10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(1):109-119. doi:10.7150/ijbs.12358 This issue Cite
Research Paper
The Complete Mitochondrial Genome of two Tetragnatha Spiders (Araneae: Tetragnathidae): Severe Truncation of tRNAs and Novel Gene Rearrangements in Araneae
Zhejiang Provincial Key Laboratory of Biometrology and Inspection and Quarantine, College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, 310018, People's Republic of China
# These authors contributed equally to this paper.
Abstract
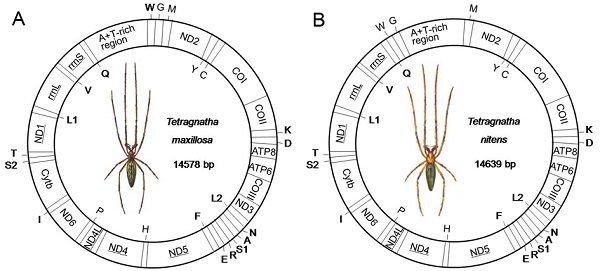
Mitogenomes can provide information for phylogenetic analysis and evolutionary biology. The Araneae is one of the largest orders of Arachnida with great economic importance. In order to develop mitogenome data for this significant group, we determined the complete mitogenomes of two long jawed spiders Tetragnatha maxillosa and T. nitens and performed the comparative analysis with previously published spider mitogenomes. The circular mitogenomes are 14578 bp long with A+T content of 74.5% in T. maxillosa and 14639 bp long with A+T content of 74.3% in T. nitens, respectively. Both the mitogenomes contain a standard set of 37 genes and an A+T-rich region with the same gene orientation as the other spider mitogenomes, with the exception of the different gene order by the rearrangement of two tRNAs (trnW and trnG). Most of the tRNAs lose TΨC arm stems and have unpaired amino acid acceptor arms. As interesting features, both trnSAGN and trnSUCN lack the dihydrouracil (DHU) arm and long tandem repeat units are presented in the A+T-rich region of both the spider mitogenomes. The phylogenetic relationships of 23 spider mitogenomes based on the concatenated nucleotides sequences of 13 protein-coding genes indicated that the mitogenome sequences could be useful in resolving higher-level relationship of Araneae. The molecular information acquired from the results of this study should be very useful for future researches on mitogenomic evolution and genetic diversities in spiders.
Keywords: Mitogenome, Araneae, Tetragnatha maxillosa, Tetragnatha nitens, Phylogeny

 Global reach, higher impact
Global reach, higher impact