10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):863-871. doi:10.7150/ijbs.24588 This issue Cite
Research Paper
Identification of Inhibitors of MMPS Enzymes via a Novel Computational Approach
1. School of Computer Science and Technology, Tianjin University, Tianjin 300350, China;
2. Tianjin University Institute of Computational Biology, Tianjin University, Tianjin 300350, China;
3. Department of Computer Science and Engineering, University of South Carolina, Columbia, SC 29208, USA;
4. School of Chemical Engineering and Technology, Tianjin University, Tianjin 300350, China.
Abstract
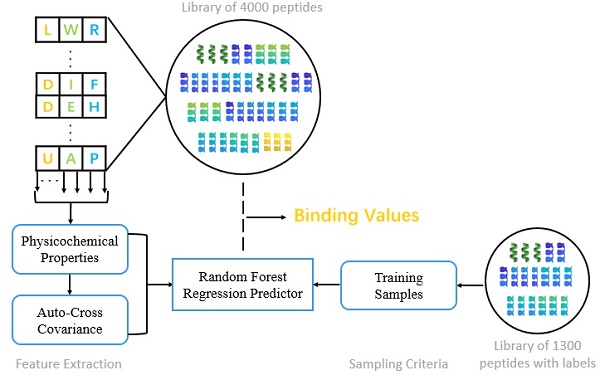
Matrix metalloproteases (MMPs) are a family of zinc-dependent proteinases that play complex and diverse roles in metabolism, which are vital for physiological development. In this paper, we present a novel method to identify peptide binding to seven matrix metalloproteases. First, we propose a novel sampling criteria for constructing a training set for each new peptide motif. Then, we select nine physicochemical properties of amino acids and compute their auto-cross covariance to effectively extract features for both natural and non-natural amino acids. Finally, we adopt random forest to predict binding values of each peptide motif respectively with seven MMPs. Our method verifies on 1300 known peptide motifs binding to seven MMPs and achieved preeminent Pearson-product-moment correlation coefficient (PCC) and root mean squared error (RMSE) on all seven MMPs, especially of 0.9181 and 9.3827 on MMP-7. We predict binding values of 4000 peptide motifs and identify peptides preferentially bind to MMP-2 and MMP-7. We herein report 4 novel inhibitor candidates of Asp-Ile-Phe, Asp-Ile-Tyr, Asp-Ile-Lys and Hser-Gly-Phe with high potency and selectivity binding to MMP-2, as well as 6 novel inhibitor candidates of Chg-Ile-Ile, Chg-Ile-Leu, Chg-Ile-Glu, Chg-Ile-Met, Chg-Val-Ile and Chg-Val-Leu selectively binding to MMP-7. Our findings facilitate the identification of inhibitors with good potency as well as desirable selectivity, providing significant insights of candidate inhibitor drugs.
Keywords: MMPs, peptide inhibitors, auto-cross covariance, random forest

 Global reach, higher impact
Global reach, higher impact