10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):872-882. doi:10.7150/ijbs.24595 This issue Cite
Research Paper
Research on folding diversity in statistical learning methods for RNA secondary structure prediction
1. College of Computer Science, Sichuan University, China
2. College of Chemistry, Sichuan University, China
3. Vice Dean of College of Computer Science, Sichuan University
4. Department of Computer Science and Information Technology, La Trobe University, Australia
*These authors contributed equally to this work and should be considered co-first authors
Abstract
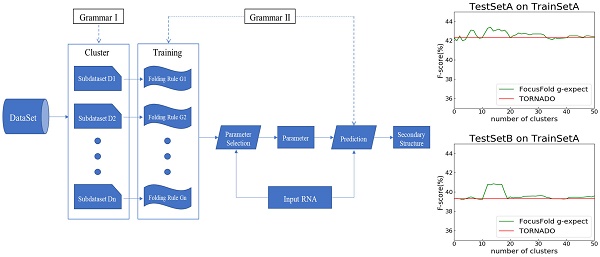
How to improve the prediction accuracy of RNA secondary structure is currently a hot topic. The existing prediction methods for a single sequence do not fully consider the folding diversity which may occur among RNAs with different functions or sources. This paper explores the relationship between folding diversity and prediction accuracy, and puts forward a new method to improve the prediction accuracy of RNA secondary structure. Our research investigates the following: 1. The folding feature based on stochastic context-free grammar is proposed. By using dimension reduction and clustering techniques, some public data sets are analyzed. The results show that there is significant folding diversity among different RNA families. 2. To assign folding rules to RNAs without structural information, a classification method based on production probability is proposed. The experimental results show that the classification method proposed in this paper can effectively classify the RNAs of unknown structure. 3. Based on the existing prediction methods of statistical learning models, an RNA secondary structure prediction framework is proposed, namely “Cluster - Training - Parameter Selection - Prediction”. The results show that, with information on folding diversity, prediction accuracy can be significantly improved.
Keywords: RNA secondary structure prediction, statistical learning model, folding diversity, stochastic context-free grammar

 Global reach, higher impact
Global reach, higher impact