ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2019; 15(7):1452-1459. doi:10.7150/ijbs.31957 This issue Cite
Research Paper
SAROTUP: a suite of tools for finding potential target-unrelated peptides from phage display data
1. School of Medicine, Guizhou University, Guiyang 550025, China
2. Center for Informational Biology, University of Electronic Science and Technology of China, Chengdu 611731, China
Abstract
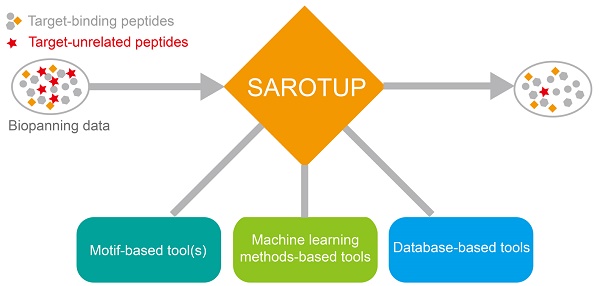
SAROTUP (Scanner And Reporter Of Target-Unrelated Peptides) 3.1 is a significant upgrade to the widely used SAROTUP web server for the rapid identification of target-unrelated peptides (TUPs) in phage display data. At present, SAROTUP has gathered a suite of tools for finding potential TUPs and other purposes. Besides the TUPScan, the motif-based tool, and three tools based on the BDB database, i.e., MimoScan, MimoSearch, and MimoBlast, three predictors based on support vector machine, i.e., PhD7Faster, SABinder and PSBinder, are integrated into SAROTUP. The current version of SAROTUP contains 27 TUP motifs and 823 TUP sequences. We also developed the standalone SAROTUP application with graphical user interface (GUI) and command line versions for processing deep sequencing phage display data and distributed it as an open source package, which can perform perfectly locally on almost all systems that support C++ with little or no modification. The web interfaces of SAROTUP have also been redesigned to be more self-evident and user-friendly. The latest version of SAROTUP is freely available at
Keywords: target-unrelated peptide, phage display, biopanning, high-throughput sequencing, computational toolkit
