10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2006; 2(3):125-132. doi:10.7150/ijbs.2.125 This issue Cite
Research Paper
Phylogenetic analysis of Amphioxus genes of the proprotein convertase family, including aPC6C, a marker of epithelial fusions during embryology
1. Laboratoire de Biologie Moléculaire de la Cellule, UMR 5161 CNRS/ENS de Lyon, Ecole Normale Supérieure de Lyon, IFR128 BioSciences Lyon-Gerland, 46, Allée d'Italie, 69364 Lyon Cedex 07, France.
2. Laboratoire Arago, UMR 7628, CNRS and University Pierre and Marie Curie, BP 44, F-66651 Banyuls sur Mer, France.
3. Marine Biology Research Division, Scripps Institution of Oceanography, University of California San Diego, La Jolla, CA 92093, USA
Received 2006-1-27; Accepted 2006-4-22;
Abstract
The proprotein convertases (PCs) comprise a family of subtilisin-like endoproteases that activate precursor proteins (including, prohormones, growth factors, and adhesion molecules) during their transit through secretory pathways or at the cell surface. To explore the evolution of the PC gene family in chordates, we made a phylogenetic analysis of PC genes found in databases, with special attention to three PC genes of the cephalochordate amphioxus, the closest living invertebrate relative to the vertebrates. Since some vertebrate PC genes are essential for early development, we investigated the expression pattern of the C isoform of the amphioxus PC6 gene (aPC6C). In amphioxus embryos and larvae, aPC6C is expressed at places where epithelia fuse. Several kinds of fusions occur: ectoderm-to-ectoderm during neurulation; mesoderm-to-ectoderm during formation of the preoral ciliated pit; and endoderm-to-ectoderm during formation of the mouth, pharyngeal slits, anus, and external opening of the club-shaped gland. Presumably, at all these sites, aPC6C is activating proteins favoring association between previously disjunct cell populations.
Keywords: amphioxus, gene expression pattern, prohormone convertases, PC6
1. Introduction
The function of proteins destined for export or for the plasma membrane surface in eukaryotic cells is usually regulated by a specific proteolysis mechanism of inactive precursors which generates biologically active molecules. This process of activation is usually performed by a family of calcium-dependent endoproteases related to the prokaryotic subtilisin and the yeast kexin enzymes that are called proprotein convertases (PCs) [1, 2] . PCs (also called kexins) are members of the subtilisin-like proteases superfamily, together with five other protein families (i.e. subtilisins, thermitases, proteinase K, lantibiotic peptidases and pyrolysins) [3] . Seven PC subfamilies have been identified, including furin/PACE [4] , PC1/PC3 [5] , PC2 [6, 7] , PC4 [8] , PACE4 [9] , PC5/PC6 [10] , and PC7/LPC/PC8 [11] . PCs have been found not only in vertebrates but also in yeast [12-14] , Hydra [15] , protostomes [16-19] , and in the invertebrate chordate amphioxus [20, 21] , which places the origin of the family very early during eukaryotic evolution. PCs share a common structure of five domains including an N-terminal signal peptide followed by a propeptide of 80-90 residues terminating with the activation canonical cleavage site R-X-R/R-R, a catalytic domain of approximately 240 residues related to subtilisin, a conserved P or Homo B domain of approximately 150 residues which function is still unknown, and a variable C-terminal domain [1, 2] , (Fig. 1). The catalytic and P-domains are highly conserved, and the C-terminal domain is variable, both in length and sequence, but contains a conserved Cys rich region and a transmembrane domain in some members of the PC family. Depending on the presence or absence of this transmembrane domain, PCs function in different subcellular compartments. The members with transmembrane domains (e.g. furin, PC6B, or PC7) appear to function primarily in the trans-Golgi network (TGN) and/or in constitutive vesicles derived from it. The other members (e.g. PC2, PC1/PC3, PACE4, PC4, PC6A) are sorted into dense-core vesicles in the regulated secretory pathway where they process a variety of prohormones or proneuropeptides [1, 2] . Two members of the PC family are expressed exclusively in neuroendocrine tissues (PC1 and PC2) and one is restricted to reproductive organs (PC4). The remaining PCs are expressed in many tissues of mammalian embryos and adults, including neuroendocrine system, liver, gut, and brain [22] . The colocalization of a PC and its substrate both at the tissue and subcellular level likely contributes to substrate selectivity. Some PCs, like PC6, undergo tissue-specific alternative splicing that in the case of PC6 generates soluble and membrane bound isoforms which are involved in the regulated secretory pathway or in the TGN compartment respectively. This indicates that alternative splicing plays a very important role in the control of the PC6 gene function.
Schematic representation of the three different splicing variants described for the amphioxus PC6 gene, aPC6A, aPC6B and aPC6C [20] . Boxes represent the coding sequence including propeptide (Pro), catalytic domain (SCD), P domain (PD), the 3' Cys rich domain (CRR) and the transmembrane domain of the B isoform (TM). The three isoforms differ in the 3' part of the CRR domain and are indicated by a grey dashed line for the A isoform or a grey line for the C isoform. The position of the 331 bp sequence of aPC6C used as a probe for the in toto in situ hybridizations is indicated with a black box.

In chordates, including the cephalochordate amphioxus, the physiological roles of PACE4 and PC6 genes during development are poorly understood, even if they are critical for a correct developmental process [23] . Amphioxus is the best available stand-in for the extinct proximate invertebrate ancestor of the vertebrates. At the anatomical level, amphioxus is vertebrate-like but simpler. In many respects amphioxus is like a stripped-down, generalized vertebrate. Indeed, it has pharyngeal gill slits, a dorsal hollow nerve cord and notochord, but lacks paired eyes, ears, limbs and neural crest. Moreover, at the genetic level, amphioxus genome also shares many characteristics with vertebrate genomes, but is less complex. The amphioxus genome has not undergone the two waves of gene duplications, that took place during vertebrate evolution and which are responsible for the presence of several duplicated vertebrate genes whereas only one pro-orthologue is present in amphioxus [24] .
To gain insight into the evolution of the PC gene family, we have studied: (i) the phylogenetic relationships of known PC genes in eukaryotes, and (ii) we have characterized the expression pattern of the isoform C of the invertebrate chordate amphioxus (Branchiostoma lanceolatum) PC6 gene (i.e. the pro-ortholoue of vertebrate PC6 and PACE4 genes). We show first, that the PC genes family is not divided into seven subfamilies as previously published [3, 20] , but only into five or six groups, and second, by using in toto in situ hybridization, we show that aPC6C is expressed in regions where there is a contact between different embryonic layers like in mouth, exterior opening of the club-shaped gland, and in anus (i. e. endoderm-ectoderm associations), in ciliated pit (ectoderm-mesoderm), or in the sealing of the ectoderm mid-dorsally after neurulation (i.e. ectoderm-ectoderm associations). These results suggest that aPC6C could be involved in epithelial fusions during embryology.
2. Materials and methods
Phylogenetic analysis
Amino acid sequences were aligned using the CLUSTAL W program [25] and manually corrected with SEAVIEW [26] . Phylogenetic trees were inferred by (1) the Neighbor-Joining method [27] with Poisson-corrected distances on amino acids, implemented in PHYLO_WIN [26] ; and (2) with PHYML [28] , a fast and accurate maximum likelihood heuristic, under the JTT substitution model [29] , with a gamma distribution of rates between sites (six categories, parameter alpha estimated by PHYML). Amino acid sites with gaps in any sequence were excluded from the calculations. The bootstrap analysis (1000 repetitions), was carried out by the method of Felsenstein [30] . Divergent sequences for which the alignment was uncertain were excluded.
Embryo collection, in situ hybridization, and histology
Ripe animals of the Mediterranean amphioxus (Branchiostoma lanceolatum) were collected in Argelès-sur-Mer (France), and gametes were obtained by heat stimulation [31] . A 331 bp fragment of B. lanceolatum aPC6 cDNA was used for the synthesis of antisense riboprobes. This fragment includes the coding sequence of the last 64 aminoacids and extends from nucleotide position 4058 to position 4389 of the previously published sequence for aPC6c [20] , and is specific to the c isoform of aPC6. Fixation, whole-mount in situ hybridization and histological sections were performed as described by Holland et al (1996) [24] .
3. Results and discussion
Phylogenetic analysis of the PC gene family
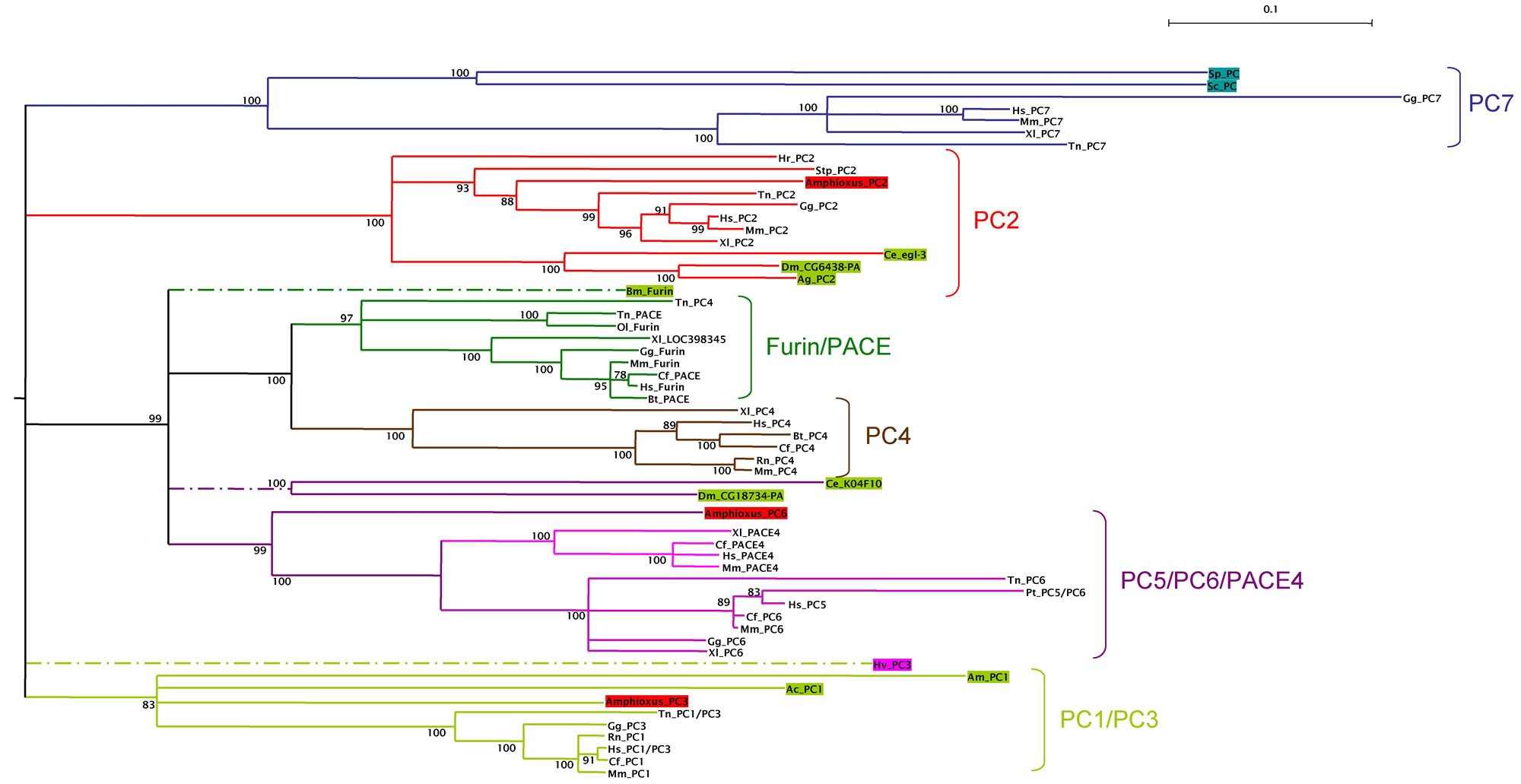
To examine the relationships between invertebrate PC and vertebrate PC genes, we constructed a phylogenetic tree with PC amino acid sequences obtained from GenBank, including all the invertebrate sequences that we found for each PC subfamily, the yeast kexins and selected vertebrate sequences from at least one mammalian, one amphibian and one fish representative. Phylogenetic trees were constructed with two different methods (ML and NJ), and were rooted with a group of invertebrate and vertebrate sequences of the the subtilisin related protein Site-1 (membrane-bound transcription factor site-1) [32] . The results obtained with both methods were similar and those obtained with the NJ method are shown in Fig. 2. From these analyses we define six orthologous PC subfamilies supported by high bootstrap values: (i) a subfamily containing vertebrate PC7 and yeast kexins; (ii) the PC2 subfamily; (iii) the furins and PACE sequences; (iv) the PC4 subfamily; (v) a family containing PC5/PC6 and PACE4 sequences and (vi) the PC1/PC3 subfamily. Different authors have divided the PC family into seven groups, since the PC5/PC6/PACE4 subfamily was previously described as two different groups. However, the use of invertebrate sequences (and particularly the invertebrate chordate amphioxus sequence of PC6), clearly shows that PACE4 and PC5/PC6 groups appeared specifically in vertebrates. These paralogous genes should have arisen from the genome duplications that occurred between the chordata and the vertebrata radiations [33] .
Developmental expression of aPC6C
Three members of the PC family are known in the cephalochordate amphioxus (B. californiense), PC2, PC3 and PC6 [20, 21] . Moreover, three different splicing variants have been described for the amphioxus PC6 gene, named aPC6A, aPC6B and aPC6C (Fig 1) [20] . However, the function of the amphioxus PCs is not yet known. As a first step in the study of the function of amphioxus PCs, we have determined the pattern of expression of the aPC6C isoform during the embryonic development of Branchiostoma lanceolatum by in situ hybridization of whole mount animals (Fig. 3). We first isolated a 331 bp cDNA fragment specific for the C isoform that includes the coding sequence for the last 64 amino acids and 192 nucleotides of the 3' non-coding region (Fig. 1). The nucleotide sequence identity between the published B. californiense sequence (accession number AAF26302) and the B. lanceolatum one is 100% (data not shown), including the 3' non-coding part. We will refer to this gene as aPC6C instead of AmphiPC6C (the customary way of referring to genes from B. floridae) because Oliva et al (2000) [20] originally used this abbreviation, even if they probably worked on B. floridae due to some confusion about the species by their biological supply company.
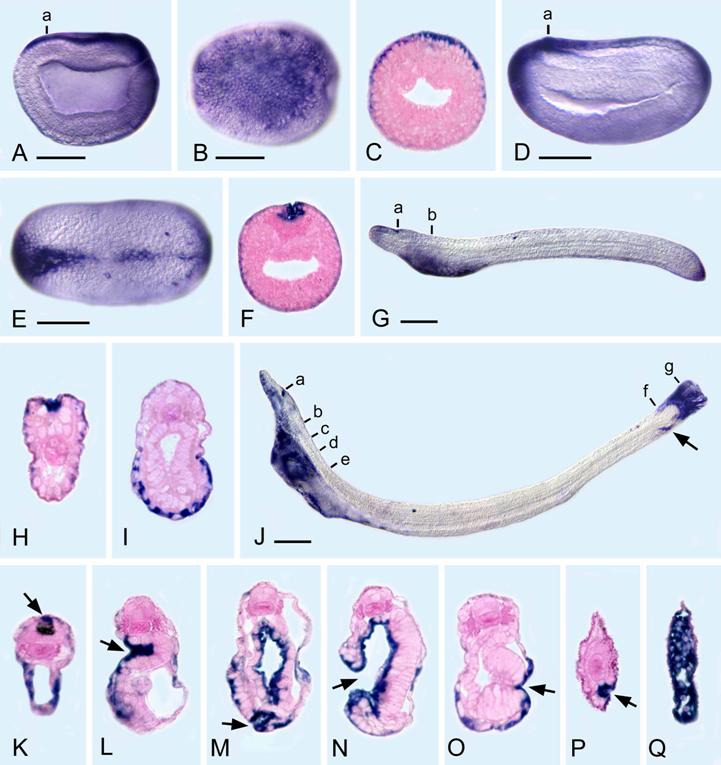
The gene expression pattern of the aPC6C isoform was examined from mid-blastula through the early larval stage. aPC6C transcripts are ubiquitously distributed up until late gastrula, but become spatially restricted thereafter. In late gastrula, aPC6C transcripts are found dorsally in the ectoderm and posteriorly around the blastopore. In mid-neurula, aPC6C is expressed in the ectoderm in the most anterior and posterior parts of the embryo as well as in ectodermal cells fusing in the dorsal midline. In the early larvae (before the mouth opens), aPC6C is still expressed in ectodermic cells posteriorly, in cells bordering the neuropore, and the ventral ectoderm of the anterior part of the body. In later larvae (with the open mouth and first gill slit), in the anterior part of the body aPC6C expression is detected in cells bordering the neuropore, in ventral ectoderm, in the cells of the ciliated pit, in the pharyngeal endoderm, in some ectodermal cells around the mouth, in cells of the club-shaped gland near its external opening and in ectodermal cells just outside the opening of the first gill slit. In the posterior part of the body aPC6C transcripts can be detected in the cells just within the anus as well as in elongated ectoderm cells comprising the tail. This expression pattern suggests that the isoform C of the amphioxus PC gene may play an important role in epithelial fusions during development. Thus, in all the body openings where ectoderm-endoderm contacts are very important (e.g. mouth, exterior opening of club-shaped gland and anus), as well as in regions where there are ectoderm-ectoderm and ectoderm-mesoderm associations (e.g. the sealing of the ectoderm mid-dorsally after neurulation and the ciliated pit respectively), aPC6C is expressed.
In vertebrates the two orthologs of the amphioxus PC6 gene (PC6 and PACE4, see fig 2) are expressed in several tissues or organs homologs to the tissues where the amphioxus PC6C gene is expressed. However, many other expression domains have also been described. Thus, in Xenopus, xPACE4 is expressed in a completely different way than the amphioxus PC6C isoform. xPACE4 is a maternal RNA unequally distributed in the oocyte. Later on, a localized expression is detected in the notochord, the brain and a subset of endodermal precursors [34] . For the second paralogue of aPC6 in Xenopus, xPC6, gene expression was studied by using a probe that recognizes all the different isoforms of the gene. Like in amphioxus, xPC6 transcripts are ubiquitously distributed until the end of gastrulation but they exhibit a dynamic expression pattern shortly thereafter. Indeed, xPC6 is mainly expressed in ectoderm-derived tissues (e.g. neural folds, neural crest, eyes, nasal placode, lateral line, otic vesicle and brain), but also in mesoderm (e.g. pronephric duct, notochord) and other mixed structures like pharyngeal arches [35] . In mouse, PC6 is first expressed in extraembryonic tissues and in the distal region of the primitive streak, a homolog of the amphioxus blastopore. Later on, expression is observed in the somites, the dorsal surface ectoderm, the vertebral cartilage primordia and in the apical ectodermal ridge of limb buds [36] .
The differences in the expression pattern between aPC6C and the vertebrate PACE4 and PC6 genes may either represent a secondary derivation in amphioxus or a function that has been coopted in vertebrates after the vertebrate-specific genes duplications. However, it is not excluded that other splicing variants, not yet characterized either in vertebrates or in amphioxus, can show closely related expression patterns different from the ones already described. A very important step for the comprehension of PC genes function during chordate development in the future will be, first, the complete characterization of the expression pattern of each splicing variant both in vertebrates as well as in amphioxus.
As we have proposed above, aPC6C could be involved in epithelial fusions during embryology. In the same way, the mouse PC6 gene shows a prominent expression in the site of fusion of the two decidual lobes, a extraembryonic structure, and in the distal region of the primitive streak [36] . Moreover, another closely related proprotein convertase in vertebrates, the furin (see fig. 2), plays a role in the ventral closure of the cardiogenic mesoderm [37] , indicating that the function of PCs in the association of different tissues may not be entirely restricted to the PC6 paralogue. Since PCs are proteases that activate many key regulatory molecules, our results suggest that similar regulators implicated in the association between different tissues can be controlled through proteolytic cleavage by using different members of the PC gene family. In vertebrates, the natural substrates of furin, PACE4 and PC6 remain poorly determined, as well as the extent to which their substrate specificities overlap under physiological conditions. However, in vitro experiments have shown that furin could be implicated in remodeling of the extracellular matrix by processing metalloproteinases [38, 39] and furin, as well as PC5/6, in the regulation of cell adhesion by processing integrins [40, 41] . The study, both in vitro and in vivo, of the substrates for each PC (including their splicing variants) under physiological conditions, as well as the degree in which different PC genes can process similar substrates in order to control perfectly the local concentration of active molecules at the place where they are required will be extremely important in the future for the comprehension of the role of different PC genes during chordate development.
Phylogeny of the PC gene family. Tree was constructed by the distance neighbor-joining method with 1000 bootstrap replicates in order to test the robustness of the branches. Bootstrap values (in %) are indicated on each branch of the tree. Root was placed using an outgroup of several subtilisin related proteins Site-1, including sequences from Arabidopsis thaliana (NP_197467), Drosophila melanogaster (NP_649337), Anopheles gambiae (XP_320328), chicken (XP_414071), human (NP_003782), and mouse (NP_062683). Accession numbers of the sequences used are: P29146, XP_541276, NP_006191, XP_520079, Q04592, XP_424841, XP_355911, NP_002561, Q9NJ15, XP_542201, NP_032819, NP_001021543, NP_727963, AAA87006, XP_424712, NP_058787, NP_038656, CAA46031, AAA87005, XP_419332, NP_002585, NP_032818, NP_001023732, NP_477318, XP_308012, NP_594835, NP_014161, NP_001025528, NP_004707, NP_032820, AAW83023, AAH84090, AAH94153, CAA47118, AAT99304, CAF99544, CAG03450, CAG06456, CAF98620, CAG01397, CAG08431, BAD11989, XP_850069, BAC97793, XP_784245, BAC05491, XP_393918, AAA27768, XP_545820, CAA92109, NP_002560, AAA37643, XP_545820, XP_585571, AAW83025, NP_060043, AAA41814, Q28193, BAA00877 and XP_542201. Invertebrate sequences are in colored boxes, yeast in blue, diploblasts in magenta, protostomes in green and deuterostomes (including amphioxus) in red. The different subfamilies are clustered with the same colored branches. Abreviations are: Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Hv, Hydra vulgaris; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Ag, Anopheles gambiae; Bm, Bombix mori; Am, Apis melifera; Ac, Aplysia californica; Hr, Halocynthia roretzi; Sp, Strongylocentrotus purpuratus; Amphioxus, Branchiostoma californiense; Gg, Gallus gallus; Xl, Xenopus laevis; Tn, Tetraodon nigroviridis; Ol, Oryzias latipes; Hs, Homo sapiens; Mm, Mus musculus; Cf, Canis familiaris; Bt, Bos taurus; Rn, Rattus norvegicus.
AmphiPC6 expression in embryos and larvae of amphioxus. All whole mounts with anterior toward left and 50 μm scale lines. Sections, counterstained pink, as seen from posterior end of body. A) Side view of late gastrula with expression in ectoderm dorsally and posteriorly. B) Dorsal view of A. C) Section through level a in A. D) Side view of early neurula with expression in ectoderm dorsally, anteriorly, and posteriorly. E) Dorsal view of D. F) Section through level a in D, showing expression in ectoderm cells fusing in the dorsal midline. G) Side view of larva shortly before the mouth opens, showing ectodermal expression in the neuropore, ventroanteriorly, and posteriorly. H) Section through level a in G showing expression in the ectoderm cells bordering the neuropore. I) Section through level b in G, showing ectodermal expression on the ventral side of the body. J) Side view of larva with an open mouth and first gill slit; the arrow indicates the position of the anus. K) Section through level a in J, showing expression in ectoderm cells ventrally and also dorsally (arrow) near the neuropore. L) Section through level b in J, showing strong expression in the cells of the ciliated pit (arrow). M) Section through c in J, showing expression in the pharyngeal endoderm, in some ectoderm cells and in cells of the club-shaped gland near its external opening (arrow). N) Section through level d in J, showing the open mouth (arrow) and expression in the pharyngeal endoderm. O) Section through level e in J, showing ectodermal expression just outside the opening of the first gill slit (arrow). P) Section through level f in J, showing expression in the cells just within the anus (arrow). Q) Section through g in J, showing expression in elongated ectoderm cells comprising the tail.
Acknowledgements
S.B. is supported by HRA Pharma, Fondation Mérieux, Fondation par la recherche Médicale and Association de Recherche contre le Cancer.
Conflict of interest
The authors have declared that no conflict of interest exists.
References
1. Rouille Y. et al. Proteolytic processing mechanisms in the biosynthesis of neuroendocrine peptides: the subtilisin-like proprotein convertases. Front Neuroendocrinol. 1995 ;16(4):322-61
2. Zhou A. et al. Proteolytic processing in the secretory pathway. J Biol Chem. 1999 ;274(30):20745-8
3. Siezen R.J, Leunissen J.A. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997 ;6(3):501-23
4. Van den Ouweland A.M. et al. Nucleotide sequence analysis of the human fur gene. Nucleic Acids Res. 1989 ;17(17):7101-2
5. Smeekens S.P and D.F Steiner. Identification of a human insulinoma cDNA encoding a novel mammalian protein structurally related to the yeast dibasic processing protease Kex2. J Biol Chem. 1990 ;265(6):2997-3000
6. Seidah N.G. et al. cDNA sequence of two distinct pituitary proteins homologous to Kex2 and furin gene products: tissue-specific mRNAs encoding candidates for pro-hormone processing proteinases. DNA Cell Biol. 1990 ;9(10):789
7. Seidah N.G. et al. cDNA sequence of two distinct pituitary proteins homologous to Kex2 and furin gene products: tissue-specific mRNAs encoding candidates for pro-hormone processing proteinases. DNA Cell Biol. 1990 ;9(6):415-24
8. Nakayama K. et al. Identification of the fourth member of the mammalian endoprotease family homologous to the yeast Kex2 protease. Its testis-specific expression. J Biol Chem. 1992 ;267(9):5897-900
9. Kiefer M.C. et al. Identification of a second human subtilisin-like protease gene in the fes/fps region of chromosome 15. DNA Cell Biol. 1991 ;10(10):757-69
10. Lusson J. et al. cDNA structure of the mouse and rat subtilisin/kexin-like PC5: a candidate proprotein convertase expressed in endocrine and nonendocrine cells. Proc Natl Acad Sci U S A. 1993 ;90(14):6691-5
11. Seidah N.G. et al. cDNA structure, tissue distribution, and chromosomal localization of rat PC7, a novel mammalian proprotein convertase closest to yeast kexin-like proteinases. Proc Natl Acad Sci U S A. 1996 ;93(8):3388-93
12. Davey J. et al. Isolation and characterization of krp, a dibasic endopeptidase required for cell viability in the fission yeast Schizosaccharomyces pombe. Embo J. 1994 ;13(24):5910-21
13. Julius D. et al. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984 ;37(3):1075-89
14. Mizuno K. et al. Yeast KEX2 genes encodes an endopeptidase homologous to subtilisin-like serine proteases. Biochem Biophys Res Commun. 1988 ;156(1):246-54
15. Chan S.J. et al. Conservation of the prohormone convertase gene family in metazoa: analysis of cDNAs encoding a PC3-like protein from hydra. Proc Natl Acad Sci U S A. 1992 ;89(15):6678-82
16. Chun J.Y. et al. The function and differential sorting of a family of aplysia prohormone processing enzymes. Neuron. 1994 ;12(4):831-44
17. Peters K, McDowall J, Rose A.M. Mutations in the bli-4 (I) locus of Caenorhabditis elegans disrupt both adult cuticle and early larval development. Genetics. 1991 ;129(1):95-102
18. Roebroek A.J. et al. cDNA sequence of a Drosophila melanogaster gene, Dfur1, encoding a protein structurally related to the subtilisin-like proprotein processing enzyme furin. FEBS Lett. 1991 ;289(2):133-7
19. Smit A.B. et al. Structural characterization of a Lymnaea putative endoprotease related to human furin. FEBS Lett. 1994 ;343(1):27-31
20. Oliva A.A Jr, Chan S.J, Steiner D.F. Evolution of the prohormone convertases: identification of a homologue of PC6 in the protochordate amphioxus. Biochim Biophys Acta. 2000 ;1477(1-2):338-48
21. Oliva A.A Jr, Steiner D.F, Chan S.J. Proprotein convertases in amphioxus: predicted structure and expression of proteases SPC2 and SPC3. Proc Natl Acad Sci U S A. 1995 ;92(8):3591-5
22. Taylor N.A, Van De Ven W.J, Creemers J.W. Curbing activation: proprotein convertases in homeostasis and pathology. Faseb J. 2003 ;17(10):1215-27
23. Essalmani R. et al. Deletion of the gene encoding proprotein convertase 5/6 causes early embryonic lethality in the mouse. Mol Cell Biol. 2006 ;26(1):354-61
24. Holland L.Z, Holland P.W.H, Holland N.D. Revealing homologies between body parts of distantly related animals by in situ hybridization to developmental genes: Amphioxus versus vertebrates. In: (ed.) Ferraris J.D, Palumbi S.R. Molecular Zoology: Advances, Strategies, and Protocols. Wiley-Liss. 1996:267-282
25. Thomson J.D, Higgins D.G, Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994 ;22:4673-4680
26. Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Computer Applications in the Biosciences. 1996 ;12:543-548
27. Saitou N. and Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987 ;4:406-425
28. Guindon S. and Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003 ;52:696-704
29. Jones D.T, Taylor W.R, Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992 ;8:275-282
30. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985 ;39:783-791
31. Fuentes M. et al. Preliminary observations on the spawning conditions of the European amphioxus (Branchiostoma lanceolatum) in captivity. J Exp Zoolog Part B Mol Dev Evol. 2004 ;302(4):384-91
32. Seidah N.G. et al. Mammalian subtilisin/kexin isozyme SKI-1: A widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc Natl Acad Sci U S A. 1999 ;96(4):1321-6
33. Dehal P, Boore J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005 ;3(10):e314
34. Birsoy B. et al. XPACE4 is a localized pro-protein convertase required for mesoderm induction and the cleavage of specific TGFbeta proteins in Xenopus development. Development. 2005 ;132(3):591-602
35. Nelsen S. et al. Proprotein convertase genes in Xenopus development. Dev Dyn. 2005 ;233(3):1038-44
36. Constam D.B, Calfon M, Robertson E.J. SPC4, SPC6, and the novel protease SPC7 are coexpressed with bone morphogenetic proteins at distinct sites during embryogenesis. J Cell Biol. 1996 ;134(1):181-91
37. Roebroek A.J. et al. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998 ;125(24):4863-76
38. Pei D, Weiss S.J. Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature. 1995 ;375(6528):244-7
39. Leighton M, Kadler K.E. Paired basic/Furin-like proprotein convertase cleavage of Pro-BMP-1 in the trans-Golgi network. J Biol Chem. 2003 ;278(20):18478-84
40. Lehmann M. et al. Lack of integrin alpha-chain endoproteolytic cleavage in furin-deficient human colon adenocarcinoma cells LoVo. Biochem J. 1996 ;317:803-9
41. Stawowy P. et al. Endoproteolytic activation of alpha(v) integrin by proprotein convertase PC5 is required for vascular smooth muscle cell adhesion to vitronectin and integrin-dependent signaling. Circulation. 2004 ;109(6):770-6
Author contact
![]() Corresponding address:
Corresponding address:

 Global reach, higher impact
Global reach, higher impact