10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(12):3781-3803. doi:10.7150/ijbs.85063 This issue Cite
Research Paper
Intergenic CircRNA Circ_0007379 Inhibits Colorectal Cancer Progression by Modulating miR-320a Biogenesis in a KSRP-Dependent Manner
1. Department of Gastrointestinal Surgery, The Third Xiangya Hospital, Central South University, Changsha, Hunan 410013, China.
2. Postdoctoral Research Station of Basic Medicine, The Third Xiangya Hospital, Central South University, Changsha, Hunan 410013, China.
3. Department of Gastrointestinal, Hernia and Enterofistula Surgery, People's Hospital of Guangxi Zhuang Autonomous Region, Nanning, Guangxi 530000, China.
4. School of Basic Medical Science, Central South University, Changsha, Hunan 410078, China.
† These authors contributed equally.
Received 2023-4-7; Accepted 2023-7-8; Published 2023-7-24
Abstract
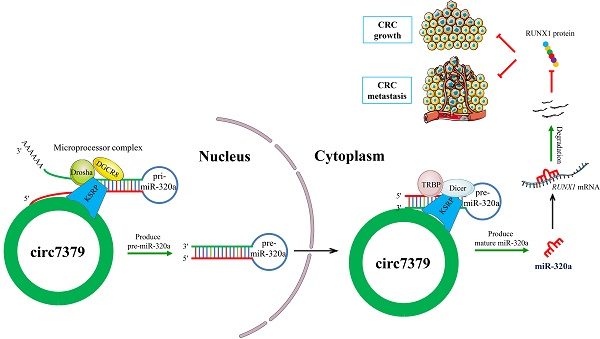
Circular RNAs (circRNAs) are covalently closed RNA structures that play multiple roles in tumorigenesis and progression. Compared with exon‒intron circRNAs, the biological functions and implications of intergenic circRNAs in human cancer are still poorly understood. Here, we performed circRNA microarray analysis and identified an intergenic circRNA, circ_0007379, that was significantly downregulated in patients with colorectal cancer (CRC). The biogenesis of circ_0007379 was mediated by reverse complementary matches (RCMs) and was negatively regulated by the RNA helicase DHX9. Functionally, circ_0007379 suppressed CRC cell growth and metastasis in cell culture as well as in patient-derived organoid and xenograft models. Mechanistically, circ_0007379 acted as a scaffold to facilitate the processing of both pri-miR-320a and pre-miR-320a in a KSRP-dependent manner, leading to miR-320a maturation and subsequent repression of transcription factor RUNX1 expression. Thus, our findings establish a previously unrecognized function of circRNA in inhibiting CRC progression.
Keywords: Intergenic circular RNAs, Circ_0007379, Colorectal cancer, Progression, KSRP, miR-320a
Introduction
Colorectal cancer (CRC) is the third most common digestive system malignancy and the second most deadly cancer worldwide [1]. Metastatic CRC associated with late diagnosis is one of the determinants of global survival disparities and substantial CRC deaths. Although targeted therapy (e.g., cetuximab and bevacizumab) and novel immunotherapy (e.g., nivolumab, pembrolizumab, and ipilimumab) have been developed and applied for the treatment of metastatic CRC patients, the therapeutic effect is not satisfactory [2]. There is still an urgent need to elucidate the molecular mechanisms underlying the initiation and progression of CRC and identify novel targets for the precision treatment of advanced CRC.
A non-coding RNA (ncRNA) is an RNA molecule that is not translated into protein, but is involved in the regulation of transcription, splicing, and translation [3]. Circular RNAs (circRNAs), as highly conserved ncRNAs, are expressed in all cells and tissues and have been implicated in various human cancers, including CRC [4, 5]. Functionally, circRNAs have the potential to act as oncogenes or tumor suppressors by acting as miRNA sponges, interacting with RNA-binding proteins, or even serving as protein templates for translation [6]. The use or targeting of naturally occurring circRNAs could be a promising alternative to existing RNA-based therapies [7]. Compared with synthetic siRNAs or antisense oligonucleotides targeting oncogenic circRNAs, restoration therapy of tumor-suppressive circRNAs can take full advantage of their advantages (e.g., high stability and specificity, and low immunogenicity) to improve therapeutic efficacy [7, 8]. Encouragingly, synthetic and engineered circRNAs that can be stably expressed in cancer cells exhibit significant anticancer activity as miRNA sponges or protein sponges in various experimental models [9-11]. Further understanding of the types and functions of circRNAs is important for the development of new anticancer strategies.
Intergenic circRNAs are non-exonic circRNAs that currently have many members due to the development of RNA sequencing technology and bioinformatics. Unlike linear RNAs, they contain two intronic circRNA segments flanked by GT-AC splicing signals that act as splicing donors and acceptors for loop junctions. The expression of circRNAs is regulated by multiple factors, including splicing factors, transcription factors, specific enzymes, and cis-acting elements. CircRNAs can be used as molecular markers for early diagnosis of digestive system tumors, providing new potential therapeutic targets [12, 13]. Although individual circRNAs have been reported as oncogenes or tumor suppressors, the key circRNAs that control CRC initiation and progression remain unclear.
In this study, we aimed to screen and identify aberrant circRNAs in CRC and explore their roles in tumor biology. We identified an intergenic circRNA (circBase ID: hsa_circ_0007379, hereafter abbreviated circ7379) that is downregulated and acts as a new tumor suppressor in CRC. We characterized the mechanism and effect of circ7379 on tumor growth and metastasis, but also demonstrated the efficacy of its application in patient-derived animal models.
Materials and methods
Patients and samples
All CRC tissues and corresponding adjacent normal tissues were obtained from patients with CRC during surgery between January 2019 and December 2019 at the Third Xiangya Hospital of Central South University. The patient inclusion criteria were as follows: 1) clear diagnosis by imaging and pathology, 2) lack of preoperative chemotherapy or radiotherapy, and 3) treatment with radical resection and the presence of complete clinicopathological records. All fresh samples were quickly frozen in liquid nitrogen and stored at -80°C until use. This study was reviewed and approved by the Ethics Committee of the Third Xiangya Hospital of Central South University (No. 2017-S141), and written informed consent forms were obtained from all patients.
Cell lines and cell culture
The human normal colonic epithelial cell line (FHC) was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Human CRC cell lines (HT29, RKO, HCT116, SW480, SW620, LoVo, HCT8 and LS123) were obtained from KeyGEN BioTECH (Nanjing, Jiangsu, China). The cell lines were cultured in appropriate medium supplemented with 10% fetal bovine serum (FBS; Biological Industries, Israel) and 1% antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin; Life Technologies, Inc., Grand Island, NY, USA). All cell lines were maintained in an incubator at 37°C in a humidified atmosphere with 5% CO2. All cell experiments were performed using mycoplasma-free cells.
CircRNA microarray analysis
A circRNA microarray analysis was carried out for the genome-wide profiling of circRNAs in CRC tissues using an Arraystar Human circRNA Array V2 (8x15K, ArrayStar, Rockville, MD, USA) by a contract service at KangChen Biotech (Shanghai, China). The sample preparation and microarray hybridization were conducted according to Arraystar's standard protocols. CircRNAs with statistically significant (P<0.05) differential expression between the two groups were identified through filtering on volcano plots. A cutoff fold change (FC) of 1.5 was applied to select the markedly downregulated circRNAs.
RNA extraction, genomic DNA (gDNA) extraction and quantitative reverse transcription-PCR (qRT‒PCR)
The total RNA was extracted from tissue and cell samples using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Nuclear and cytoplasmic RNA were isolated from cells using a Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Thorold, ON, Canada; Product # 21000, 37400). The concentration and purity of all RNA samples were subsequently measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The corresponding cDNAs were generated using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan). gDNA was extracted from cells using a Genomic DNA Kit (Omega Bio-Tek, Guangzhou, China) following the manufacturer's instructions. qRT‒PCR was performed on a LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland) using a KOD SYBR® qPCR Mix Kit (TOYOBO, Osaka, Japan) according to the manufacturer's protocol. The relative quantification of circRNA and messenger RNA (mRNA) expression was carried out by the 2-ΔΔCt method, and GAPDH was used as an internal control. All primer pairs were designed and synthesized by Tsingke Biotechnology Co., Ltd. (Beijing, China), and the sequences of the forward and reverse primers are listed in Supplemental Table 1.
RNase R and actinomycin D assays
For the RNase R treatment assay, the total RNA (2 μg/group) from HCT116 and SW480 cells was incubated with 3 U/μg RNase R (Epicenter Technologies, Madison, WI, USA) for 30 min at 37°C and then purified by an RNeasy MinElute Cleaning Kit (Qiagen, Germantown, MD, USA). Subsequently, the abundance of circ7379 and GAPDH mRNA was analyzed by qRT‒PCR.
For the actinomycin D assay, HCT116 and SW480 cells were exposed to 5 µg/mL actinomycin D (Sigma‒Aldrich, St. Louis, MO, USA) to block gene transcription. Then, the cells were harvested at the indicated time points (0 h, 4 h, 8 h, 12 h, and 24 h), and the total RNA was extracted. The relative expression levels of circ7379 were measured by qRT‒PCR, and its half-life was determined.
RNA fluorescence in situ hybridization (FISH)
Cy5-labeled circ7379 probes (TTTGCAACTC+TGTGATTTCTCATCATC+TGCACTCTGGATG) were designed and synthesized by GenePharma (Suzhou, China). FISH assays were carried out using an RNA FISH Kit (GenePharma, Suzhou, China) according to the manufacturer's protocol. In brief, HCT116 and SW480 cells were seeded into confocal dishes (Biosharp, Hefei, China) and grown to 30%-50% confluence. The cells were washed with PBS and then fixed with 4% formaldehyde for 15 min at room temperature. The fixed cells were permeabilized in PBS containing 0.5% Triton X-100 for 15 min at room temperature and then blocked with 1× blocking solution for 30 min at 37°C. Subsequently, the cells were hybridized with the corresponding probes at 37°C for 12-16 h in a humidified chamber. After adding DAPI working solution and incubating for 15 min, the cells were scanned and imaged under a confocal laser scanning microscope (Zeiss, Oberkochen, Germany).
Construction of plasmids, small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs) and stable cell lines
The full-length sequence of circ7379 was amplified and cloned into a circRNA-specific overexpression lentiviral vector, GV689 (GeneChem, Shanghai, China), which contained two homology arms upstream and downstream of the circRNA sequence to promote circRNA cyclization. DHX9, RUNX1 and KSRP overexpression plasmids (GV367) were also purchased from GeneChem (Shanghai, China). Three siRNAs targeting the BSJ sites of circ7379 were designed and synthesized by GenePharma (Suzhou, China). SiRNAs targeting DHX9, RUNX1, and KSRP were also purchased from GenePharma (Suzhou, China). Lentiviruses containing shRNAs targeting circ7379 and RUNX1 were purchased from GeneChem (Shanghai, China). Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was used for the cell transfection according to the manufacturer's instructions. After the transfection, stable HCT116 and SW480 cells were selected with puromycin (3 μg/mL; Gibco, Grand Island, NY, USA) for one week, and the surviving cells were continuously cultured as stable cells. The siRNA and shRNA sequences are listed in Supplemental Table 2.
Cell counting kit-8 (CCK-8) assay
Treated cells were seeded into 96-well plates with five replicates per condition. Ten microliters of CCK-8 reagent (Dojindo Laboratories, Kumamoto, Japan) were added to each well at 0 h, 24 h, 48 h, and 72 h. Then, the cells were incubated at 37°C for 2 h. The absorbance values at 450 nm (A450 values) were measured to determine the cell viability using a BioTek microplate reader (ELX800, BioTek Instruments, Inc., USA).
Plate colony formation assay
Treated cells were seeded into 6-well plates with three replicates per group. The cultures were maintained for 10-14 days, and the medium was replaced every 4 days. After washing with PBS, the colonies were fixed with 4% paraformaldehyde for 30 min and then stained with 0.25% crystal violet for 15 min at room temperature. Finally, the colonies were imaged and counted.
Transwell assays
Transwell assays were performed to evaluate cell migration and invasion using 24-well Transwell chambers with polycarbonate membranes (pore size, 8 μm; Corning Inc., Tewksbury, MA, USA). For the cell migration assays, treated cells resuspended in 200 μL of serum-free medium were seeded into the upper compartment of each chamber, and 600 μL of complete medium supplemented with 20% FBS were added to the lower compartment.
For the cell invasion assays, the filter membranes were first precoated with BD MatrigelTM solution (BD BioSciences, Bedford, MA, USA) and incubated at 37°C for 30 min. After culture for 36 h, the cells on the upper surface of the filter membrane were gently removed with a cotton swab. Then, the cells that migrated through or invaded the membrane were fixed with 4% paraformaldehyde for 30 min and stained with 0.25% crystal violet for 15 min at room temperature. Finally, an inverted Olympus BX51 microscope (Olympus, Tokyo, Japan) was used to acquire images at 100× magnification in 5 randomly chosen fields, and ImageJ software was utilized to count the cells in the compartments.
High-throughput RNA sequencing (RNA-seq)
High-throughput RNA-seq was performed to identify the downstream target genes of circ7379 using the Illumina NovaSeq™ 6000 platform (LC-Bio Technology Co., Ltd., Hangzhou, China) by a contract service at LC-Bio (Hangzhou, China). The sample preparation and high-throughput RNA-seq were conducted according to LC-Bio's standard protocols. The differentially expressed mRNAs were identified by the R package edgeR or DESeq2 as those with |log2FC| ≥ 1 and P < 0.05, and Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the differentially expressed mRNAs were performed.
Western blot (WB) analysis and antibodies
The total protein was isolated from tissues or cells using a total protein extraction kit (KeyGEN BioTECH, Nanjing, China) following the manufacturer's protocol. Equal amounts of protein were separated by 10% SDS-PAGE and then transferred onto PVDF membranes (Millipore, CA, USA). After blocking with 5% skim milk for 2 h at room temperature, the PVDF membranes were incubated first with specific primary antibodies at 4°C overnight and then with a secondary antibody at room temperature for 1 h. Finally, the signals on the membranes were visualized using an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, NE, USA). The following antibodies were used in this study: anti-DHX9 (Proteintech, #17721-1-AP; 1:1000 dilution), anti-RUNX1 (Proteintech, #25315-1-AP; 1:1000 dilution), anti-KSRP (Bioworld Technology, #BS9961 M; 1:1000 dilution), anti-Flag (Affinity Biosciences, #T0053; 1:1000 dilution), anti-Drosha (ZEN-Bioscience, #381855; 1:1000 dilution), anti-Dicer (Proteintech, #20567-1-AP; 1:1000 dilution) and anti-GAPDH (Proteintech, #10494-1-AP; 1:5000 dilution).
Immunofluorescence (IF) analysis
Cells were seeded into confocal dishes (Biosharp, Hefei, China) and grown to 30%-50% confluence. The cells were rinsed with PBS and fixed with 4% paraformaldehyde for 30 min prior to permeabilization using 0.5% Triton X-100 in PBS for 15 min at room temperature. After blocking with 3% BSA for 30 min, the cells were incubated with a primary antibody at 4°C overnight. Subsequently, the cells were incubated with a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Beyotime, Shanghai, China) for 1 h, and the nuclei were stained with DAPI for 15 min at room temperature. Finally, images were acquired under a confocal laser scanning microscope (Zeiss, Oberkochen, Germany) and analyzed with ZEN Imaging Software 2.6 (blue edition).
RNA-binding protein (RBP) immunoprecipitation (RIP)
RIP assays were performed using an EZ-Magna RNA-Binding Protein Immunoprecipitation Kit (Merck, KGaA, Darmstadt, Germany; 17-701) following the manufacturer's instructions. In brief, approximately 2×107 cells were collected and lysed in 100% RIP lysis buffer supplemented with proteinase and RNase inhibitors. The cell lysates were then incubated with RIP buffer containing magnetic beads conjugated to antibodies or IgG (NC) at 4°C overnight. On the following day, the RNA‒protein-bead complexes were washed six times and resuspended in buffer supplemented with RNase-free DNase and proteinase K. RNA was extracted and purified by the phenol:chloroform:isoamyl alcohol (25:24:1) method. Finally, the immunoprecipitated RNAs were subjected to qRT‒PCR to calculate enrichment normalized to input.
RNA pulldown assay
The RNA pulldown assay was performed using a Pierce™ Magnetic RNA‒Protein Pull-Down Kit (Pierce Biotechnology, Rockford, IL, USA; 20164) following the manufacturer's protocol. In brief, 1×107 cells were harvested, lysed and sonicated. Biotin-labeled circ7379-specific probes or NC probes were incubated with 50 µL of prewashed streptavidin magnetic beads for 30 min at room temperature to generate probe-coated beads, which were then incubated with the cell lysates overnight at 4°C with rotation. Subsequently, the complexes were washed with washing buffer and incubated with 50 µL of elution buffer for 30 min at 37°C with agitation. Finally, the RNAs retained on the beads were isolated and further analyzed by qRT-PCR, while the proteins in the precipitated complexes were analyzed by a WB analysis using an anti-KSRP antibody.
Dual luciferase reporter assay
A dual luciferase reporter assay was performed to evaluate the direct binding between miR-320a and the RUNX1 3' untranslated region (3'UTR). Wild-type (WT) or mutant fragments of the RUNX1 3'UTR were cloned into the GV272 plasmid (GeneChem, Shanghai, China) downstream of the firefly luciferase reporter gene. Cells were seeded into 24-well plates and cultured for 24 h. Then, we cotransfected the miR-320a mimic or NC mimic, the firefly luciferase reporter plasmid, and the Renilla luciferase reporter plasmid, which served as an internal control, into cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). We also cotransfected the wild-type or mutant firefly luciferase reporter plasmid with the miR-320a or NC inhibitor into cells. Forty-eight hours later, the firefly luciferase and Renilla luciferase activities were detected using a Dual-Luciferase® Reporter Assay System (Promega Corporation, Madison, WI, USA; E1910) according to the manufacturer's protocol and an EnVision® Xcite Multimode Plate Reader (PerkinElmer, USA).
Animal experiments
Male BALB/c athymic nude mice (4-6 weeks of age, 18-20 g) were provided by the Department of Laboratory Animals of Central South University (Changsha, Hunan, China). The mice were housed in sterile individual ventilated cages (IVCs) in a barrier facility, maintained under a 12 h light/dark cycle, and provided sterilized food and water ad libitum. To establish the xenograft models, 5 × 106 transfected cells were suspended in 100 μL of Matrigel solution and subcutaneously injected into the left armpit of each mouse (n=5 mice/group). Tumor formation in the mice was observed every 3 days, and the tumor volumes and mouse weights were measured weekly. The tumor volume was calculated as 0.5×length×width2. After 4 weeks, the mice were euthanized, and the tumors were removed for further analysis.
To establish the pulmonary metastasis models, 2.5 × 106 treated cells were suspended in 100 μL of PBS and injected into the lateral tail vein of the recipient mice (n=10 mice/group). The mice were observed every 3 days, and mouse weights were measured weekly. Six to eight weeks after the first injection, the mice were euthanized, and lung tissues were harvested. Finally, the metastatic nodules formed in the corresponding organs were counted and analyzed by hematoxylin and eosin (H&E) staining. All animal experiments were conducted with the approval of the Department of Laboratory Animals of Central South University (Changsha, Hunan, China) (No. 2019sydw0235) and complied with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
H&E and immunohistochemical (IHC) staining
Paraffin-embedded tissues were sectioned at 5 μm thickness and then stained with H&E. The tissue sections were deparaffinized in xylene and rehydrated using a graded ethanol series. To quench endogenous peroxidase activity, the sections were immersed in a 0.3% peroxidase-methanol solution for 30 minutes. For the antigen retrieval, the sections were pretreated with citrate buffer for 15 minutes at 100°C in a microwave oven. The sections were hybridized with a primary antibody at 4°C overnight at a dilution of 1:100 and visualized using an UltraVision Quanto Detection System HRP DAB Kit (Thermo Scientific, Shanghai, China) according to the manufacturer's protocols. The stained sections were counterstained with hematoxylin, and photomicrographs were captured under an Olympus BX51 microscope (Olympus, Tokyo, Japan).
CRC patient-derived organoid (PDO) model
According to the standard operating procedures (SOPs) for PDOs provided in the NCI Patient-Derived Models Repository (PDMR) and previously published protocols [14], we successfully established PDO models from human colorectal adenocarcinoma samples. Then, lentiviral particles carrying the circ7379 vector and the NC vector were transfected into the organoids as previously described. The growth of the organoids was observed daily by microscopy, and the diameter of the organoids was measured using a scale bar.
CRC patient-derived xenograft (PDX) model
In accordance with the SOPs for the PDXs provided by the PDMR and previously published protocols [15], we also successfully established PDX models from human colorectal adenocarcinoma samples. Subsequently, passage 3 tumor tissues were used to perform the in vivo tumor suppression experiments. In brief, we first subcutaneously implanted P2 tumor fragments of equal size into the contralateral side of the lower back of NOD-SCID mice to maintain the homogeneity of the two groups. When the tumors were 50-100 mm3, lentiviral particles (1×107 TU) carrying the circ7379 vector and the NC vector were injected into the tumors on the right side and left side, respectively, of each mouse. Each group included nine mice, which harbored tumors originating from three CRC patients (3 mice/patient). The tumor diameter was measured weekly, and the tumor volume was calculated as described above. Four weeks later, all mice were euthanized, and the tumor tissues were collected for further analysis.
Statistical analysis
All experiments were repeated at least three times, and data from one representative experiment are presented. All data are presented as the mean ± standard deviation (S.D.) of at least three biological replicates. The statistical significance of the differences was evaluated by a two-tailed Student's t test or a two-way analysis of variance (ANOVA) as indicated in the corresponding figure legends. The correlations among gene expression in CRC patients were determined by a Pearson correlation analysis. All statistical analyses were performed using GraphPad Prism version 8.0.1 (GraphPad Software, Inc., San Diego, CA, USA). Differences with P < 0.05 were considered statistically significant and are noted by asterisks (*, P <0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001).
Results
Downregulated circ7379 is associated with CRC progression
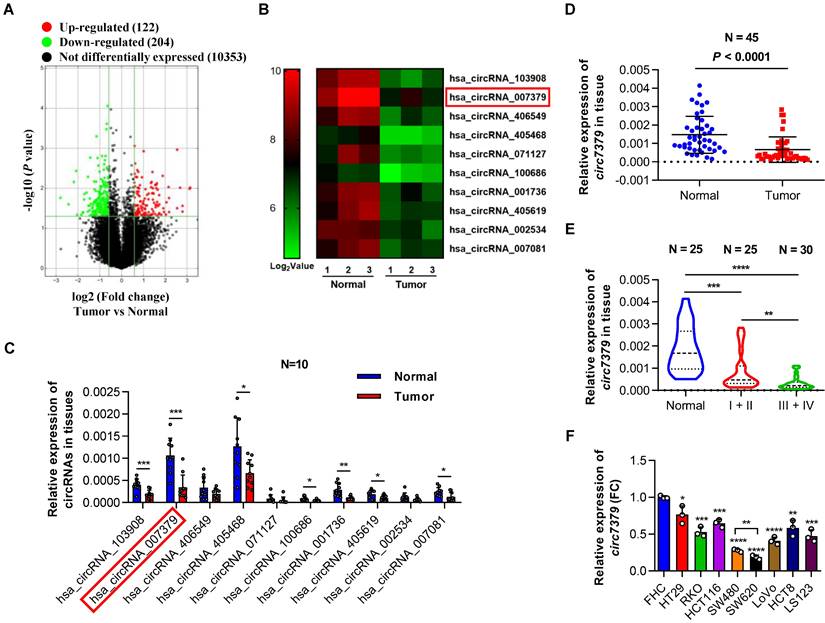
To characterize the circRNA expression profiles in CRC tissues, we compared three pairs of CRC tissues and corresponding adjacent normal tissues using circRNA microarrays. This assay revealed that 204 circRNAs (P<0.05 and FC >1.5) were significantly downregulated in the CRC tumor tissues (Figure 1A). The top 10 downregulated circRNAs (Figure 1B and Supplementary Table 3) were further verified in 10 pairs of CRC tissues and matched adjacent normal tissues using qRT-PCR. Notably, circ7379 was the most downregulated circRNA with relative high abundance (Figure 1C). According to the microarray analysis, the expression level of circ7379 in the adjacent normal tissues ranked 16th among 204 downregulated circRNAs, which was also higher than that of some known downregulated circRNAs in CRC, such as circLPAR1 [16], circPLCE1 [17], circ_0002138 [18] and circTADA2A [19] (Figure S1A). The down-regulation trend of circ7379 in CRC tumor tissues compared with normal tissue samples was further confirmed in another cohort study of 45 patients (Figure 1D).
Next, we examined the relationship between circ7379 expression in tumor tissues and clinical stage of CRC. Compared to stage I+II group, the expression of circ7379 was further downregulated in tumor tissues of stage III+IV group (Figure 1E). To further analyze the correlation between the circ7379 expression level in CRC tissues and the clinicopathological features of CRC patients, 55 CRC patients were stratified into a low circ7379 expression group (n=28) and a high circ7379 expression group (n=27) based on the median circ7379 expression level. Statistical analysis revealed that a low expression of circ7379 in CRC tissues was significantly associated with a larger tumor size (P=0.037), greater invasion depth (P=0.004), and an advanced TNM stage (P=0.044) (Supplementary Table 4), highlighting circ7379 as a biomarker for predicting CRC progression.
To further investigate the differential expression of circ7379, we detected circ7379 in 8 human CRC cell lines (HT29, RKO, HCT116, SW480, SW620, LoVo, HCT8 and LS123) and a normal human colon mucosal epithelial cell line (FHC) (Figure 1F). Consistent with the expression levels in the circRNA microarray (Figure S1A), the content of circ7379 in FHC cells was significantly higher than that of tumor suppressor circRNAs (Figure S1B). Circ7379 expression was lower in SW620 cells (with a high metastatic potential) than that in SW480 cells (derived from the primary lesion in the same patient) (Figure 1F), suggesting that the circ7379 expression level may be related to the metastatic potential of CRC cell lines. Altogether, these clinical findings establish a strong link between circ7379 downregulation and CRC progression.
Circ7379 is an intergenic circRNA localized in the nucleus and cytoplasm
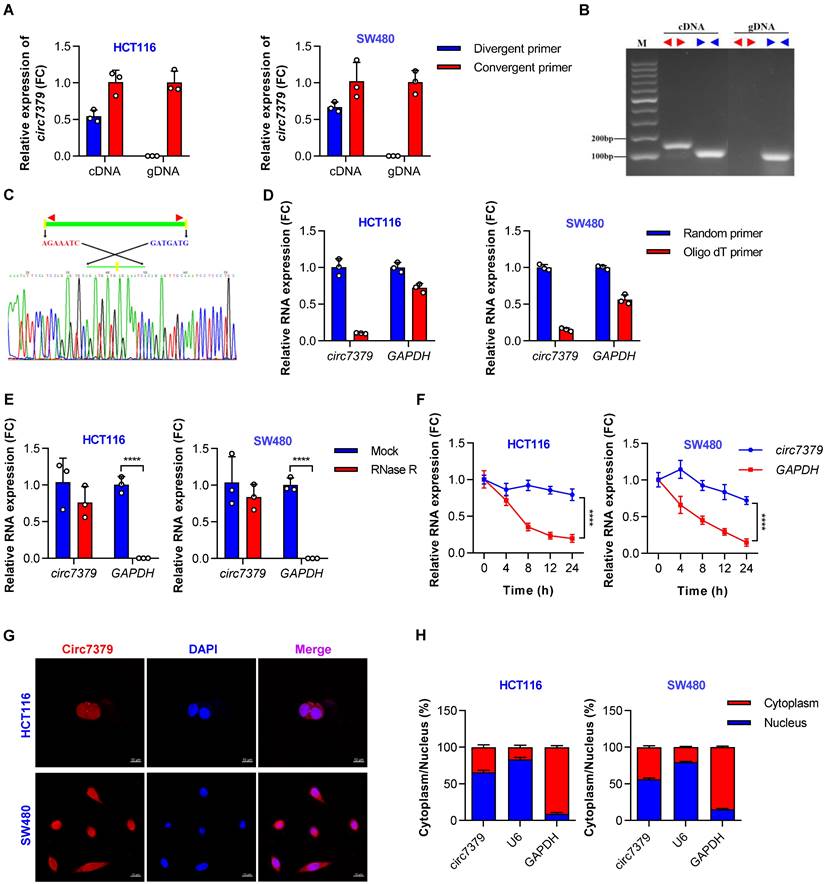
According to the circBase database and circBank database, circ7379 (circBase ID: hsa_circ_0007379; circBank ID: hsa_circ_chr14_00334) was marked as an intergenic circRNA generated from chr14:35020919-35024118 with a splice length of 3199 nucleotides (nt). Given that circ7379 has not been studied before, we conducted a series of experiments to verify its existence and circularization. First, divergent and convergent primers were designed, and the backsplice junction of circ7379 was amplified using divergent primers (Figure S2A). The backsplice junction of circ7379 could be amplified from only cDNA, but not from gDNA (Figures 2A and 2B), suggesting that circ7379 is generated by backsplicing of pre-mRNA after gDNA transcription. The sequence of the backsplice junction was confirmed by Sanger sequencing (Figure 2C), which was consistent with the circBase database annotation and the circ7379 probe used in the circRNA microarray analysis. Furthermore, circ7379 was barely detected, but linear glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA detection was unchanged when the random hexamer primers were replaced with oligo (dT)18 primers (Figure 2D), indicating that circ7379 does not contain a poly(A) tail. Finally, the RNase R (Figure 2E) and actinomycin D treatment assays (Figure 2F) confirmed that circ7379 was relatively resistant to RNase R digestion and had a longer half-life than the tested linear mRNAs (e.g., GAPDH). Collectively, these findings demonstrate that circ7379 is a typical circRNA in CRC cells.
Downregulated circ7379 is associated with CRC progression. (A) Volcano plot showing the expression profile of 3 CRC tissues and matched normal tissues. Conditions for screening differences: │Fold Change│>1.5, P<0.05. The red points in the plot indicate significantly upregulated circRNAs, and the green points indicate significantly downregulated circRNAs. (B) Heatmap showing the top 10 downregulated circRNAs in CRC tissues. High values are shown in red, while low values are shown in green. Each column indicates one sample, and each row indicates one circRNA. (C) qRT‒PCR showing the expression of selected circRNAs in 10 pairs of CRC tissues and adjacent normal tissues. (D) qRT‒PCR showing the expression of circ7379 in 45 pairs of CRC tissues and adjacent normal tissues. (E) qRT‒PCR showing the expression of circ7379 in 25 paracancerous normal tissues, 25 stage I+II CRC tissues, and 30 stage III+IV CRC tissues. (F) qRT‒PCR showing the expression of circ7379 in a normal colon cell line (FHC) and a series of CRC cell lines (HT29, RKO, HCT116, SW480, SW620, LoVo, HCT8 and LS123). The data are shown as the mean ± SD. The P values were determined by a two-tailed paired (C and D) or unpaired Student's t test (E and F); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S1.
Regarding the cellular distribution of circ7379, the FISH assay showed that circ7379 (red) was localized in the nucleus and cytoplasm of CRC cells (Figure 2G). The qRT-PCR assay also revealed that 55%-65% of circ7379 was localized in the nucleus, while 35%-45% was localized in the cytoplasm (Figure 2H). Thus, circ7379 may function in both the nucleus and cytoplasm.
The biogenesis of circ7379 is mediated by RCMs and negatively regulated by DHX9
Reverse complementary matches (RCMs) are a hallmark of circRNA biogenesis in animals [20]. Thus, we obtained the upstream sequence (1000 nt) and downstream sequence (1000 nt) of circ7379 from circBase and aligned these two sequences with BLAST to identify all possible RCMs. A highly matched RCM with 81% identity over 221 nt was identified in each sequence (Figure S3A), and they were termed usRCM (upstream RCM) and dsRCM (downstream RCM), respectively (Figure 3A). We then fused these RCMs to the sequence of circ7379 and cloned the constructs into a GV367 vector (without homology arms) (wild-type, WT) (Figures 3B and S3B). Similarly, we constructed a series of mutant-type constructs, including MT#1 (deletion of usRCM), MT#2 (deletion of dsRCM), and MT#3 (deletion of both RCMs) (Figure 3B). After transient transfection, qRT-PCR results showed that only the WT vector, but not MT#1-3, significantly overexpressed circ7379 (Figure 3C), suggesting that usRCM and dsRCM are responsible for the biogenesis of circ7379.
Circ7379 is an intergenic circRNA localized in the nucleus and cytoplasm. (A) qRT‒PCR showing the amplification of circ7379 from cDNA or gDNA of CRC cell lines using divergent primers and convergent primers. cDNA, complementary DNA; gDNA, genomic DNA. (B) Agarose gel electrophoresis of PCR products showing the existence and circulation of circ7379 in CRC cell lines. (C) Sanger sequencing showing the back-splice junction of circ7379. (D) RT‒PCR using random primers or oligo dT primers showing the circular characteristics of circ7379. GAPDH was used as a control for a linear RNA transcript. (E) qRT‒PCR showing the expression of circ7379 and GAPDH mRNA after treatment with RNase R in CRC cell lines. (F) qRT‒PCR showing the expression of circ7379 and GAPDH mRNA after treatment with actinomycin D at the indicated time points in CRC cell lines. (G) RNA fluorescence in situ hybridization (FISH) showing the location of circ7379 in CRC cell lines. Nuclei were stained with DAPI. Scale bar, 10 µm. (H) Cytoplasmic and nuclear mRNA fractionation experiments showing the location of circ7379 in CRC cell lines. U6 and GAPDH were used as positive controls in the nucleus and cytoplasm, respectively. The data are shown as the mean ± SD. The P values were determined by a two-tailed unpaired Student's t test (D and E) or two-way ANOVA (F); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S2.
The biogenesis of circ7379 is mediated by RCMs and negatively regulated by DHX9. (A) Schematic representation of the following: back-splice junction (BSJ), upstream RCM (usRCM) and downstream RCM (dsRCM) of circ7379. RCM, reverse complementary match. (B) Schematic representation of the construction of the wild-type (WT) and the series of mutant-type (MT) vectors used for circ7379 overexpression. The red cross indicates deletion. (C) qRT‒PCR showing the expression of circ7379 after the transfection of CRC cell lines with different vectors. (D) qRT‒PCR showing the expression of circ7379 after the transfection of the negative control siRNAs (si-NC) or DHX9 siRNAs (si-DHX9) in CRC cell lines. (E) qRT‒PCR showing the enrichment of usRCM and dsRCM in a representative anti-DHX9 RIP assay in CRC cells. IgG was used as a control. (F) qRT‒PCR showing the expression of circ7379 after the transfection of the circ7379 vector or DHX9 vector or cotransfection of circ7379 vector and DHX9 vector in CRC cells. (G) qRT‒PCR showing the expression of DHX9 mRNA in 20 pairs of CRC tissues and adjacent normal tissues. (H) Circ7379 expression was negatively correlated with DHX9 expression in CRC tissues. The data are shown as the mean ± SD. The P values were determined by a two-tailed paired (G) or unpaired Student's t test (D, E and F), two-way ANOVA (C), or Pearson correlation analysis (H); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S3.
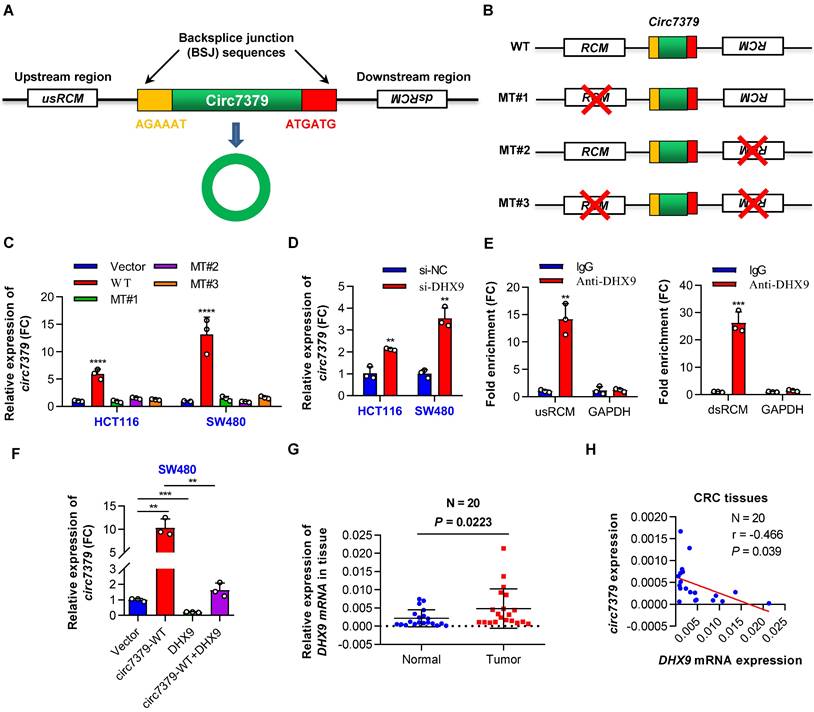
Double-stranded RNA (dsRNA)-specific adenosine deaminase (ADAR) enzymes [20] and ATP-dependent RNA helicase A (DHX9) [21] can suppress the biogenesis of circRNAs that rely on base pairing between inverted repeats. We next investigated whether the expression of circ7379 is regulated by ADAR or DHX9. circ7379 was significantly upregulated upon the knockdown of DHX9 (Figure 3D), but not the knockdown of ADAR (Figures S3C and S3D). The anti-DHX9 RIP assay further revealed a significant enrichment of usRCM and dsRCM (Figure 3E), suggesting a high interaction probability between DHX9, usRCM, and dsRCM. In contrast, the overexpression of DHX9 inhibited the expression of circ7379 and reversed the upregulation of circ7379 achieved by the WT vector mentioned above (Figures 3F and S3E). Additionally, the mRNA expression levels of DHX9 in CRC tissues were increased compared with those in the paired normal tissues (Figure 3G), and the expression level of circ7379 was negatively correlated with the mRNA level of DHX9 (Figure 3H). Thus, a high DHX9 expression may be responsible for the downregulation of circ7379 in CRC by preventing base pairing between RCMs.
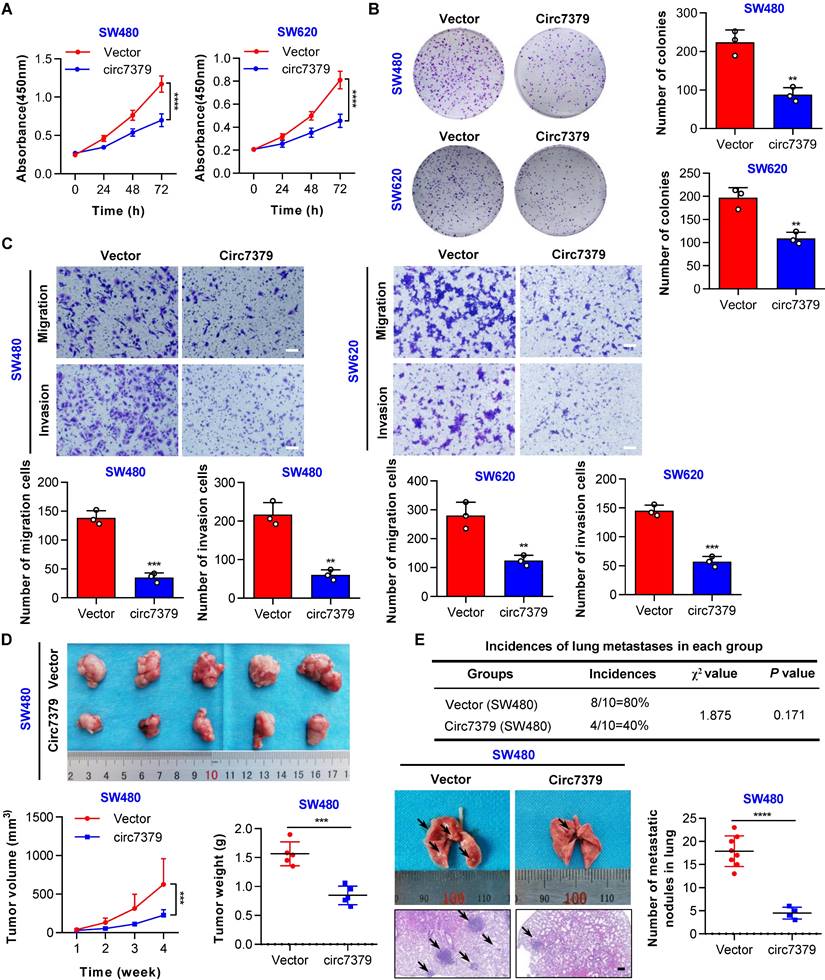
Circ7379 inhibits the growth and metastasis of CRC cells in vitro and in vivo
To explore the functional role of circ7379 in CRC cells, gain- and loss-of-function experiments were performed. First, we overexpressed circ7379 in two CRC cell lines with relatively low expression of circ7379 (SW480 and SW620) by lentiviral plasmid delivery and confirmed the overexpression efficiency by qRT-PCR (Figure S4A). The CCK-8 and plate colony formation assays revealed that circ7379 overexpression suppressed the proliferation of CRC cells (Figures 4A and 4B). The transwell assays showed that circ7379 was a repressor of the migration and invasion of SW480 and SW620 cells (Figure 4C). Second, we inhibited the expression of circ7379 by using different siRNAs that specifically targeted the BSJ site of circ7379 in two CRC cell lines (HT29 and HCT-116) with relatively high expression of circ7379 (Figure. S4B). Functional studies showed that silencing of circ7379 enhanced the proliferation, migration and invasion abilities of HT29 and HCT116 cells (Figure S4C-S4E).
Subsequently, we investigated the role of circ7379 in the growth and metastasis of CRC cells using in vivo xenograft models and metastasis models. Notably, tumors derived from SW480 cells stably overexpressing circ7379 were smaller (Figure 4D), whereas tumors derived from HCT116 cells transduced with lentiviral shRNA targeting circ7379 were larger than control group (Figure S4F). In addition, compared with the mice in the corresponding control group, the incidence of lung metastases in mice in the circ7379 overexpression group was significantly decreased (Figure 4E), but the incidence of lung metastases in the mice in the circ7379-silenced group was increased (Figure S4G). Fewer and smaller nodules were observed in the lungs of the mice in the circ7379 overexpression group (Figure 4E), while more and larger nodules were observed in the lungs of the mice in the circ7379 silencing group than in the corresponding control groups (Figure S4G). Taken together, these results support the tumor suppressive effect of circ7379 in vitro and in vivo.
Circ7379 exerts its anticancer effect by inhibiting RUNX1 expression
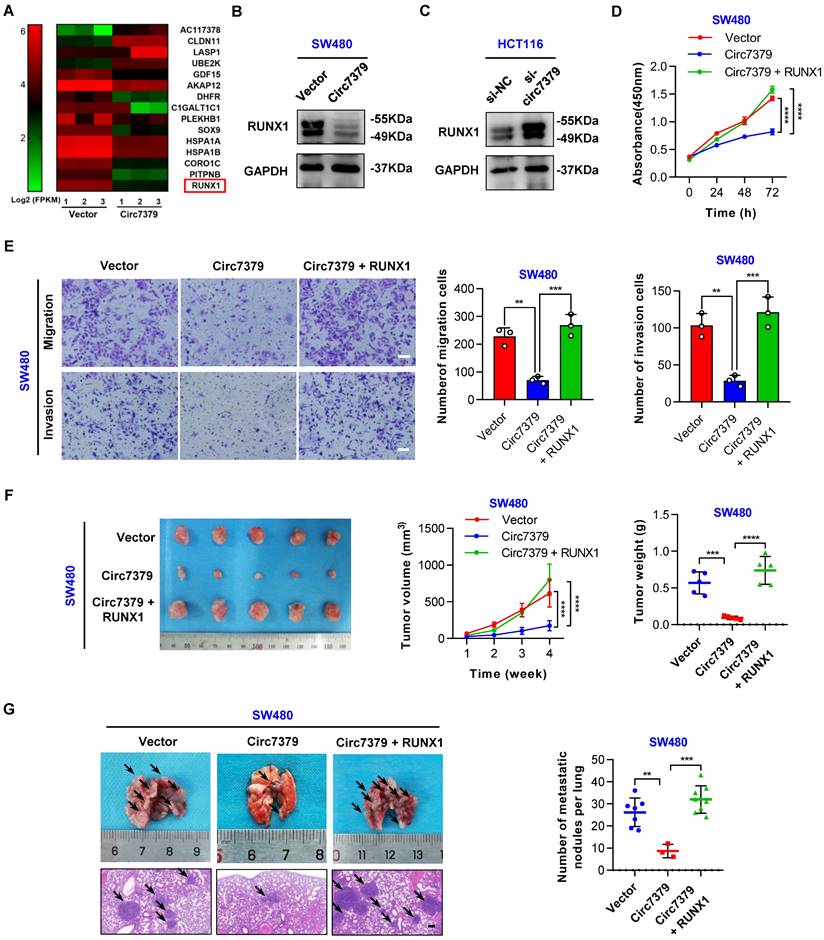
To elucidate the molecular mechanism by which circ7379 regulates cell growth and metastasis, we carried out high-throughput RNA-seq of three pairs of SW480 cells transduced with different lentiviral plasmids (circ7379 and NC) to identify the target genes of circ7379. 166 genes were found to be differentially expressed (|log2FC|>1, P<0.05) after circ7379 overexpression in SW480 cells. Among them, 64 genes were upregulated, and 102 genes were downregulated (Figure S5A). GO functional enrichment analysis revealed that differentially expressed genes were significantly associated with cell proliferation and cell migration (Figure S5B). Subsequently, we selected the 15 most dysregulated genes (|log2FC| ≥ 1.3, P<0.05) with relatively high expression abundances (FPKM >10) (Figure 5A) and verified their mRNA expression in SW480 and HCT116 cells. Of note, RUNX1 was the most downregulated gene in SW480 cells with circ7379 overexpression (Figure S5C) and the most upregulated gene in HCT116 cells with circ7379 depletion (Figure S5D). Western blot also confirmed the inhibitory effect of circ7379 on RUNX1 protein expression (Figures 5B and 5C). The qRT-PCR and WB analysis further observed that RUNX1 expression was upregulated in CRC tissues and cells (Figures S5E, S5F, S5G and S5H). Importantly, the expression of RUNX1 mRNA in CRC tissues and cells were negatively correlated with those of circ7379 (Figure S5I), supporting that RUNX1 expression is regulated by cellular levels of circ7379.
Next, we determined whether circ7379-mediated downregulation of RUNX1 is required for tumor growth and metastasis. The CCK-8 (Figure 5D) and transwell (Figure 5E) assays showed that the overexpression of RUNX1 reversed the inhibitory effect of circ7379 overexpression on the proliferation, migration and invasion of SW480 cells. In contrast, silencing RUNX1 negated the “oncogenic” effect of circ7379 deficiency in HCT116 cells (Figures S5J and S5 K). To further confirm these in vitro results, in vivo rescue experiments were performed with xenograft and metastasis models. Indeed, small subcutaneous tumors were observed in the mice in the SW480-circ7379 group (Figure 5F), whereas the tumors in the mice in the SW480-circ7379+RUNX1 group were markedly larger (Figure 5F). As a control, the expression levels of RUNX1 were markedly decreased in the tumors in the SW480-circ7379 group compared with those in the SW480-circ7379+RUNX1 group (Figure S5 L). Experiments in an animal model of CRC lung metastasis also showed that the reduction of lung metastasis mediated by ectopic circ7379 was successfully reversed by RUNX1 overexpression (Figures 5G and S5M). Consistently, IHC staining revealed that overexpression of circ7379 reduced the expression of RUNX1 in mouse lung micrometastases, and overexpression of RUNX1 reversed the inhibitory effect of circ7379 on RUNX1 (Figure S5N). Therefore, circ7379 inhibits CRC growth and metastasis by downregulating the expression of RUNX1.
Circ7379 inhibits the growth and metastasis of CRC cells in vitro and in vivo. (A) CCK-8 assay showing the proliferation ability of CRC cell lines after the transfection of the control vector or circ7379 overexpression vector. (B) Plate clone formation assay showing the clone formation ability of CRC cell lines after the transfection of the control vector or circ7379 overexpression vector. (C) Transwell assay showing the migration and invasion abilities of CRC cell lines after the transfection of the control vector or circ7379 overexpression vector. Scale bar, 100 µm. (D) In vivo xenograft models showing the tumorigenesis ability of CRC cells after the transfection of the control vector or circ7379 overexpression vector. Top, images of tumors in mice in each group (n=5 mice/group). Bottom (left), tumor growth curves in mice in each group. Bottom (right), tumor weights in mice in each group. (E) In vivo pulmonary metastasis models showing the metastatic ability of CRC cells after the transfection of the control vector or circ7379 overexpression vector. Top, incidences of lung metastases in mice in each group (n=10 mice/group). Bottom (left), representative lung and representative H&E staining of lung metastatic lesions (black arrow). Scale bar, 200 µm. Bottom (right), the number of metastatic nodules formed in the lungs of the mice in each group. The data are shown as the mean ± SD. The P values were determined by a two-tailed unpaired Student's t test (B, C, D, and E), two-way ANOVA (A and D), or chi-square test (E); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S4.
Circ7379 exerts its anticancer effect by inhibiting RUNX1 expression. (A) Heatmap showing the 15 most dysregulated genes in SW480 cells overexpressing circ7379. High values are shown in red, while low values are shown in green. Each column indicates one sample, and each row indicates one gene. (B) Representative Western blot of RUNX1 in CRC cells after control vector or circ7379 vector transfection. GAPDH was used as a loading control. (C) Representative Western blot of RUNX1 in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379). (D) CCK-8 assay showing the proliferation ability of CRC cells after the transfection of the control vector or circ7379 vector or cotransfection of circ7379 + RUNX1 vectors. (E) Transwell assay showing the migration and invasion abilities of CRC cells after the transfection of the control vector or circ7379 vector or cotransfection of circ7379 + RUNX1 vectors. Scale bar, 100 µm. (F) In vivo xenograft models showing the tumorigenesis ability of CRC cells after the transfection of the control vector or circ7379 vector or cotransfection of circ7379 + RUNX1 vectors. Top (left), tumor growth curves in mice in each group. Top (right), tumor weights in mice in each group. Bottom, images of tumors in mice in each group (n=5 mice/group). (G) In vivo pulmonary metastasis models showing the metastatic ability of CRC cells after the transfection of the control vector or circ7379 vector or cotransfection of circ7379 + RUNX1 vectors. Left, representative lung and representative H&E staining of lung metastatic lesions (black arrow). Scale bar, 200 µm. Right panel, the number of metastatic nodules formed in the lungs in mice in each group (n=10 mice/group). The data are shown as the mean ± SD. The P values were determined by a two-tailed unpaired Student's t test (E, F and G) or two-way ANOVA (D and F); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S5.
Circ7379 inhibits RUNX1 expression by interacting with KSRP
Previous work has shown that miRNA-responsive element (MRE)-enriched circRNAs can act as miRNA sponges [22]. We identified a maximum of four binding sites for hsa-miR-661, hsa-miR-1827 and hsa-miR-940 in circ7379 (Supplementary Table 5). To determine whether circ7379 regulates targets as a miRNA sponge in CRC cells, we conducted an anti-AGO2 RIP assay. The results showed that ciRS-7 (a circRNA that binds AGO2) [22, 23] was enriched by an anti-AGO2 antibody, but not circ7379 or circNDUFB2 (a circRNA the does not bind AGO2) [24] (Figure S6A). We further performed RNA pulldown analysis using biotin-labeled circ7379-specific RNA probes. The results showed that none of the three miRNAs (hsa-miR-661, hsa-miR-1827 and hsa-miR-940) were enriched in the circ7379 probe group compared with the control probe group (Figure S6B), suggesting that circ7379 may not act as a miRNA sponge for these miRNAs in CRC cells. However, whether circ7379 can sponge other miRNAs remains an open question.
Other work has shown that circRNAs, including open reading frames (ORFs) with internal ribosome entry site (IRES) elements and AUG sites, may be translated under certain circumstances, resulting in unique peptides [25]. Although circ7379 is predicted to include a putative ORF (size 291) with three IRES elements, it has weak coding potential compared with other translated circRNAs, including circZNF609 [26], circFBXW7 [27], circSHPRH [28], circFNDC3B [29], circCOL6A3 [30] and circARHGAP35 [31] (Supplementary Table 6). These data suggest that circ7379 regulates RUNX1 expression and controls CRC cell functions through a molecular mechanism independent of miRNA sponging and protein translation.
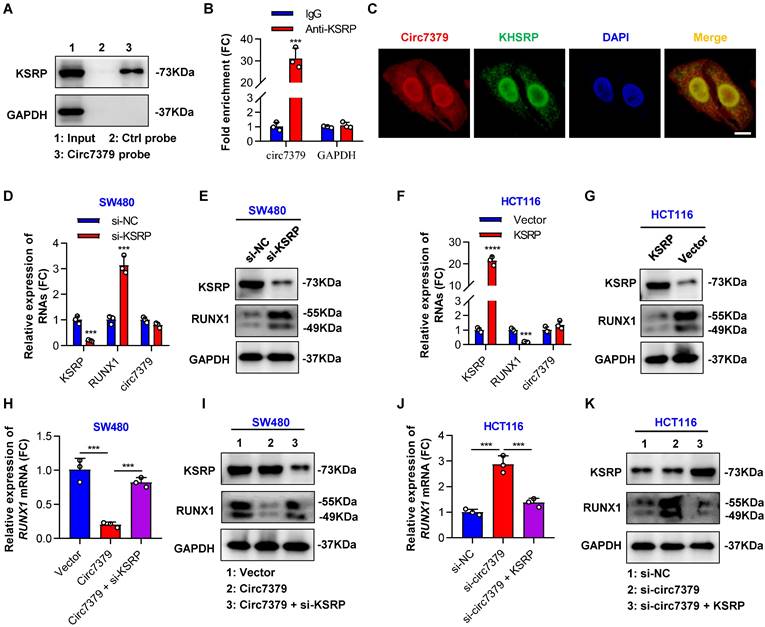
Because circRNAs can indirectly regulate gene expression by interacting with RBPs and enhancing their functions [32], we attempted to identify proteins that may interact with circ7379. We first searched for potential circ7379-binding proteins by screening the catRAPID online database. Far upstream element-binding protein 2 (FUBP2) was predicted to be the most likely interacting protein, with the highest Z score (0.61) and discriminative power (92%), followed by LN28B and ELAV1 (Figure S6C). The analysis using RNA‒Protein Interaction Prediction (RPISeq) database also showed that FUBP2 had the highest interaction probability based on both random forest (RF: 0.80) and support vector machine (SVM: 0.94) classifiers (Figure S6D). FUBP2, also known as KSRP, is encoded by the KHSRP gene. qRT‒PCR assay found that silencing KSRP, rather than LN28B or ELAV1, increased RUNX1 mRNA expression (Figure S6E). Direct interaction between circ7379 and KSRP was confirmed by RNA pull-down analysis (Figure 6A) and the RIP assay (Figure 6B). Furthermore, the colocalization (yellow) of endogenously expressed circ7379 (red) and KSRP (green) in the nucleus and cytoplasm was observed by the FISH-IF assay (Figure 6C). These different assays suggest that circ7379 and KSRP can form RNA-protein complexes.
Subsequently, we investigated the role of KSRP in the circ7379-driven inhibition of RUNX1 expression. We demonstrated that the knockdown of KSRP led to elevated levels of RUNX1 mRNA and RUNX1 protein in SW480 cells (Figures 6D and 6E), while the overexpression of KSRP resulted in the downregulation of RUNX1 expression in HCT116 cells (Figures 6F and 6G). Notably, knockdown of KSRP or overexpression of KSRP had little effect on the expression levels of circ7379 in CRC cells (Figures 6D and 6F), suggesting that KSRP did not regulate circ7379 biogenesis in CRC cells. Additionally, overexpression of circ7379 or knockdown of circ7379 had no effect on the expression levels of KSRP (Figures 6I and 6K), suggesting that circ7379 did not regulate KSRP levels in CRC cells. Finally, we designed rescue experiments to confirm that KSRP is indispensable in circ7379-mediated inhibition of RUNX1 expression (Figures 6H, 6I, 6J and 6K).
Circ7379 binds the KH4 domain of KSRP through the GUCC motif
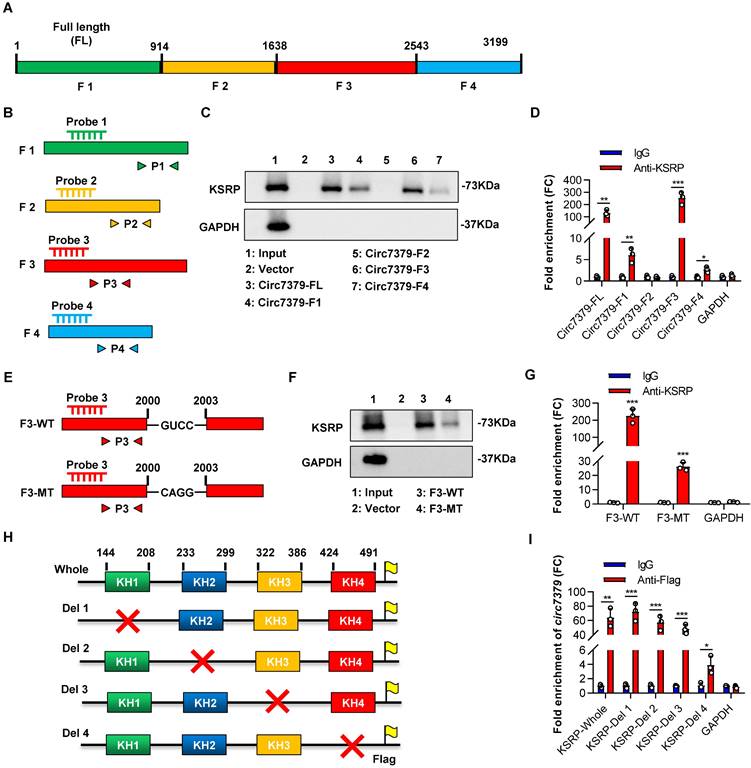
We further investigated the exact motif of circ7379 that directly interacts with KSRP. Based on the potential binding regions of KSRP predicted by the MEME databases and catRAPID databases (Figures S7A and S7B), we constructed a vector containing the full length (FL) of circ7379 and four vectors encoding the corresponding circ7379 fragments: F1 (1-913), F2 (914-1638), F3 (1639-2542), and F4 (2543-3199) (Figure 7A). We also designed specific probes and primers for the full length and corresponding fragments of circ7379 (Figure 7B). After transient transfection, qRT-PCR showed that these vectors resulted in significant overexpression of the corresponding circ7379 fragments in CRC cells (Figure S7C). Then, using specific probes against the four circ7379 fragments for RNA pulldown assays, we found that the F3 probe could significantly pull down KSRP (Figure 7C). The RIP assay also confirmed that F3 was the most enriched fragment in the complexes precipitated with the anti-KSRP antibody (Figure 7D). These data indicate that circ7379 binds KSRP mainly in the region between nt 1979-2285, which is consistent with the predictions from MEME and catRAPID databases (Figures S7A and S7B).
To better delineate the interaction between circ7379 and KSRP, we searched the RNA-Binding Protein DataBase (RBPDB) to predict the exact binding sites and identified “GUCC” as the possible binding motif for KSRP (Figure S7D). We identified eight “GUCC” motifs, one of which was in the region between nt 1979 and 2285 of circ7379, could be a putative KSRP binding motif (Figure S7E). Mutation of the “GUCC” motif (nt 2000-2003) (Figure 7E) significantly reduced the ability of F3 to bind KSRP as evidenced by RNA pulldown and RIP assays (Figures 7F and 7G).
Furthermore, we sought to identify the domain of KSRP that contributes to its interaction with circ7379, as its structure contains four RNA-binding KH domains that are essential for the function of the KSRP protein (Figure S7F). All four domains have binding potential to circ7379, especially KH3 and KH4, as predicted by the catRAPID database (Figure S7G). We then constructed expression plasmids for Flag-tagged whole KSRP and four truncation mutants of KSRP (with truncation of the individual KH domains) (Figure 7H) and verified the expression of these KSRP truncation mutants by western blot analysis (Figure S7H). Anti-Flag RIP assays revealed that the truncation mutants containing the KH4 domain of KSRP specifically bound circ7379, while the truncation mutant lacking the KH4 domain had weak ability to bind circ7379 (Figure 7I), suggesting that this domain is essential for the binding of KSRP to circ7379.
Circ7379 inhibits RUNX1 expression by interacting with KSRP. (A) Representative Western blot showing the enrichment of KSRP upon circ7379 pull-down in CRC cell lysates. GAPDH was used as a negative control. (B) qRT‒PCR showing the enrichment of circ7379 in a representative anti-KSRP RIP assay in CRC cells. IgG was used as a control. (C) Immunofluorescence (IF)-FISH assay showing the colocalization of circ7379 with KSRP protein both in the nucleus and the cytoplasm. Scale bar, 10 µm. (D) qRT‒PCR showing the expression of KSRP and RUNX1 mRNAs, and circ7379 in CRC cells under control conditions (si-NC) or upon KSRP knockdown (si-KSRP). (E) Representative Western blot of KSRP and RUNX1 proteins in CRC cells under control conditions (si-NC) or upon KSRP knockdown (si-KSRP). GAPDH was used as a loading control. (F) qRT‒PCR showing the expression of KSRP and RUNX1 mRNAs, and circ7379 in CRC cells after control vector or KSRP vector transfection. (G) Representative Western blot of KSRP and RUNX1 proteins in CRC cells after control vector or KSRP vector transfection. (H) qRT‒PCR showing the expression of RUNX1 mRNA in CRC cells after the transfection of the control vector or circ7379 vector or cotransfection of circ7379 vector + si-KSRP. (I) Representative Western blot of KSRP and RUNX1 proteins in CRC cells after the transfection of the control vector or circ7379 vector or cotransfection of circ7379 vector + si-KSRP. (J) qRT‒PCR showing the expression of RUNX1 mRNAs in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379) or si-circ7379 + KSRP vector cotransfection. (K) Representative Western blot of KSRP and RUNX1 proteins in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379) or si-circ7379 + KSRP vector cotransfection. The data are shown as the mean ± SD. The P values were determined by a two-tailed unpaired Student's t test; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S6.
Circ7379 binds the KH4 domain of KSRP through the GUCC motif. (A) Schematic representation of the following: full length (FL), fragment #1, fragment #2, fragment #3, and fragment #4 of circ7379. (B) Schematic representation of the design of specific probes and primers for the corresponding fragments of circ7379. (C) Representative Western blot showing the enrichment of KSRP upon circ7379-corresponding fragment pull-down in CRC cell lysates. GAPDH was used as a negative control. (D) qRT‒PCR showing the enrichment of circ7379 corresponding fragments in a representative anti-KSRP RIP assay in CRC cells. IgG was used as a control. (E) Schematic representation of the construction of wild-type (WT) fragment #3 and mutant-type (MT) fragment #3 vectors for RNA overexpression. (F) Representative Western blot showing the enrichment of KSRP upon wild-type fragment #3 or mutant-type fragment #3 pull-down in the CRC cell lysate. GAPDH was used as a negative control. (G) qRT‒PCR showing the enrichment of wild-type fragment #3 or mutant-type fragment #3 in a representative anti-KSRP RIP assay in CRC cells. IgG was used as a control. (H) Schematic representation of the construction of expression plasmids for Flag-tagged whole KSRP and four truncation mutants of KSRP (with truncation of individual KH domains). The red cross indicates deletion. (I) qRT‒PCR showing the enrichment of circ7379 in a representative anti-Flag RIP assay in CRC cells. IgG was used as a control. The data are shown as the mean ± SD. The P values were determined by a two-tailed unpaired Student's t test; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S7.
Circ7379 and KSRP synergistically regulate miR-320a biogenesis
The major function of KSRP is to serve as a component of both the Drosha complex and Dicer complex, regulating the biogenesis of a subset of miRNAs in mammalian cells [33]. We speculated that circ7379 and KSRP may collaboratively modulate the biogenesis of certain miRNAs to further inhibit RUNX1 expression in CRC cells. To validate this hypothesis, we first identified potential target miRNAs using a bioinformatics analysis. The intersection of the miRNAs positively related to KSRP in the TCGA database with the miRNAs predicted to target RUNX1 in the TargetScan and ENCORI databases identified two overlapping miRNAs (hsa-miR-320a and hsa-miR-1276) (Figure 8A). Then, we verified that miR-320a, rather than miR-1276, was a common target of circ7379 and KSRP in CRC cells (Figures S8A and S8B).
Circ7379 and KSRP synergistically regulate miR-320a biogenesis. (A) Venn diagram showing the intersection between miRNAs positively related to KSRP in the TCGA database and miRNAs predicted to target RUNX1 in the TargetScan and ENCORI databases. (B) qRT‒PCR showing the expression of pri-miR-320a, pre-miR-320a, and miR-320a in CRC cells after control vector or circ7379 vector transfection. (C) qRT‒PCR showing the expression of pri-miR-320a, pre-miR-320a, and miR-320a in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si- circ7379). (D) qRT‒PCR showing the expression of pri-miR-320a, pre-miR-320a, and miR-320a in CRC cells after the transfection of the control vector, circ7379 vector, KSRP siRNAs (si-KSRP), or cotransfection of circ7379 vector + si-KSRP. (E) qRT‒PCR showing the expression of pri-miR-320a, pre-miR-320a, and miR-320a in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379), KSRP overexpression, or si-circ7379 + KSRP vector cotransfection. (F) qRT‒PCR showing the enrichment of pri-miR-320a and pre-miR-320a upon circ7379 pull-down in CRC cell lysates. U6 was used as a negative control. (G) qRT‒PCR showing the enrichment of pri-miR-320a and pre-miR-320a in a representative anti-KSRP RIP assay in CRC cells. IgG was used as a control. (H) qRT‒PCR showing the enrichment of pri-miR-320a and pre-miR-320a after anti-KSRP RIP in CRC cells under control conditions (Vector) or upon circ7379 overexpression (Circ7379). (I) qRT‒PCR showing the enrichment of pri-miR-320a and pre-miR-320a after anti-KSRP RIP in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379). (J) Co-IP assay showing the interaction between KSRP and Drosha and the interaction between KSRP and Dicer in CRC cells in an RNA-independent manner. This experiment was performed by incubating KSRP immunoprecipitates with or without RNase A (0.1 mg/mL) at 37°C for 20 min. (K) Representative Western blot showing the enrichment of KSRP, Drosha, and Dicer proteins after circ7379 pull-down in CRC cells under control conditions (si-NC) or upon KSRP knockdown (si-KSRP). (L) qRT‒PCR showing the enrichment of pri-miR-320a and pre-miR-320a after anti-Drosha and anti-Dicer RIP, respectively, in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379). The data are shown as the mean ± SD. The P values were determined by a two-tailed unpaired Student's t test or one-way ANOVA (D and E); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S8.
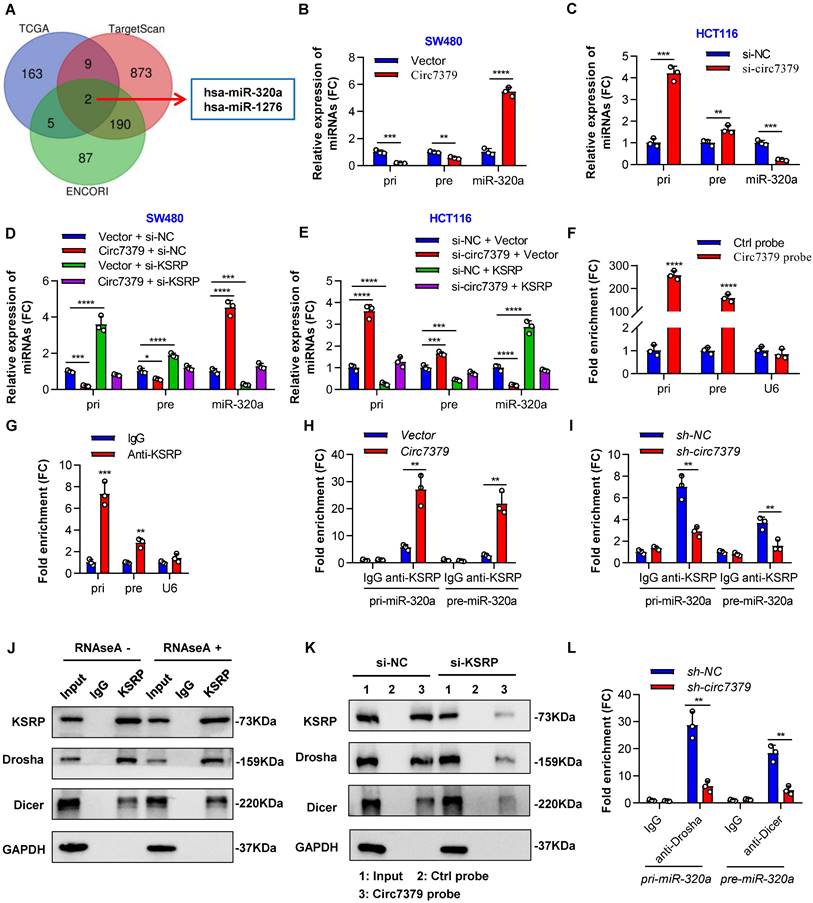
To examine whether circ7379/KSRP-driven upregulation of miR-320a is mediated through the modulation of primary miR-320a (pri-miR-320a) and/or precursor miR-320a (pre-miR-320a) processing, we detected the expression of pri-miR-320a and pre-miR-320a using specific PCR primers. The levels of pri-miR-320a and pre-miR-320a were decreased in the cells overexpressing circ7379, but the levels of miR-320a were robustly increased (Figure 8B). Moreover, the opposite results were observed in cells with circ7379 depletion (Figure 8C). Rescue experiments demonstrated that circ7379 modulated the processing of pri-miR-320a and pre-miR-320a in a KSRP-dependent manner (Figures 8D and 8E).
How circ7379 and KSRP modulate the processing of pri-miR-320a and pre-miR-320a? NcRNAs, including lncRNAs and circRNAs, can interact with specific target RNAs through high-complementarity base pairing and perform regulatory functions [34, 35]. Therefore, we compared the RNA sequence of circ7379 with those of pri-miR-320a and pre-miR-320a using BLAST and identified two highly complementary regions in pri-miR-320a and one in pre-miR-320a (Figure S8C). RNA pulldown analysis using biotin-labeled circ7379-specific RNA probes revealed that both pri-miR-320a and pre-miR-320a were enriched in the circ7379 probe group compared with the control probe group (Figure 8F). Notably, knockdown of KSRP had little effect on pri-miR-320a or pre-miR-320a binding to circ7379 (Figure S8D), suggesting that circ7379 may interact with pri-miR-320a or pre-miR-320a independent of KSRP.
The binding between KSRP and both pri-miR-320a and pre-miR-320a was demonstrated by RIP assay (Figure 8G). Moreover, the overexpression of circ7379 increased the enrichment of pri-miR-320a and pre-miR-320a in the complexes precipitated with the anti-KSRP antibody (Figure 8H); in contrast, the circ7379 depletion attenuated the interactions between KSRP and pri-miR-320a or pre-miR-320a (Figure 8I). Taken together, our data indicate that circ7379 acts as a scaffold to enhance the interaction between KSRP and pri-miR-320a or pre-miR-320a.
Given that the maturation of miRNAs heavily relies on the efficient work of the Drosha complex and Dicer complex [36, 37], we further hypothesised that the circ7379/KSRP complex may facilitate Drosha binding to pri-miR-320a and Dicer binding to pre-miR-320a, thus promoting their processing. To confirm this speculation, we performed an endogenous Co-IP assay and found that KSRP interacted with Drosha and Dicer in an RNA-independent manner (Figure 8J). RNA pulldown assay revealed that knocking down KSRP decreased the amount of Drosha and Dicer that were pulled down by the circ7379 probe (Figure 8K). Similarly, the circ7379 depletion attenuated the interaction between pri-miR-320a and Drosha and the interaction between pre-miR-320a and Dicer (Figure 8L). These results indicate that circ7379 could directly bind pri-miR-320a and pre-miR-320a and indirectly recruit Drosha and Dicer through KSRP, promoting the processing of both pri-miR-320a and pre-miR-320a and leading to a robustly increased level of mature miR-320a.
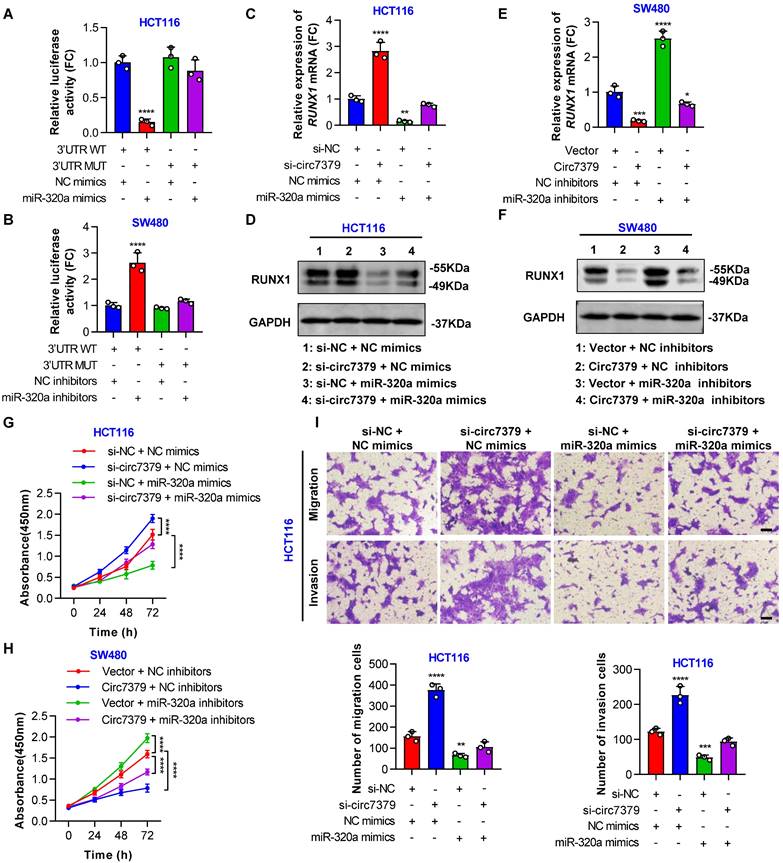
Circ7379 inhibits RUNX1 expression by upregulating miR-320a
In subsequent studies, we attempted to clarify the exact role of miR-320a in circ7379-driven RUNX1 downregulation and circ7379-induced CRC inhibition. First, we transfected mimics or inhibitors of miR-320a into CRC cells and performed qRT-PCR to confirm the overexpression or silencing of miR-320a (Figures S9A and S9B). Functionally, ectopic miR-320a expression in CRC cells reduced the mRNA and protein levels of RUNX1 (Figures S9C and S9D), while the knockdown of miR-320a led to an increased expression of RUNX1 (Figures S9E and S9F).
In addition, a conserved 7mer-m8 site at nt 713-719 in the RUNX1 3'UTR, which may serve as a miRNA response element (MRE) for miR-320a, was predicted by TargetScan (Figure S9G). To prove this prediction, we cloned a wild-type fragment of the RUNX1 3'UTR containing the miR-320a binding sites and a mutant fragment of the RUNX1 3'UTR into the GV272-basic luciferase reporter vector separately (Figure S9H). The dual luciferase reporter assay showed that the miR-320a mimic decreased luciferase activity in the wild-type group, but not in the mutant group (Figure 9A). Moreover, the luciferase activity driven by the wild-type vector, but not that driven by the mutant vector, was increased by the miR-320a inhibitor (Figure 9B), suggesting that RUNX1 is a target gene of miR-320a. Importantly, the increased RUNX1 expression induced by the knockdown of circ7379 was abolished by miR-320a mimics (Figures 9C and 9D), and the decreased RUNX1 expression resulting from the overexpression of circ7379 was also partially reversed by miR-320a inhibitors (Figures 9E and 9F). These results demonstrated that circ7379 inhibits the expression of RUNX1 mainly by upregulating miR-320a.
Subsequently, we designed a series of functional rescue experiments using the miR-320a mimic and inhibitor to further confirm that circ7379 inhibits CRC cell proliferation and metastasis through the circ7379/miR-320a/RUNX1 axis. Indeed, the miR-320a mimic abrogated the proliferation-, migration- and invasion-promoting effects induced by the knockdown of circ7379 in HCT116 cells (Figures 9G and 9H). Similarly, the inhibitory effects of circ7379 overexpression in SW480 cells were also partially reversed by the miR-320a inhibitor (Figures 9I and S9I). In summary, these data demonstrate that circ7379 serves as a pivotal regulator of miR-320a to downregulate RUNX1 expression, leading to the inhibition of the proliferation, migration and invasion of CRC cells.
Circ7379 inhibits RUNX1 expression by upregulating miR-320a. (A) The luciferase activities of CRC cells cotransfected with WT or MUT RUNX1 3'UTR luciferase reporter vector and control miRNA mimics or miR-320a mimics. (B) The luciferase activities of CRC cells cotransfected with WT or MUT RUNX1 3'UTR luciferase reporter vector and control miRNA inhibitors or miR-320a inhibitors. (C) qRT‒PCR showing the expression of RUNX1 mRNA in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379), miR-320a overexpression (miR-320a mimics) or si-circ7379 + miR-320a mimic cotransfection. (D) Representative Western blot of RUNX1 protein in CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379), miR-320a overexpression (miR-320a mimics) or si-circ7379 + miR-320a mimic cotransfection. (E) qRT‒PCR showing the expression of RUNX1 mRNA in CRC cells after the transfection of the control vector, circ7379 vector, or miR-320a inhibitors or cotransfection of circ7379 vector + miR-320a inhibitors. (F) Representative Western blot of RUNX1 protein in CRC cells after the transfection of the control vector, circ7379 vector, or miR-320a inhibitors or cotransfection of circ7379 vector + miR-320a inhibitors. (G) CCK-8 assay showing the proliferation ability of CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379), miR-320a overexpression (miR-320a mimics) or si-circ7379 + miR-320a mimic cotransfection. (H) CCK-8 assay showing the proliferation ability of CRC cells after the transfection of the control vector, circ7379 vector, or miR-320a inhibitors or cotransfection of circ7379 vector + miR-320a inhibitors. (I) Transwell assay showing the migration and invasion abilities of CRC cells under control conditions (si-NC) or upon circ7379 knockdown (si-circ7379), miR-320a overexpression (miR-320a mimics) or si-circ7379 + miR-320a mimic cotransfection. Scale bar, 100 µm. The data are shown as the mean ± SD. The P values were determined by a one-way or two-way ANOVA (G and I); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. See also Figure S9.
Exogenous circ7379 inhibits CRC growth in PDO and PDX models. (A) The growth of CRC PDOs after the transfection of the control vector or circ7379 lentiviral vector. Left, representative images of CRC PDOs at the indicated time points in each group; scale bar, 50 µm. Right panel, growth curves of CRC PDOs in each group. (B) qRT‒PCR showing the expression of circ7379, miR-320a, and RUNX1 mRNA in CRC PDOs after the transfection of the control vector or circ7379 lentiviral vector. (C) Immunofluorescence (IF) showing the expression of RUNX1 protein in CRC PDOs after the transfection of the control vector or circ7379 lentiviral vector. Scale bar, 20 µm. (D) The growth of CRC PDXs after intratumoral injection of the control vector or circ7379 lentiviral vector. Top, images of tumors in mice in each group (n=3 mice/group). Bottom, tumor weights of CRC PDXs in each group. (E) qRT‒PCR showing the expression of circ7379, miR-320a, and RUNX1 mRNA in CRC PDXs after intratumoral injection of the control vector or circ7379 lentiviral vector. (F) Immunohistochemistry (IHC) showing the expression of RUNX1 protein in CRC PDXs after intratumoral injection of the control vector or circ7379 lentiviral vector. Scale bar, 50 µm (left), 20 µm (right). The data are shown as the mean ± SD. The P values were determined by a two-tailed unpaired Student's t test or two-way ANOVA (A); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
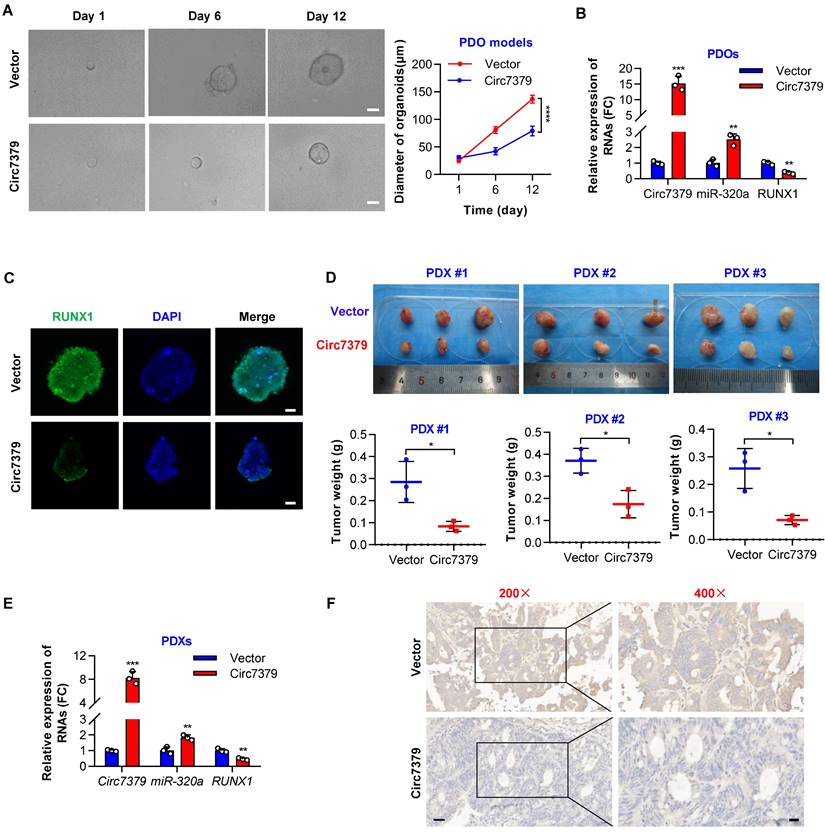
Exogenous circ7379 inhibits CRC growth in PDO and PDX models
To further investigate the potential clinical implications of circ7379 in CRC treatment, we successfully established CRC patient-derived organoid (PDO) and xenograft (PDX) models. We first overexpressed circ7379 in PDOs using lentiviral vectors and found that ectopic circ7379 inhibited the growth of PDOs (Figure 10A). qRT-PCR results showed that the expression levels of circ7379 and miR-320a were increased in PDOs in the circ7379 overexpression group compared with those in the NC vector group, while the expression level of RUNX1 mRNA was decreased (Figure 10B). Consistently, the expression levels of RUNX1 protein were decreased in PDOs in the circ7379 overexpression group compared to the control group as evidenced by the IF assay (Figure 10C).
Additionally, we overexpressed circ7379 in three PDXs by intratumoral injection of lentiviral particles carrying the circ7379 vector and found that circ7379 overexpression suppressed tumor growth in mice (Figure 10D). As expected, increased levels of circ7379 and miR-320a and decreased levels of RUNX1 mRNA was observed in the circ7379 overexpression plasmid-treated group (Figure 10E). Furthermore, the IHC analysis showed that the level of RUNX1 protein was also decreased in the circ7379 overexpression plasmid-treated group compared to the control group (Figure 10F), indicating that ectopic circ7379 effectively reduced PDX tumor growth by blocking RUNX1 expression. In summary, exogenous circ7379 inhibits the growth of CRC in CRC patient-derived tumor models.
Discussion
Recent advances in RNA manufacturing and cell delivery have enabled the development of RNA-based therapeutics for a wide range of applications. In this study, we first identified an intergenic circRNA, circ7379, that was downregulated in CRC. We then confirmed that the biogenesis of intergenic circ7379 was mediated by RCMs and negatively regulated by DHX9. Importantly, we revealed that circ7379 inhibited CRC growth and metastasis by modulating miR-320a maturation and blocking RUNX1 expression in a KSRP-dependent manner. Finally, our experimental models supported circ7379 as a promising RNA treatment molecule for CRC.
Intergenic circRNAs constitute a special subclass of circRNAs in terms of genomic distribution, and their expression in tumors is cell type specific. For example, approximately 5.0% of dysregulated circRNAs are intergenic circRNAs in bladder cancer [38], whereas the percentage may be as high as 35.3% in gastric cancer [39]. Intergenic circRNA abundance was relatively low in CRC compared with gastric cancer, ranging from 1.5% to 14.0% [40, 41]. In our circRNA microarray analyses, two (4.3%) of 46 intergenic circRNAs were differentially expressed in CRC tissues, and circ7379 was verified to be the most downregulated circRNA. Notably, circ7379 was also one of the most downregulated circRNAs in CRC tissues as determined by RNA-Seq [16] and other microarrays (e.g., GSE121895). We further found that circ7379 had a relatively high abundance in both adjacent normal tissues and normal FHC cells, suggesting its vital role in normal physiological processes. Furthermore, a low circ7379 level was associated with CRC tumorigenesis and unfavorable clinicopathological characteristics in CRC patients.
Regarding the function of circ7379 in normal physiology, we speculated that circ7379 may act as a tumor suppressor gene, which inhibits the transformation of normal intestinal epithelial cells into abnormal cells or even cancer cells. Notably, several circRNAs have been reported to be involved in regulating the homeostasis of the intestinal epithelium. For example, Xiao et al. surveyed circRNAs required for intestinal epithelial repair after acute injury and identified circular homeodomain-interacting protein kinase 3 (circHIPK3) as a major regulator that promotes homeostasis of the intestinal epithelium by reducing microRNA 29b (miR-29b) function [42, 43]. Another study by Zhu et al. found that deletion of the immune cell-associated circular RNA circPan3 (originating from the Pan3 gene transcript) in Lgr5+ intestinal stem cells (ISCs) impairs their self-renewal capacity and the regeneration of gut epithelium in a manner dependent on immune cells [44, 45]. More recently, Guo et al. identified a circular RNA, circBtnl1, that suppresses the self-renewal capacity of ISCs and epithelial regeneration via disruption of Atf4 mRNA stability [46]. However, whether circ7379 plays a role in intestinal epithelium homeostasis remains elusive, and we will focus on this issue in our future work.
The mechanism of circRNA biogenesis in CRC and the regulatory process of circRNA biogenesis remain rather elusive [5]. In this study, we identified highly matched reverse complementary sequences in the upstream and downstream flanking regions of circ7379 and confirmed that these two RCMs are responsible for the biogenesis of circ7379. Our study further revealed that highly expressed DHX9 can bind the upstream and downstream flanking regions of circ7379 and prevent base pairing between reverse complementary sequences, resulting in the downregulation of circ7379. A previous study showed that PI3KK-mediated phosphorylation of DHX9 near the substrate binding sites may impair the capacity of DHX9 to resolve RNA pairing, which, in turn, promotes oxaliplatin-induced expression of circCCDC66 in CRC cells [47]. However, in contrast to this proposed model, our findings may provide a novel and alternative mechanism to explain the link between downregulated circRNAs and high expression of DHX9 in CRC.
Aberrant intergenic lncRNAs (lincRNAs) affect many aspects of CRC biology, including tumorigenesis, stemness, metastasis, and chemoradiotherapy resistance. For example, LINC00460 [48], LINC00152 [49], and LINRIS [50], which are highly expressed in CRC, promote CRC progression and/or confer resistance to oxaliplatin (L-OHP)-induced apoptosis. In this study, we found that the overexpression of intergenic circ7379 suppressed the proliferation and metastasis of CRC cells in vitro and in vivo. Subsequently, we performed high-throughput RNA-seq and identified RUNX1 as the target gene of circ7379. Our rescue experiments also revealed that circ7379 affects the malignant phenotype of CRC cells by downregulating RUNX1 expression. Thus, these findings provide new insights into the role of intergenic circRNAs in CRC initiation and progression.
Although the competitive endogenous RNA (ceRNA) hypothesis has been proposed as the most common mechanism by which circRNAs perform their functions, this sponge function of circRNAs has been questioned partially because the physiological changes in the expression of most individual circRNAs do not compromise miRNA activity [51, 52]. In contrast, circRNAs can interact with RBPs to control transcription and splicing or scaffolding or sequestering macromolecules to interfere with miRNA activities or signaling pathways [53]. Inspired by these findings, we identified that circ7379 can bind the KH4 domain of KSRP via the fifth GUCC motif (sites 2000-2003). Our study further revealed that circ7379 acted as a scaffold to promote the processing of both pri-miR-320a and premiR-320a in a KSRP-dependent manner, leading to a robustly increased level of mature miR-320a. These structural and functional findings establish a key circRNA-miRNA-mRNA regulatory pathway to suppress CRC cell growth.
Generally, the biogenesis of mature miRNAs involves the transcription of miRNA genes as pri-miRNAs, processing of pri-miRNAs into pre-miRNAs, exporting of pre-miRNAs to the cytoplasm, and processing of pre-miRNAs into mature miRNAs [36]. The processing and maturation of miRNAs heavily rely on the efficient activity of the microprocessor complex in the nucleus and the Dicer complex in the cytoplasm [37]. The microprocessor complex, comprising mainly the RNase III enzyme Drosha and a DGCR8 dimer, cleaves the pri-miRNA to liberate a pre-miRNA containing approximately 70 nt, while Dicer, in concert with TRBP, generates a miRNA duplex of approximately 22 nt from the pre-miRNA [54]. Our present work shows that KSRP can directly interact with Drosha and Dicer in CRC cells. Specifically, we demonstrated that circ7379 can directly bind pri-miR-320a and pre-miR-320a through high-complementarity base pairing and indirectly recruit the Drosha complex in the nucleus and the Dicer complex in the cytoplasm through KSRP to facilitate the processing of both pri-miR-320a and pre-miR-320a and promote the maturation of miR-320a, which targets RUNX1. Thus, circRNAs may regulate miRNA biogenesis in CRC at the posttranscriptional level [55, 56]. Our work indicated that intergenic circRNAs act as an accessory component of both the microprocessor complex and the Dicer complex, extending the understanding of the regulatory functions of intergenic circRNAs in physiological and pathological processes.
Another unique advantage of the current study is that we were able to confirm the hypothesis by using clinically relevant PDO and PDX models. PDO and PDX models have recently emerged as robust preclinical models with the potential to predict clinical outcomes in patients [57, 58]. However, few studies investigating circRNAs have applied PDO or PDX models to further substantiate their findings [39, 59]. Here, we found that the ectopic overexpression of circ7379 suppressed the growth of CRC in PDO and PDX models. Researchers have designed and constructed artificial circRNAs using enzymatic ligation in vitro, and these synthetic circRNAs can be stably expressed in cancer cells and efficiently function as miRNA sponges (e.g., miR-21 and miR-93) or protein sponges (e.g., hnRNP L) [9-11]. We will attempt to synthesize circ7379 in vitro in the future to further explore the clinical value for CRC treatment alone or combined with photodynamic therapies or sonodynamic therapies.
However, our study has limitations that should be mentioned. First, the mechanism by which circ7379 is exported from the nucleus to the cytoplasm remains unclear, although we found that this location change in circ7379 promotes the processing of pre-miR-320a. Second, although several endonucleases (e.g., RNase P and RNase L) can cleave circRNAs internally under certain conditions [60, 61], we did not further explore the mechanism underlying circ7379 decay.
Conclusions
We provide the first evidence that intergenic region-derived circ7379 inhibits the proliferation and metastasis of CRC via the KSRP/miR-320a/RUNX1 axis. These findings could not only increase the understanding of circRNAs regulating tumor development, but also provide potential resources for monitoring and treating CRC.
Abbreviations
ATCC, American Type Culture Collection; BLAST, Basic Local Alignment Search Tool; CCK-8, cell counting kit-8; cDNA, complementary DNA; ceRNA, competitive endogenous RNA; circRNA, circular RNA; CRC, colorectal cancer; FC, fold change; FISH, fluorescence in situ hybridization; gDNA, genomic DNA; GO, Gene Ontology; IF, immunofluorescence; KEGG, Kyoto Encyclopedia of Genes and Genomes; KSRP, KH-type splicing regulatory protein; lincRNA, long intergenic non-coding RNA; miRNA, microRNA; MRE, miRNA reaction element; ncRNA, non-coding RNA; PDO, patient-derived organoid; PDX, patient-derived xenograft; RAP, RNA antisense purification; RBP, RNA binding protein; RCM, reverse complementary match; RIP, RNA-binding protein immunoprecipitation; RUNX1, RUNX family transcription factor 1; shRNA, short hairpin RNA; siRNA, small interfering RNA.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank Prof. Daolin Tang (Department of Surgery, University of Texas Southwestern Medical Center) for his critical reading of the manuscript; Dr. Zhi Lin (Department of Pediatrics, The Third Xiangya Hospital, Central South University,) for advice and useful discussions.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82172833), Natural Science Foundation of Hunan Province (No. 2019JJ50906, No. 2019JJ80030, and No. 2023JJ40894), Hunan Provincial Science and Technology Plan Project (No. 2022RC1218), the Natural Science Foundation of Changsha City (No. kq2208361), the Wisdom Accumulation and Talent Cultivation Project of the Third Xiangya Hospital of Central South University (No. YX202107), and the College Students' Innovative Entrepreneurial Training Plan Program of the Third Xiangya Hospital of Central South University (No. S2021105331000, No. S2022105330470).
Ethics approval and consent to participate
This study was reviewed and approved by the Ethics Committee of the Third Xiangya Hospital of Central South University, and written informed consent forms were obtained from all patients.
Consent for publication
All authors consent to publication.
Availability of data and material
All the data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request.
Author contributions
FL and LL co-designed and performed most of the experiments. CX, MM, ZW, and BL collected and analyzed the data. ZL and MY performed bioinformatical analyses. FZ, ZN, CZ, and BY coordinated and performed animal experiments. SL, LW, BT, KY, YG, and MC helped in the experimental part. JC, XL, and GH interpreted the data. FL, LL, CL, and YZ wrote the manuscript with inputs from all the authors. CL and YZ co-designed, supervised, and coordinated the work. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
2. Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK. et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. Journal of the National Comprehensive Cancer Network: JNCCN. 2021;19:329-59
3. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nature reviews Cancer. 2021;21:22-36
4. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nature reviews Clinical oncology. 2022;19:188-206
5. Long F, Lin Z, Li L, Ma M, Lu Z, Jing L. et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Molecular cancer. 2021;20:26
6. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nature reviews Molecular cell biology. 2020;21:475-90
7. Liu X, Zhang Y, Zhou S, Dain L, Mei L, Zhu G. Circular RNA: An emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. Journal of controlled release: official journal of the Controlled Release Society. 2022;348:84-94
8. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nature reviews Drug discovery. 2021;20:629-51
9. Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G. et al. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Molecular therapy Nucleic acids. 2018;13:312-21
10. Wang Z, Ma K, Cheng Y, Abraham JM, Liu X, Ke X. et al. Synthetic circular multi-miR sponge simultaneously inhibits miR-21 and miR-93 in esophageal carcinoma. Laboratory investigation; a journal of technical methods and pathology. 2019;99:1442-53
11. Schreiner S, Didio A, Hung LH, Bindereif A. Design and application of circular RNAs with protein-sponge function. Nucleic Acids Res. 2020;48:12326-35
12. Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185:2016-34
13. Lin C, Ma M, Zhang Y, Li L, Long F, Xie C. et al. The N(6)-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Molecular cancer. 2022;21:80
14. Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D. et al. Isolation and in vitro expansion of human colonic stem cells. Nature medicine. 2011;17:1225-7
15. Woo XY, Giordano J, Srivastava A, Zhao ZM, Lloyd MW, de Bruijn R. et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nature genetics. 2021;53:86-99
16. Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W. et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Molecular cancer. 2022;21:49
17. Chen Z, Chen H, Yang L, Li X, Wang Z. CircPLCE1 facilitates the malignant progression of colorectal cancer by repressing the SRSF2-dependent PLCE1 pre-RNA splicing. Journal of cellular and molecular medicine. 2021;25:7244-56
18. Ruan H, Deng X, Dong L, Yang D, Xu Y, Peng H. et al. Circular RNA circ_0002138 is down-regulated and suppresses cell proliferation in colorectal cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;111:1022-8
19. Li Z, Yao H, Wang S, Li G, Gu X. CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. J Exp Clin Cancer Res. 2020;39:160
20. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell reports. 2015;10:170-7
21. Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G. et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115-9
22. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
23. Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T. et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res. 2017;23:3918-28
24. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H. et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nature communications. 2021;12:295
25. He L, Man C, Xiang S, Yao L, Wang X, Fan Y. Circular RNAs' cap-independent translation protein and its roles in carcinomas. Molecular cancer. 2021;20:119
26. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37.e9
27. Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X. et al. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Molecular therapy Nucleic acids. 2019;18:88-98
28. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805-14
29. Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang J. et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Molecular cancer. 2020;19:71
30. Zhang C, Zhou X, Geng X, Zhang Y, Wang J, Wang Y. et al. Circular RNA hsa_circ_0006401 promotes proliferation and metastasis in colorectal carcinoma. Cell Death Dis. 2021;12:443
31. Li Y, Chen B, Zhao J, Li Q, Chen S, Guo T. et al. HNRNPL Circularizes ARHGAP35 to Produce an Oncogenic Protein. Advanced science (Weinheim, Baden-Wurttemberg, Germany). 2021;8:2001701
32. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Molecular cancer. 2020;19:172
33. Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A. et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010-4
34. Wang FW, Cao CH, Han K, Zhao YX, Cai MY, Xiang ZC. et al. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J Clin Invest. 2019;129:727-43
35. Rossi F, Beltran M, Damizia M, Grelloni C, Colantoni A, Setti A. et al. Circular RNA ZNF609/CKAP5 mRNA interaction regulates microtubule dynamics and tumorigenicity. Molecular cell. 2022;82:75-89.e9
36. Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews Molecular cell biology. 2014;15:509-24
37. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nature reviews Cancer. 2015;15:321-33
38. Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L. et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res. 2018;24:6319-30
39. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z. et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Molecular cancer. 2019;18:20
40. Tian Y, Xu Y, Wang H, Shu R, Sun L, Zeng Y. et al. Comprehensive analysis of microarray expression profiles of circRNAs and lncRNAs with associated co-expression networks in human colorectal cancer. Funct Integr Genomics. 2019;19:311-27
41. Zhou C, Liu HS, Wang FW, Hu T, Liang ZX, Lan N. et al. circCAMSAP1 Promotes Tumor Growth in Colorectal Cancer via the miR-328-5p/E2F1 Axis. Molecular therapy: the journal of the American Society of Gene Therapy. 2020;28:914-28
42. Xiao L, Ma XX, Luo J, Chung HK, Kwon MS, Yu TX. et al. Circular RNA CircHIPK3 Promotes Homeostasis of the Intestinal Epithelium by Reducing MicroRNA 29b Function. Gastroenterology. 2021;161:1303-17.e3
43. Kulkarni DH, Newberry RD. A Novel Role of Circular RNA in Intestinal Epithelial Repair. Gastroenterology. 2021;161:1108-10
44. Zhu P, Zhu X, Wu J, He L, Lu T, Wang Y. et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nature immunology. 2019;20:183-94
45. Soveg F, von Moltke J, Savan R. CIRCling the wagons to protect intestinal stem cells. Nature immunology. 2019;20:114-6
46. Guo H, Zhang J, Jiang Z, Zhu X, Yang J, Mu R. et al. Noncoding RNA circBtnl1 suppresses self-renewal of intestinal stem cells via disruption of Atf4 mRNA stability. The EMBO journal. 2023;42:e112039
47. Lin YC, Yu YS, Lin HH, Hsiao KY. Oxaliplatin-Induced DHX9 Phosphorylation Promotes Oncogenic Circular RNA CCDC66 Expression and Development of Chemoresistance. Cancers (Basel). 2020 12
48. Meng X, Sun W, Yu J, Zhou Y, Gu Y, Han J. et al. LINC00460-miR-149-5p/miR-150-5p-Mutant p53 Feedback Loop Promotes Oxaliplatin Resistance in Colorectal Cancer. Molecular therapy Nucleic acids. 2020;22:1004-15
49. Ou C, Sun Z, He X, Li X, Fan S, Zheng X. et al. Targeting YAP1/LINC00152/FSCN1 Signaling Axis Prevents the Progression of Colorectal Cancer. Advanced science (Weinheim, Baden-Wurttemberg, Germany). 2020;7:1901380
50. Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX. et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Molecular cancer. 2019;18:174
51. Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766-76
52. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nature reviews Genetics. 2016;17:272-83
53. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503-17
54. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nature reviews Molecular cell biology. 2019;20:5-20
55. Zhi Q, Wan D, Ren R, Xu Z, Guo X, Han Y. et al. Circular RNA profiling identifies circ102049 as a key regulator of colorectal liver metastasis. Molecular oncology. 2021;15:623-41
56. Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX. et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Molecular cancer. 2020;19:60
57. Durinikova E, Buzo K, Arena S. Preclinical models as patients' avatars for precision medicine in colorectal cancer: past and future challenges. J Exp Clin Cancer Res. 2021;40:185
58. Yoshida GJ. Applications of patient-derived tumor xenograft models and tumor organoids. Journal of hematology & oncology. 2020;13:4
59. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD. et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nature communications. 2019;10:4695
60. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK. et al. Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74:494-507.e8
61. Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK. et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell. 2019;177:865-80.e21
Author contact
![]() Corresponding authors: linchangweiedu.cn (C.L.); yzhangxy3edu.cn (Y.Z.).
Corresponding authors: linchangweiedu.cn (C.L.); yzhangxy3edu.cn (Y.Z.).

 Global reach, higher impact
Global reach, higher impact