10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2006; 2(3):95-103. doi:10.7150/ijbs.2.95 This issue Cite
Review
Hox genes are not always Colinear
Department of Zoology, University of Oxford, UK
Received 2006-2-15; Accepted 2006-4-15; Published 2006-5-5
Abstract
The deuterostomes are the clade of animals for which we have the most detailed understanding of Hox cluster organisation. With the Hox cluster of amphioxus (Branchiostoma floridae) we have the best prototypical, least derived Hox cluster for the group, whilst the urochordates present us with some of the most highly derived and disintegrated clusters. Combined with the detailed mechanistic understanding of vertebrate Hox regulation, the deuterostomes provide much of the most useful data for understanding Hox cluster evolution. Considering both the prototypical and derived deuterostome Hox clusters leads us to hypothesize that Temporal Colinearity is the main constraining force on Hox cluster organisation, but until we have a much deeper understanding of the mechanistic basis for this phenomenon, and know how widespread across the Bilateria the mechanism(s) is/are, then we cannot know how the Hox cluster of the last common bilaterian operated and what have been the major evolutionary forces operating upon the Hox gene cluster.
Keywords: Amphioxus, echinoderm, hemichordate, deuterostomes, Temporal Colinearity.
1. Introduction
The Hox genes are a set of transcription factor-encoding genes that pattern the anterior-posterior body axis of animals. They tend to have a very distinctive organisation in the genome, being arranged in gene clusters in which the order of the genes within the cluster corresponds to (or is Colinear with) some aspect of the gene expression. Spatial Colinearity was first recognised by Ed Lewis [1] in Drosophila melanogaster, whereby the domains of action of each homeotic locus of the Bithorax complex (BX-C) are staggered along the anterior-posterior axis in an order that matches their position along the chromosome. Such Spatial Colinearity is also observed in the Antennapedia Complex (ANT-C), which together with the BX-C constitutes the Hox cluster of D. melanogaster. The phenomenon was also extended to the Hox clusters of vertebrates soon after the discovery of the homeobox; the homeobox being the major motif of the Hox genes, that is 180bp long and encodes a sequence-specific DNA-binding domain [2, 3, 4].
In addition to Spatial Colinearity, Temporal Colinearity is also observed in some Hox clusters; principally those of vertebrates in which the genes at the 3'/Anterior end of the cluster become transcriptionally active first, and then progressively genes towards the 5'/Posterior end of the cluster are initiated later [5]. This temporal control of the mammalian Hox clusters is correlated with modulation of their chromatin conformation [6]. Temporal and Spatial Colinearity can be mechanistically linked, with the time of gene initiation determining the axial limits of expression [7]. However the two phenomena are also separable. Drosophila exhibits Spatial Colinearity without any obvious Temporal Colinearity, as does the larvacean urochordate Oikopleura dioica [8] (although see below), and distinct regulatory regions that drive Temporal and Spatial Colinearity have recently been discovered in mice [9].
A third form of Colinearity is Quantitative. In mouse limb development the proximity to a digit enhancer determines the levels of expression of a gene, the gene closest to the enhancer being the most highly expressed [10, 11]. As with Temporal Colinearity potentially resulting in Spatial Colinearity, Quantitative Colinearity could similarly lead to Spatial Colinearity as well. The organisation of the Hox cluster and the position of the genes within it are thus intimately linked to the transcriptional regulation of the genes.
Hox gene Colinearity in some shape or form seems to be widespread, and so is usually assumed to be the major reason that the genes remain clustered. Hox genes from flies to humans clearly are homologous, and even orthologous in most instances, and undoubtedly they were clustered in the last common ancestor of the bilaterian animals (and probably even deeper in animal evolution - before the origin of the Cnidaria) [12]. If we can discern what the homologous features of Hox cluster organisation, function and regulation are across the animal kingdom, then we can discover how these genes were arranged and were operating in an organism that lived more than 525 Million Years Ago, from which all subsequent animal lineages diverged.
Here we outline how an understanding of both prototypical and divergent organisms is essential in order to distinguish the most fundamental ancestral characters, taking the deuterostomes as our example (see figure 1). With such an approach ourselves and others have proposed that the mechanism(s) producing Temporal Colinearity is/are potentially the principal constraining force on Hox cluster organisation [7, 13, 14, 15], because the various derived animal lineages that have broken up their Hox clusters almost invariably possess a mode of development in which there is a lack of opportunity for Temporal Colinearity to exist. The disintegrating clusters of Drosophilids, nematodes and urochordates correlate with the rapid embryogenesis of these animals, which can still however retain elements of Hox Spatial Colinearity, thus showing that the mechanisms generating Spatial Colinearity do not absolutely require Hox gene clustering. The mechanisms generating Temporal Colinearity may well thus be the principal reason that Hox gene clusters are maintained as clusters. However a much closer examination of these mechanisms is required in order to discover whether these similarities are truly homologous, and conversely how some lineages 'escape' from these 'fundamental' constraints during their divergence and diversification.
Hox gene cluster organisation of deuterostomes. The amphioxus Hox cluster is, at present, the only deuterostome Hox cluster that is complete and not rearranged. Horizontal lines represent the chromosome, so that the Ciona cluster is broken into five separate contigs, the Oikopleura cluster has completely disintegrated, and the nature of the Hox gene linkage in the hemichordate and 'other echinoderms' is unknown. Boxes represent individual Hox genes, and their colour denotes their relationships (red = Anterior group/Hox1-2; yellow = Group 3; green = Central group/Hox4-8; blue = Posterior group/Hox9+). Full vertical arrows denote clear orthology relationships, and dashed arrows represent putative orthology relationships that are less certain due to the paucity of diagnostic residues by which the different Central group members can be distinguished. The Posterior group genes are bracketed rather than connected by individual arrows to denote the ambiguity in their orthology relationships, due to Deuterostome Posterior Flexibility and possibly some independent duplication events (see text). X denotes gene loss. For presentation purposes Ciona Hox10 is drawn with the other Posterior group Hox genes, but in reality it is located on the same chromosome and in amongst Hox1-6 (see text for details). Oikopleura Hox4 is followed by a question mark to indicate the uncertainty of its identity due to its position in phylogenetic trees [8]. The genes in the 'other echinoderms' row are an amalgamation of sequences from Crinoids, Ophiuroids and Asteroids [27, 53, 57], and total fourteen different genes, which could potentially indicate the total number of Hox genes for the ancestral deuterostome. The dashed-outline boxes are genes only known from partial fragments.
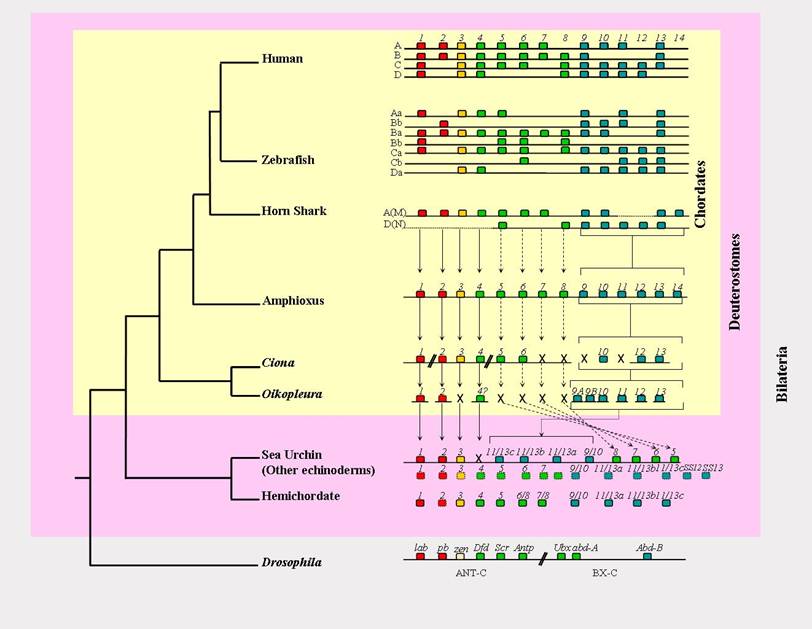
2. An echinoderm Hox cluster: intact but scrambled, and no Temporal Colinearity
Echinoderms are the sister group of hemichordates within the Deuterostome clade, and together as the Ambulacraria form a sister group to chordates [16, 17, 18, 19] (see figure 1). The most distinctive morphological feature of echinoderms is the pentaradial symmetry of the adults, which contrasts with the bilateral symmetry of all other triploblastic animals. This is clearly a derived feature of echinoderms, and along with several further echinoderm synapomorphies (e.g water vascular system, calcareous mesodermally-derived exoskeleton), make it difficult to compare the echinoderm body-plan to other bilaterian, triploblastic animals. Echinoderms do however possess Hox genes, and so examination of their organisation and function in these unusual animals can reveal how the Hox cluster changes in such a derived lineage, and conversely which aspects of Hox biology remain conserved relative to other triploblast animals.
Since Hox genes are integral to regional differentiation along the body axis in animal development it was thought that studying Hox gene expression patterns could give insights into the evolutionary origins of the echinoderm radial body plan. Several Hox genes and Hox fragments were cloned from different sea urchin and starfish species [20, 21, 22, 23, 24, 25, 26, 27]. Expression has been examined most extensively in the sea urchin, Stronglyocentrotus purpuratus. The Hox genes SpHox7 and SpHox11/13b are expressed during embryogenesis, whilst SpHox2, SpHox3, SpHox5, SpHox8, SpHox9/10 and SpHox11/13a are not detectable until adult rudiment formation [29, 29, 30]. These differential activation times, with an 'Anterior' and a 'Posterior' Hox gene being expressed before the intervening genes are activated clearly contravenes Temporal Colinearity (even taking into account the bizarre organisation of the sea urchin Hox cluster, see below).
The expression of some of the Hox genes during adult rudiment development does however seem to be consistent with Spatial Colinearity [31]. Dealing with a subset of the urchin Hox cluster Arenas-Mena et al [31] showed that the expression domains of five Hox genes (SpHox7, SpHox8, SpHox9/10, SpHox11/13a and SpHox11/13b) form a nested pattern in the adult rudiment, such that the staggered boundaries of different Hox domains wrap around the oral-aboral axis of the animal, the different domains or Hox codes potentially specifying each of the five points of the pentaradially symmetrical body-plan. There are further Hox genes in addition to those that exhibit Spatial Colinearity in the adult rudiment, and as of yet we do not know how their expression relates to the genes examined so far. This expression information will be of particular interest given the organisation of the sea urchin Hox cluster.
Sea urchins have a single Hox cluster, which at first was thought to be composed of 10 genes, spanning approximately 0.5Mb, with a gene order matching that of vertebrates [32]. However, more recently fully sequenced bacterial artificial chromosomes containing S. purpuratus Hox genes revealed that in fact the sea urchin has an unusual, rearranged cluster of eleven genes. The four Posterior Hox genes (SpHox9/10, SpHox11/13a, SpHox11/13b and SpHox11/13c) are embedded in between the three Anterior genes (SpHox1, SpHox2 and SpHox3), and the four Central genes (SpHox5, SpHox6, SpHox7 and SpHox8) (Figure 1) [33]. Before this unusual arrangement was known, the sea urchin Hox cluster seemed rather paradoxical in the context of hypotheses that Temporal Colinearity is the principal reason for cluster maintenance. It stood out as an intact cluster that lacked Temporal Colinearity, as distinct from the various other organisms that lack Hox gene Temporal Colinearity and which also have broken and disintegrating Hox clusters (reviewed in [14]). With these new insights from Cameron et al [33] it is now clear that although the sea urchin Hox genes do not exhibit Temporal Colinearity from within an intact cluster, this does in fact correlate with an extremely derived cluster organisation after all.
Nevertheless, the sea urchin still does have a Hox cluster, in contrast to the other derived lineages with regards to Hox gene organisation, which have lost the clustering of their Hox genes (Drosophilids, nematodes, schistosomes and urochordates (reviewed in [8, 14, 34]). Perhaps then the sea urchin Hox cluster is maintained due to enhancer sharing amongst the genes, but without some form of a global cluster regulatory mechanism that results in Colinearity in such Hox clusters as those of mice (reviewed in [35]).
3. Hemichordates: bilaterian ambulacrarians and the potential for a basal deuterostome Hox cluster
Morphological comparison of Saccoglossus kowalevskii, a hemichordate, and chordates initially placed both in the same phylum, in large part due to the deuterostomic embryology and the presence of the stomochord, with its hypothesised homology with the notochord of chordates proper. Hemichordates are however very distinct from the chordates in many respects, and became established as their own phylum [36]. The phylum is composed of two classes: Enteropneusts (Acorn worms) and Pterobranchs (here we will ignore the controversial third class, Planctosphaeoidea) [37]. 18S rDNA phylogenetic analyses place hemichordates as the sister group of echinoderms, and these two phyla together are the Ambulacraria, which in turn is the sister group of chordates [16, 18]. The hemichordates provide us with an ambulacrarian animal that is more easily compared to the chordates than are the echinoderms. Both the hemichordates and chordates are bilaterally symmetrical deuterostomes, with gill slits, and clear (and homologous) anterior-posterior and dorsal-ventral body axes [37, 38, 39].
Several Hox genes have been cloned from two different hemichordate species. A total of eleven Hox genes are described in the literature from S. kowalevskii and Ptychodera flava [38, 40]; Hox1, Hox3, Hox4, Hox5, Hox6/8, Hox7/8, Hox9/10, Hox11/13a, Hox11/13b and Hox11/13c. A partial homeobox of Hox2 was also obtained from S. kowalevskii by PCR, but the sequence was not extended and the entire homeodomain is not available [41]. The genomic organisation of hemichordate Hox genes is still unknown. Some expression information is however available for S. kowalevskii Hox1, 3, 4, 7/8 and one of the 11/13 genes [38]. Interestingly, Hox genes are expressed in a staggered anterior-posterior fashion and are strongly expressed during gastrulation. Lowe et al [38] showed that the expression of Hox genes is colinear with the gene number identity. However given the oddities of the echinoderm cluster organisation we must await the resolution of the hemichordate Hox cluster organisation before it is possible to confidently talk about Colinearity. The hemichordate data is tantalizing, and the resolution of the Hox cluster organisation, along with further expression data, could well be exceptionally illuminating with regards to understanding the Hox cluster of the ancestral deuterostome.
One aspect of deuterostome Hox evolution where the hemichordate data have already changed our perspective is with regard to the phenomenon of Deuterostome Posterior Flexibility. This phenomenon was first described from an analysis of the Posterior Hox genes of amphioxus (Branchiostoma floridae), and describes how the orthology relationships amongst the Hox9-14 genes of deuterostomes are not resolved in molecular phylogenetic trees. This is in stark contrast to the behaviour of the Anterior Hox genes and the Posterior Hox genes of protostome taxa in such trees [42]. The initial hypothesis was that such a lack of resolution could be due to higher rates of sequence evolution in the deuterostome Posterior Hox genes, relative to the Posterior genes of protostomes or the Anterior Hox genes. Now with additional data, particularly from the hemichordates, some cross-phyla orthology relationships do seem to be appearing after all, between some hemichordate and echinoderm Posterior Hox genes [40]. The hemichordate Hox11/13a gene resolves robustly with the echinoderm Hox11/13a gene. The Hox11/13b and c genes, however, have a slightly more ambiguous relationship in the phylogenetic trees; the b and c genes group robustly together in a b/c clade, but hemichordate 11/13b does not group with the echinoderm 11/13b any more clearly than it does with echinoderm 11/13c, and the same is true for hemichordate 11/13c. Yet another topology is exhibited by the Hox9/10 genes, which is the type of relationship that is consistent with the Deuterostome Posterior Flexibility hypothesis. Namely the hemichordate Hox9/10 gene does not resolve with the echinoderm Hox9/10 gene [33, 40]. In this context Cameron et al [33] proposed that the Posterior Hox genes of Ambulacraria and chordates may have resulted from two independent sets of tandem duplications. Evidence of such independent duplications however is not clearly apparent from the phylogenetic trees, as the ambulacrarian genes do not form one clade with the chordate genes forming another [33, 40]. The course of deuterostome Posterior Hox evolution may thus have taken a mixture of routes, producing the diversity of tree topologies described above. Potentially some independent tandem duplications along the ambulacrarian and chordate lineages could account for the robust resolution of some echinoderm and hemichordate orthologies (e.g. Hox11/13a), possibly with further lineage-specific duplications (e.g. a Hox11/13b-c ancestral gene duplicating independently on the hemichordate and echinoderm lineages - although this is not clearly supported or refuted by the phylogenetic trees), whilst Deuterostome Posterior Flexibility still accounts for many of the unresolved Posterior gene relationships across the deuterostomes as a whole (e.g. lack of resolution of ambulacrarian Hox9/10 genes, and the uncertainty of the Hox11/13b-c gene relationships).
4. Urochordates: a case of extreme cluster disintegration and the implications for Colinearity
The subphylum Urochordata contains the ascidians such as Ciona intestinalis, the larvaceans such as O. dioica, the salps (thaliaceans), and the sorberaceans [37]. Together with cephalochordates the urochordates are invertebrate sister groups to vertebrates in the phylum Chordata. Until recently it was generally accepted that cephalochordates group with vertebrates to the exclusion of urochordates (eg. [43, 44]). Now phylogenetic analyses of multiple protein sequence alignments suggest a closer relationship between urochordates and vertebrates than between the latter and cephalochordates [45, 46].
Not withstanding the need to resolve the true phylogenetic position of urochordates within the Chordata, ascidians are important to try to understand deuterostome Hox evolution. Ascidian larvae exhibit prototypical features of the chordate body plan; most notably the possession of a notochord [47]. C. intestinalis belongs to the Ascidiacea class and is the most intensely studied urochordate. Nine Hox genes are present in Ciona (CiHox1,2,3,4,5,6,10,12 and 13) [48,49]. Clearly, several Hox genes have been lost along the Ciona lineage [49]; at least representatives of Hox7, Hox8, Hox9 and Hox11. In addition to this extensive gene loss the arrangement of C. intestinalis Hox genes is unusual. Using fluorescent in situ hybridization (FISH) Ikuta and colleagues [50] showed that Hox genes map on two chromosomes instead of being together on one, and those that are on the same chromosome have an unusual, rearranged gene order, with some large gaps between them that contain many non-Hox genes [48, 50]. This derived, disintegrated organisation of the Hox genes correlates with only remnants of Temporal and Spatial Colinearity persisting (reviewed in [51]).
Dispersal of the Hox genes has occurred to an even greater extent in the larvacean urochordate, O. dioica [8]. In contrast with Ciona however, the remnants of Spatial Colinearity do seem to have been conserved (although we cannot, strictly speaking, talk of Colinearity when there is not a cluster and an ordered array of Hox genes along the chromosome for Hox expression to be Colinear with!). Oikopleura has ten Hox genes (Hox1, 2, 4-1, 4-2, 9A, 9B, 10, 11, 12 and 13), which exhibit spatially coordinated expression in the notochord, tail, muscle, nerve cord and epidermis [8]. The expression domains occur in an anterior-posterior sequence, according to the gene number, i.e. Hox1 is expressed the most anteriorly and Hox13 the most posteriorly. This data does however have to be interpreted with some care. In the molecular phylogenetic trees only the Hox1, Hox2 and Hox13 sequences group robustly with their orthologues from other animals. The remaining Oikopleura Hox genes (Hox4, 9A, 9B, 10, 11 and 12) do not resolve with the paralogy groups of the same number. For example the Oikopleura Hox4 gene resides as the sister group to all of the Central Hox genes of other bilaterians, from Hox4 to Hox8/AbdA [8]. Also the names of Hox9A and Hox9B should not be taken to mean that these two genes are both Hox9 genes that have originated by an Oikopleura-specific duplication. The two sequences do not group together in the phylogenetic tree, as duplicates would be expected to do, and do not resolve with any particular Posterior Hox paralogy group (this being consistent with Deuterostome Posterior Flexibility as outlined above). Notwithstanding this ambiguity over the naming of the Oikopleura Hox genes it is clear that their expression forms a staggered array of domains along the anterior-posterior axis, even though the genes do not exist in a cluster. With regards to the temporal aspect of the expression Seo et al [8] showed that all of the genes start to express at the same time. So Oikopleura provides the most extreme case of Hox cluster disintegration that we know of to date, and this correlates with a lack of a temporal staggering of Hox initiation, but the retention of a spatial staggering.
5. The Amphioxus Hox cluster: the prototypical chordate (and deuterostome?) Hox cluster
Amphioxus (Branchiostoma floridae) has a single, intact Hox cluster consisting of 14 genes, AmphiHox1 - AmphiHox14 (see Figure 1) [42,52]. This Hox cluster is the only one characterised for a deuterostome so far, that is not only intact but has also not lost any genes, and has retained these genes in their ancestral order. All of the vertebrate Hox clusters have lost some genes. The urochordates mentioned above, as well as breaking up their Hox clusters, have also lost genes (see Figure 1), and the sea urchin cluster has lost its Hox4 gene (which is present in other echinoderms [27]), as well as some Posterior Hox genes judging from comparisons to other echinoderms which possess more Posterior Hox genes than the four of the S. purpuratus cluster [53] (see figure 1).
Along with this prototypical organisation of the amphioxus Hox cluster the preliminary data concerning Colinearity also seem to be consistent with the prototypical nature of amphioxus. Wada et al [54] have examined the expression of the first four genes of the amphioxus Hox cluster, and found that they are activated in a temporal progression from 8-9 hours of development for AmphiHox1 through to 14 hours for AmphiHox4. Intriguingly although AmphiHox1, 3 and 4 also exhibit Spatial Colinearity, AmphiHox2 is an exception. The expression domain of AmphiHox2 is initially in the mesoderm, and not in the neural tube [54]. Amphioxus thus exhibits a prototypical Hox cluster organisation for the chordates and deuterostomes, with Temporal Colinearity at least for its Anterior region, but with exceptions to Spatial Colinearity.
The fourteenth amphioxus Hox gene, AmphiHox14, was a surprising find when it was first isolated [42]. It was initially unclear whether the gene was an amphioxus-specific innovation or was revealing to us that the ancestral condition for the vertebrate Hox clusters (with 13 paralogy groups) had not been reconstructed properly from the tetrapod and fish data available at that time. The discovery of Hox14 genes in two separate vertebrates, the Coelocanth and the Horn Shark, has since resolved the issue, at least to some extent [55], since it implies that the vertebrate ancestor had a fourteenth Hox gene as well. It is likely that these vertebrate Hox14 genes are orthologous to AmphiHox14, but as yet this is not formally proven. The position in the Hox cluster and the common organisation of the cephalochordate and vertebrate Hox14 genes, with an intron in the same place in the homeobox, is consistent with the amphioxus and fish genes being orthologous. However the topology of the molecular phylogenetic trees for these genes does not reveal a robust grouping of the cephalochordate and vertebrate Hox14 sequences. So formally they could still have originated by independent tandem duplications. A possible way to resolve this uncertainty would be to see whether the most basal extant vertebrates, the agnathan fish (lampreys and hagfish), have a Hox14 gene, and if so whether its sequence can then break the long branches separating the amphioxus and vertebrate genes, and produce a clearer resolution of the orthology relationships [56]. Whether a Hox14 gene was also present in the deuterostome ancestor is another intriguing possibility. Other Ambulacrarian Hox clusters besides the derived cluster of the sea urchin are one possible approach that could resolve this. Intriguingly when the Hox data from across the echinoderms is considered, there are fourteen different Hox genes present in the phylum as a whole (See Figure 1) [57]. But given the phenomenon of Deuterostome Posterior Flexibility (see above) the relationships between the ambulacrarian and chordate Posterior Hox sequences may remain incompletely resolved even with further basal deuterostome Hox clusters.
6. Vertebrates: multiple clusters with gene loss, and the potential to find the ancestral Colinearity mechanism(s)
During the early stages of vertebrate evolution large-scale, probably whole-genome, duplications occurred. Further whole-genome duplications have occurred in certain lineages, such as the teleost fish and then the salmonid fish [58]. This has resulted in vertebrates possessing multiple Hox gene clusters, such as the four of mammals and the seven of zebrafish (caused by duplications to eight clusters, followed by loss of one entire cluster), and possibly fourteen in Trout [59]. During the course of the diversification of the vertebrates, numerous Hox genes have been lost on the different lineages [60]. The result is that there is not a single vertebrate Hox gene cluster that possesses a member of every paralogy group; every vertebrate Hox cluster is incomplete. This is presumably made possible due to the extensive redundancy and cross-regulation between the genes of different clusters [61, 62], as well as the swapping of functions between paralogues of different vertebrate lineages [63].
There are several forms of Colinearity in the vertebrate Hox genes, and the different forms, as well as separate aspects of each form, are distinguishable as separate regulatory regions in and around the Hox cluster [35, 64]. Even one particular form of Colinearity, such as Temporal Colinearity, seems to be operating in several contexts (e.g. axial mesoderm, nervous system, and early and late limb bud development), with potentially separate regulatory regions directing the process in each different context [9, 65]. Can we possibly navigate through such complexity to discover an ancestral mechanism, which is homologous to systems used in amphioxus (and possibly other deuterostomes – although the various derived deuterostome clusters may have lost such an ancestral mechanism), and even across the Bilateria (see figure 1) Although the Hox genes of the ancestral bilaterian were clearly clustered, were they expressed in a Colinear fashion, and if so was there a general, pan-cluster mechanism operating to produce this Colinearity which has since been conserved in at least some animal lineages (and awaits our discovery)?
Temporal Colinearity seems the most likely form of Colinearity to be acting in a global fashion over Hox clusters, and hence constraining them in well-ordered clusters [8, 14, 15, 35]. The mechanism(s) by which Temporal Colinearity is brought about is still a mystery, even in such an intensively investigated system as the mouse. Regulatory regions involved in at least some aspects of Temporal Colinearity are however known [35]. Whether these regions are responsible for producing Temporal Colinearity in aspects of embryology that are basal for the chordates, such as body axis development, rather than solely involved in vertebrate innovations such as the limb buds, remains to be resolved (Deschamps and van Nes [64] suggest the ELCR limb control region could also be functioning in early embryogenesis (Figure No 8 in reference [64]), whilst Spitz et al [65] suggest the body axis Colinearity mechanisms are driven from within the cluster). This endeavour will require not only the clarification of how the vertebrate Hox genes are regulated and operating in the various aspects of vertebrate body-axis development (e.g. paraxial and lateral mesoderm, as well as neuroectoderm), but also clarification of the precise degree of homology between the axial development of vertebrates and invertebrate chordates such as amphioxus, which may use mechanisms since lost by the vertebrates (e.g. engrailed stripes [66]) in addition to conserved, homologous processes (e.g. RA regulation of the Hox genes, [67, 68]).
Chromatin modulation has long been hypothesised to be involved in Hox regulation, and has recently been shown to be intimately linked to the Temporal Colinearity of vertebrate Hox activation [6]. Whether this chromatin modulation is a cause or effect of Hox regulation needs to be resolved [64], but it is another good candidate for hunting down an ancestral mechanism that may have been present not only in the deuterostome ancestor, but also in the bilaterian ancestor as well – since the chromatin modulating complexes of the Polycomb group (PcG) and Trithorax group (TrxG) proteins are involved in Hox control in vertebrates and insects [69]. The form of this PcG/TrxG control is different between vertebrates and insects, and may be involved at different points in Hox control (both initiation and memory [69]), but at least we know of some homologous regulatory proteins controlling homologous target genes. The Cdx genes are another strong candidate [64, 70], as also are the Wnt genes [71, 72, 73]. The mechanistic link between these proteins and the Hox genes of prototypical, non-derived taxa will be of great interest, and may be one of the best routes to understanding how the bilaterian ancestor controlled its Hox genes, and whether Hox Colinearity was produced by a pan-cluster process that has since been conserved across some bilaterian lineages, and elaborated upon during the divergence and differentiation of others.
7. Conclusion
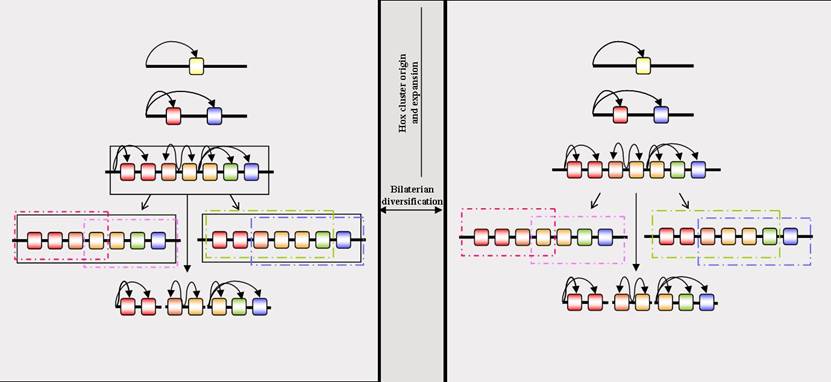
Temporal Colinearity is potentially the most important form of Colinearity with regards to constraining the Hox cluster as an intact, ordered array (with no taxon contradicting this view any more since the resolution of the sea urchin Hox cluster organisation). The comparative approach, encompassing both prototypical and derived taxa, is essential, both in coming to this conclusion and in understanding how to proceed further. A mechanistic understanding of Temporal Colinearity is required, not only in a model system such as the mouse, but also in other taxa such as amphioxus, as well as protostomes that are less derived than the current models of Drosophila and nematodes. This should then reveal whether the Hox cluster arose and then was constrained by a pan-cluster mechanism before the divergence of the bilaterian lineages, or alternatively that the time-span between the origin of the Hox cluster and the divergence of the bilaterian lineages was so brief that there was simply not enough time for the cluster to break-up. In this later scenario, since the divergence of the bilaterian lineages, some bilaterian Hox clusters have finally accumulated viable rearrangements that break them up, whilst others have since become constrained by the evolution of lineage-specific pan-cluster regulatory mechanisms (see Figure 2). Such a deep mechanistic understanding is essential to discern whether an apparently similar phenomenon, such as Temporal Colinearity, that can be observed in different lineages really is homologous, rather than resulting from the independent evolution of separate mechanisms that result in an apparently similar effect; is Temporal Colinearity homologous across the chordates, deuterostomes and Bilateria?
Is there a homologous global Colinearity mechanism across the bilaterians? The left-hand panel represents the evolution of a global Colinearity mechanism prior to the divergence of the bilaterian lineages, and its subsequent conservation in some, but not all, lineages. The right-hand panel represents the lack of a global Colinearity mechanism prior to the divergence of the bilaterians, but the retention of a Hox cluster up to this point due to the origin of the genes by tandem duplication leading to extensive enhancer-sharing amongst the genes, and a consequent reduced opportunity for viable breaks in the cluster. Rectangles represent long-range Colinearity mechanisms, of which there can be several different, lineage-specific types. The black rectangle in the left-hand panel represents a global Colinearity mechanism potentially present in the bilaterian ancestor and conserved in some lineages. Where this mechanism is lost (the central lineage of the left-hand panel) the Hox cluster can disintegrate (broken horizontal line). Curved arrows denote enhancer/gene interactions. Multiple arrows originating from the same point represent enhancers that are shared by multiple Hox genes. Such enhancer-sharing will restrict the number of locations within a Hox cluster at which viable breaks are possible, and slow the rate at which Hox clusters can disintegrate. If the time between the origin of the Hox cluster and the divergence of the bilaterian lineages is relatively short, such a restriction by enhancer-sharing may be sufficient to account for the retention of a Hox cluster even in the absence of a global regulatory mechanism (right-hand panel). Subsequently evolving Colinearity mechanisms in disparate bilaterian lineages will then not be homologous.
Acknowledgements
The authors would like to thank Renata Freitas and the Oxford Evo-Devo group for helpful discussions, and Joanne Ferrier for proof-reading. Work in the authors' laboratory is funded by the BBSRC.
Conflict of interest
The authors have declared that no conflict of interest exist
References
1. Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978 ;276(5688):565-70
2. McGinnis W, Garber R.L, Wirz J, Kuroiwa A, Gehring W. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984 ;37:403-408
3. Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989 ;8(5):1497-505
4. Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989 ;57:367-378
5. Dolle P, Izpisua-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989 ;342(6251):767-72
6. Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004 ;18(10):1119-30
7. Duboule D. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev Suppl. 1994 :135-42
8. Seo HC, Edvardsen RB, Maeland AD. et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004 ;431(7004):67-71
9. Tarchini B, Duboule D. Control of Hoxd genes' collinearity during early limb development. Dev Cell. 2006 ;10:93-103
10. Kmita M, Fraudeau N, Herault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002 ;420(6912):145-50
11. Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003 ;113(3):405-17
12. Ferrier DEK, Holland PWH. Ancient origins of the Hox gene cluster. Nature Reviews Genetics. 2001 ;2:33-38
13. Ferrier DEK, Holland PWH. Ciona intestinalis ParaHox genes: evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Molecular Phylogenetics and Evolution. 2002 ;24:412-417
14. Ferrier DE, Minguillon C. Evolution of the Hox/ParaHox gene clusters. Int J Dev Biol. 2003 ;47(7-8):605-11
15. Patel NH. Evolutionary biology: time, space and genomes. Nature. 2004 ;431(7004):28-9
16. Halanych KM. The phylogenetic position of the pterobranch hemichordates based on 18S rDNA sequence data. Mol Phylogenet Evol. 1995 ;4(1):72-6
17. Bromham LD, Degnan BM. Hemichordates and deuterostome evolution: robust molecular phylogenetic support for a hemichordate + echinoderm clade. Evol Dev. 1999 ;1(3):166-71
18. Cameron CB, Garey JR, Swalla BJ. Evolution of the chordate body plan: new insights from phylogenetic analyses of deuterostome phyla. Proc Natl Acad Sci U S A. 2000 ;97(9):4469-74
19. Furlong RF, Holland PW. Bayesian phylogenetic analysis supports monophyly of ambulacraria and of cyclostomes. Zoolog Sci. 2002 ;19(5):593-9
20. Dolecki GJ, Wannakrairoj S, Lum R. et al. Stage-specific expression of a homeo box-containing gene in the non-segmented sea urchin embryo. EMBO J. 1986 ;5(5):925-30
21. Di Bernardo M, Russo R, Oliveri P, Melfi R, Spinelli G. Expression of homeobox-containing genes in the sea urchin (Parancentrotus lividus) embryo. Genetica. 1994 ;94(2-3):141-50
22. Ruddle FH, Bentley KL, Murtha MT, Risch N. Gene loss and gain in the evolution of the vertebrates. Dev Suppl. 1994 :155-61
23. Popodi E, Kissinger JC, Andrews ME, Raff RA. Sea urchin Hox genes: insights into the ancestral Hox cluster. Mol Biol Evol. 1996 ;13(8):1078-86
24. Mito T, Endo K. A PCR survey of hox genes in the sea star, Asterina minor. Mol Phylogenet Evol. 1997 ;8(2):218-24
25. Morris VB, Brammall J, Byrne M, Frommer M. Hox-type and non-Hox homeobox gene sequences in genomic DNA of the sea urchin Holopneustes purpurescens. Gene. 1997 ;201(1-2):107-10
26. Morris VB, Brammall J, Byrne M, Frommer M. cDNA hox sequences 3' of the homeobox isolated from the sea urchin Holopneustes purpurescens are definitive for sea urchin Hox orthologues. DNA Seq. 2002 ;13(4):185-93
27. Long S, Martinez P, Chen WC, Thorndyke M, Byrne M. Evolution of echinoderms may not have required modification of the ancestral deuterostome HOX gene cluster: first report of PG4 and PG5 Hox orthologues in echinoderms. Dev Genes Evol. 2003 ;213(11):573-6
28. Angerer LM, Dolecki GJ, Gagnon ML, Lum R, Wang G, Yang Q, Humphreys T, Angerer RC. Progressively restricted expression of a homeo box gene within the aboral ectoderm of developing sea urchin embryos. Genes Dev. 1989 ;3(3):370-83
29. Dobias SL, Zhao AZ, Tan H, Bell JR, Maxson R. SpHbox7, a new Abd-B class homeobox gene from the sea urchin Strongylocentrotus purpuratus: insights into the evolution of hox gene expression and function. Dev Dyn. 1996 ;207(4):450-60
30. Arenas-Mena C, Martinez P, Cameron RA, Davidson EH. Expression of the Hox gene complex in the indirect development of a sea urchin. Proc Natl Acad Sci U S A. 1998 ;95(22):13062-7
31. Arenas-Mena C, Cameron AR, Davidson EH. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development. 2000 ;127(21):4631-43
32. Martinez P, Rast JP, Arenas-Mena C, Davidson EH. Organization of an echinoderm Hox gene cluster. Proc Natl Acad Sci USA. 1999 ;96(4):1469-74
33. Cameron RA, Rowen L, Nesbitt R, Bloom S, Rast JP, Berney K, Arenas-Mena C, Martinez P, Lucas S, Richardson PM, Davidson EH, Peterson KJ, Hood L. Unusual gene order and organization of the sea urchin hox cluster. J Exp Zoolog B Mol Dev Evol. 2006 ;306(1):45-58
34. Pierce RJ, Wu W, Hirai H, Ivens A, Murphy LD, Noel C, Johnston DA, Artiguenave F, Adams M, Cornette J, Viscogliosi E, Capron M, Balavoine G. Evidence for a dispersed Hox gene cluster in the platyhelminth parasite Schistosoma mansoni. Mol Biol Evol. 2005 ;22(12):2491-503
35. Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003 ;301(5631):331-3
36. Hyman LH. The invertebrates: smaller coelomate groups. New York: McGraw-Hill. 1959
37. Brusca RC, Brusca GJ. Invertebrates. Sunderland, MA: Sinauer. 1990
38. Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003 ;113(7):853-65
39. Rychel AL, Smith SE, Shimamoto HT, Swalla BJ. Evolution and Development of the Chordates: Collagen and Pharyngeal Cartilage. Mol Biol Evol. 2006 ;23(3):541-549
40. Peterson KJ. Isolation of Hox and Parahox genes in the hemichordate Ptychodera flava and the evolution of deuterostome Hox genes. Mol Phylogenet Evol. 2004 ;31(3):1208-15
41. Pendleton JW, Nagai BK, Murtha MT, Ruddle FH. Expansion of the Hox gene family and the evolution of chordates. Proc Natl Acad Sci U S A. 1993 ;90(13):6300
42. Ferrier DEK, Minguillón C, Holland PWH, Garcia-Fernàndez J. The amphioxus Hox cluster: deuterostome posterior flexibility and Hox14. Evol Devel. 2000 ;2:284-93
43. Wada H, Satoh N. Details of the evolutionary history from invertebrates to vertebrates, as deduced from the sequences of 18S rDNA. Proc Natl Acad Sci U S A. 1994 ;91(5):1801-4
44. Winchell CJ, Sullivan J, Cameron CB, Swalla BJ, Mallatt J. Evaluating hypotheses of deuterostome phylogeny and chordate evolution with newLSU and SSU ribosomal DNA data. Mol Biol Evol. 2002 ;19(5):762-76
45. Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol Biol Evol. 2005 ;22(5):1246-53
46. Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005 ;22(11):2275-84
47. Satoh N, Jeffery WR. Chasing tails in ascidians: developmental insights into the origin and evolution of chordates. Trends Genet. 1995 ;11(9):354-9
48. Dehal P, Satou Y, Campbell RK. et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002 ;298:2157-67
49. Spagnuolo A, Ristoratore F, Di Gregorio A, Aniello F, Branno M, Di Lauro R. Unusual number and genomic organization of Hox genes in the tunicate Ciona intestinalis. Gene. 2003 ;309(2):71-9
50. Ikuta T, Saiga H. Organization of Hox genes in ascidians: present, past, and future. Dev Dyn. 2005 ;233(2):382-9
51. Ikuta T, Yoshida N, Satoh N, Saiga H. Ciona intestinalis Hox gene cluster: Its dispersed structure and residual colinear expression in development. Proc Natl Acad Sci U S A. 2004 ;101(42):15118-23
52. Garcia-Fernàndez J, Holland PWH. Archetypal organization of the amphioxus Hox gene cluster. Nature. 1994 ;370:563-566
53. Mito T, Endo K. PCR survey of Hox genes in the crinoid and ophiuroid: evidence for anteriorconservation and posterior expansion in the echinoderm Hox gene cluster. Mol Phylogenet Evol. 2000 ;14(3):375-88
54. Wada H, Garcia-Fernandez J, Holland PW. Colinear and segmental expression of amphioxus Hox genes. Dev Biol. 1999 ;213(1):131-41
55. Powers TP, Amemiya CT. Evidence for a Hox14 paralog group in vertebrates. Curr Biol. 2004 ;14(5):R183-4
56. Ferrier DE. Hox genes: Did the vertebrate ancestor have a Hox14? Curr Biol. 2004 ;14(5):R210-1
57. Long S, Byrne M. Evolution of the echinoderm Hox gene cluster. Evol Dev. 2001 ;3(5):302-11
58. Jaillon O, Aury JM, Brunet F. et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the earlyvertebrate proto-karyotype. Nature. 2004 ;431(7011):946-57
59. Moghadam HK, Ferguson MM, Danzmann RG. Evidence for Hox gene duplication in rainbow trout (Oncorhynchus mykiss): a tetraploid model species. J Mol Evol. 2005 ;61(6):804-18
60. Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends Genet. 2005 ;21(8):421-4
61. Condie BG, Capecchi MR. Mice with targeted disruptions in the paralogous genes hoxa-3 and hoxd-3 reveal synergistic interactions. Nature. 1994 ;370(6487):304-7
62. Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003 ;301(5631):363-7
63. McClintock JM, Carlson R, Mann DM, Prince VE. Consequences of Hox gene duplication in the vertebrates: an investigation of the zebrafish Hox paralogue group 1 genes. Development. 2001 ;128(13):2471-84
64. Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005 ;132(13):2931-42
65. Spitz F, Gonzalez F, Peichel C, Vogt TF, Duboule D, Zakany J. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 2001 ;15(17):2209-14
66. Holland LZ, Kene M, Williams NA, Holland ND. Sequence and embryonic expression of the amphioxus engrailed gene (AmphiEn): the metameric pattern of transcription resembles that of its segment-polarity homolog in Drosophila. Development. 1997 ;124(9):1723-32
67. Manzanares M, Wada H, Itasaki N, Trainor PA, Krumlauf R, Holland PW. Conservation and elaboration of Hox gene regulation during evolution of the vertebrate head. Nature. 2000 ;408(6814):854-7
68. Schubert M, Yu JK, Holland ND, Escriva H, Laudet V, Holland LZ. Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development. 2005 ;132(1):61-73
69. Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004 ;38:413-43
70. Copf T, Schroder R, Averof M. Ancestral role of caudal genes in axis elongation and segmentation. Proc Natl Acad Sci U S A. 2004 ;101(51):17711-5
71. Hunter CP, Harris JM, Maloof JN, Kenyon C. Hox gene expression in a single Caenorhabditis elegans cell is regulated by a caudal homolog and intercellular signals that inhibit wnt signalling. Development. 1999 ;126(4):805-14
72. Holland LZ. Heads or tails? Amphioxus and the evolution of anterior-posterior patterning in deuterostomes. Dev Biol. 2002 ;241(2):209-28
73. Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005 ;433(7022):156-60
Author contact
![]() Corresponding address:
Corresponding address:

 Global reach, higher impact
Global reach, higher impact