Dazl genomic organization and its continuous and dynamic distribution throughout germline development in gynogenetic gibel carp. J Exp Zool Part B (Mol Dev Evol). 2009;312B:855-71
11. Wu N, Yue HM, Chen B, Gui JF. Histone H2A has a novel variant in fish oocytes. Biol Reprod. 2009;81:275-83
12. Yue HM, Li Z, Wu N, Liu Z, Wang Y, Gui JF. Oocyte-specific H2A variant H2af1o is required for cell synchrony before mid-blastula transition in early zebrafish embryos. Biol Reprod. 2013;89:82
13. Gui JF, Zhu ZY. Molecular basis and genetic improvement of economically important traits in aquaculture animals. Chin Sci Bull. 2012;57:1751-60
14. Zhou L, Wang Y, Gui JF. Genetic evidence for gonochoristic reproduction in gynogenetic silver crucian carp (Carassius auratus gibelio Bloch) as revealed by RAPD assays. J Mol Evol. 2000;51:498-506
15. Yang L, Gui JF. Positive selection on multiple antique allelic lineages of transferrin in the polyploid Carassius auratus. Mol Biol Evol. 2004;21:1264-77
16. Gui JF, Zhou L. Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploid Carassius auratus gibelio. Sci China Life Sci. 2010;53:409-15
17. Wang ZW, Zhu HP, Wang D, Jiang FF, Guo W, Zhou L. et al. A novel nucleo-cytoplasmic hybrid clone formed via androgenesis in polyploid gibel carp. BMC Res notes. 2011;4:82
18. Jiang FF, Wang ZW, Zhou L, Jiang L, Zhang XJ, Apalikova OV. et al. High male incidence and evolutionary implications of triploid form in northeast Asia Carassius auratus complex. Mol Phylogenet Evol. 2013;66:350-9
19. Zhai YH, Zhou L, Wang Y, Wang ZW, Li Z, Zhang XJ. et al. Proliferation and resistance difference of a liver-parasitized myxosporean in two different gynogenetic clones of gibel carp. Parasitol Res. 2014;113:1331-41
20. Liu JX, Zhai YH, Gui JF. Molecular characterization and expression pattern of AFPIV during embryogenesis in gibel carp (Carassiu auratus gibelio). Mol Biol Rep. 2009;36:2011-8
21. Doxey AC, Yaish MW, Griffith M, McConkey BJ. Ordered surface carbons distinguish antifreeze proteins and their ice-binding regions. Nat Biotechnol. 2006;24:852-5
22. Ewart K, Lin Q, Hew C. Structure, function and evolution of antifreeze proteins. Cell Mol Life Sci. 1999;55:271-83
23. Deng G, Andrews DW, Laursen RA. Amino acid sequence of a new type of antifreeze protein, from the longhorn sculpin Myoxocephalus octodecimspinosis. FEBS Lett. 1997;402:17-20
24. Gauthier SY, Scotter AJ, Lin FH, Baardsnes J, Fletcher GL, Davies PL. A re-evaluation of the role of type IV antifreeze protein. Cryobiology. 2008;57:292-6
25. Lee JK, Kim YJ, Park KS, Shin SC, Kim HJ, Song YH. et al. Molecular and comparative analyses of type IV antifreeze proteins (AFPIVs) from two Antarctic fishes, Pleuragramma antarcticum and Notothenia coriiceps. Comp Biochem Physiol B Biochem Mol Biol. 2011;159:197-205
26. Deng G, Laursen RA. Isolation and characterization of an antifreeze protein from the longhorn sculpin, Myoxocephalus octodecimspinosis. Biochim Biophys Acta. 1998;1388:305-14
27. Goetz FW, McCauley L, Goetz GW, Norberg B. Using global genome approaches to address problems in cod mariculture. Ices J Mar Sci. 2006;63:393-9
28. Breton TS, Anderson JL, Goetz FW, Berlinsky DL. Identification of ovarian gene expression patterns during vitellogenesis in Atlantic cod (Gadus morhua). Gen Comp Endocr. 2012;179:296-304
29. Celik Y, Drori R, Pertaya-Braun N, Altan A, Barton T, Bar-Dolev M. et al. Microfluidic experiments reveal that antifreeze proteins bound to ice crystals suffice to prevent their growth. Proc Natl Acad Sci USA. 2013;110:1309-14
30. Fletcher GL, Hew CL, Davies PL. Antifreeze proteins of teleost fishes. Annu Rev Physiol. 2001;63:359-90
31. Cheng C-HC, Detrich HW. Molecular ecophysiology of Antarctic notothenioid fishes. Philos T Roy Soc B. 2007;362:2215-32
32. Geng FS, Zhou L, Gui JF. Construction and characterization of a BAC library for Carassius auratus gibelio, a gynogenetic polyploid fish. Anim Genet. 2005;36:535
33. Xu H, Lim M, Dwarakanath M, Hong Y. Vasa identifies germ cells and critical stages of oogenesis in the Asian seabass. Int J Biol Sci. 2014;10:225-35
34. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725-9
35. Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104-5
36. Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). Eugene: University of Oregon Press. 1995
37. Huang W, Zhou L, Li Z, Gui JF. Expression pattern, cellular localization and promoter activity analysis of ovarian aromatase (Cyp19a1a) in protogynous hermaphrodite red-spotted grouper. Mol Cell Endocrinol. 2009;307:224-36
38. Mei J, Zhang QY, Li Z, Lin S, Gui JF. C1q-like inhibits p53-mediated apoptosis and controls normal hematopoiesis during zebrafish embryogenesis. Dev Biol. 2008;319:273-84
39. Mei J, Yue HM, Li Z, Chen B, Zhong JX, Dan C. et al. C1q-like factor, a target of miR-430, regulates primordial germ cell development in early embryos of Carassius auratus. Int J Biol Sci. 2014;10:15-24
40. Li Z, Korzh V, Gong Z. Localized rbp4 expression in the yolk syncytial layer plays a role in yolk cell extension and early liver development. BMC Dev Biol. 2007;7:117
41. Lou QY, He JY, Hu L, Yin Z. Role of lbx2 in the noncanonical Wnt signaling pathway for convergence and extension movements and hypaxial myogenesis in zebrafish. BBA-Mol Cell Res. 2012;1823:1024-32
42. Zhong JX, Zhou L, Li Z, Wang Y, Gui JF. Zebrafish Noxa promotes mitosis in early embryonic development and regulates apoptosis in subsequent embryogenesis. Cell Death Differ. 2014;21:1013-24
43. Cheng JC, Miller AL, Webb SE. Organization and function of microfilaments during late epiboly in zebrafish embryos. Dev Dyn. 2004;231:313-23
44. Köppen M, Fernandez BG, Carvalho L, Jacinto A, Heisenberg CP. Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development. 2006;133:2671-81
45. Wang Y, Zhou L, Li Z, Li WH, Gui JF. Apolipoprotein C1 regulates epiboly during gastrulation in zebrafish. Sci China Life Sci. 2013;56:975-84
46. Carvalho L, Heisenberg CP. The yolk syncytial layer in early zebrafish development. Trends Cell Biol. 2010;20:586-92
47. Strahle U, Jesuthasan S. Ultraviolet irradiation impairs epiboly in zebrafish embryos: evidence for a microtubule-dependent mechanism of epiboly. Development. 1993;119:909-19
48. Solnica-Krezel L, Driever W. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 1994;120:2443-55
49. Bakkers J, Kramer C, Pothof J, Quaedvlieg NEM, Spaink HP, Hammerschmidt M. Has2 is required upstream of Rac1 to govern dorsal migration of lateral cells during zebrafish gastrulation. Development. 2004;131:525-37
50. Zhu S, Liu L, Korzh V, Gong Z, Low BC. RhoA acts downstream of Wnt5 and Wnt11 to regulate convergence and extension movements by involving effectors Rho Kinase and Diaphanous: Use of zebrafish as an in vivo model for GTPase signaling. Cell Signal. 2006;18:359-72
51. Ohno S. Evolution by gene duplication. New york: Springer-Verlag. 1970
52. Li J, Yuan Z, Zhang Z. Revisiting the contribution of cis-elements to expression divergence between duplicated genes: the role of chromatin structure. Mol Biol Evol. 2010;27:1461-6
53. Tong Y, Zheng K, Zhao SF, Xiao GX, Luo C. Sequence divergence in the 3'-untranslated region has an effect on the subfunctionalization of duplicate genes. J Exp Zool Part B (Mol Dev Evol). 2012;318B:531-44
54. Decembrini S, Andreazzoli M, Vignali R, Barsacchi G, Cremisi F. Timing the generation of distinct retinal cells by homeobox proteins. PLoS Biol. 2006;4:e272
55. Xia JH, Liu JX, Zhou L, Li Z, Gui JF. Apo-14 is required for digestive system organogenesis during fish embryogenesis and larval development. Int J Dev Biol. 2008;52:1089-98
56. Choudhury M, Yamada S, Komatsu M, Kishimura H, Ando S. Homologue of mammalian apolipoprotein A-II in non-mammalian vertebrates. Acta Bioch Bioph Sin. 2009;41:370-8
57. Zhang T, Yao S, Wang P, Yin C, Xiao C, Qian M. et al. Apoa-II directs morphogenetic movements of zebrafish embryo by preventing chromosome fusion during nuclear division in yolk syncytial layer. J Biol Chem. 2011;286:9514-25
58. Seth A, Machingo QJ, Fritz A, Shur BD. Core fucosylation is required for midline patterning during zebrafish development. Dev Dyn. 2010;239:3380-90
59. Chen SR, Kimelman D. The role of the yolk syncytial layer in germ layer patterning in zebrafish. Development. 2000;127:4681-9
60. Ebert AM, McAnelly CA, Srinivasan A, Linker JL, Horne WA, Garrity DM. Ca2+ channel-independent requirement for MAGUK family CACNB4 genes in initiation of zebrafish epiboly. Proc Natl Acad Sci USA. 2008;105:198-203
61. Lachnit M, Kur E, Driever W. Alterations of the cytoskeleton in all three embryonic lineages contribute to the epiboly defect of Pou5f1/Oct4 deficient MZspg zebrafish embryos. Dev Biol. 2008;315:1-17
62. Du S, Draper BW, Mione M, Moens CB, Bruce A. Differential regulation of epiboly initiation and progression by zebrafish Eomesodermin A. Dev Biol. 2012;362:11-23
Author contact
![]() Corresponding author: Tel: +86-27-68780707; Fax: +86-27-68780123; E-mail: jfguiac.cn.
Corresponding author: Tel: +86-27-68780707; Fax: +86-27-68780123; E-mail: jfguiac.cn.

 Global reach, higher impact
Global reach, higher impact