10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2006; 2(4):194-196. doi:10.7150/ijbs.2.194 This issue Cite
Letter To The Editor
Does WAVE1 contain a GoLoco/GPR motif?
Department of Pharmacology, CB# 7365, 1107 Mary Ellen Jones Building, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7365 USA
Received 2006-7-5; Accepted 2006-7-19; Published 2006-7-25
Heterotrimeric G-protein alpha subunits (Gα) are molecular switches regulated by the binding and hydrolysis of guanosine 5-triphosphate [1]. Non-receptor proteins that modulate the nucleotide binding and hydrolysis activities of Gα proteins have recently become of considerable interest [2, 3]. One class of Gα modulating proteins is defined by the presence of a GoLoco motif(s) in their primary sequence [4]. The GoLoco motif was discovered as a region of sequence homology within novel Gα-binding proteins [5-7], and is also referred to as the G-protein regulatory (GPR) motif [5]. The GoLoco motif is a peptide of 20-40 amino acids that binds Gαi/o•GDP and prevents the spontaneous release of GDP by Gα, thus acting as a guanine nucleotide dissociation inhibitor (GDI) (reviewed in [4]).
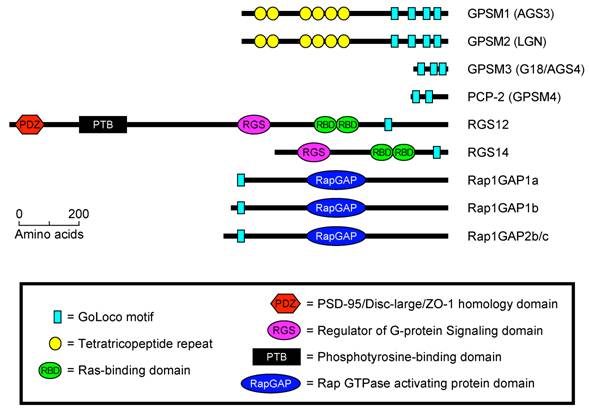
To date, GoLoco/GPR motifs are known to exist in four well-defined families of proteins (Fig. 1). (1) Single GoLoco motifs are present in the RGS and RBD domain containing proteins RGS12, RGS14, and Drosophila Loco (Fig. 1). (2) PCP-2/GPSM4 and GPSM3/G18 contain two or three GoLoco motifs respectively, but no other currently identifiable functional domains or sequence motifs (Fig. 1). (3) Single GoLoco motifs are present in the N-termini of RapGAP domain-containing proteins (Rap1GAP1a, -b and Rap1GAP2b, -c) (Fig. 1). (4) The GoLoco motif is found in multiple arrays in the tetratricopeptide-repeat (TPR) domain containing proteins PINS (Partner of Inscuteable), GPSM2/LGN, and GPSM1/AGS3 (Fig. 1).
Song et al., recently described the presence of a GoLoco/GPR motif in the actin polymerization regulating protein WAVE1 [8]. A synthetic peptide corresponding to this putative WAVE1 GoLoco motif was found to have GDI activity for Gαi1,2,3 in vitro. Furthermore, in coimmunoprecipitation experiments full length WAVE1 was found to interact with Gαi3•GDP but not Gαi3•GTPγS in both COS-7 cell lysates and rat brain extracts [8].
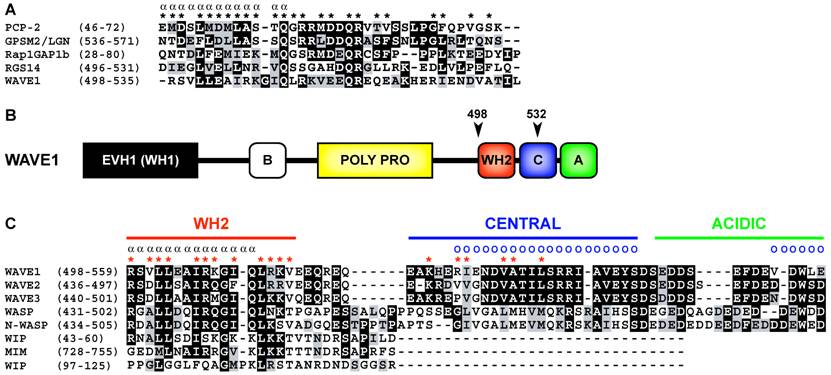
Primary sequence analysis of WAVE1 (a.k.a. SCAR1) indicates that, in many ways, does conform to the GoLoco motif consensus sequence (Fig. 2A). WAVE1 contains a predicted alpha helical region that is terminated by a glutamine residue. Subsequently a [R]-[K]-[aliphatic]-[DE]-[DE]-[Q]-[R] motif characteristic of many GoLoco motifs is present. However, upon further analysis of the primary sequence of WAVE1 it can be observed that the predicted GoLoco/GPR motif overlaps the WASP homology 2 (WH2) domain (Fig. 2B) [8]. The C-terminal domain architecture consisting of a WH2-domain, central domain, and acidic domain (also referred to as the VCA region) is characteristic of proteins such as WAVE1-3, WASP, and N-WASP that stimulate Arp2/3-dependent actin nucleation [10]. The WH2 and central regions interact with actin monomers [11, 12] whereas Arp2/3 binds to the central and acidic domains of WAVE/WASP proteins [13] (binding sites are illustrated in Fig 2C). The structure and function of the WH2-central-acidic cassette in actin nucleating proteins is highly conserved throughout evolution [14].
As the WH2 domain of WAVE1 directly overlies the predicted GoLoco/GPR motif described by Song et al. [8] (Fig 2B, C), logic states that WAVE1 can either contain a WH2 domain or a GoLoco motif but not both. Available biochemical and structural evidence strongly suggests that WAVE1 contains a functional WH2 domain that binds actin monomers with low micromolar affinity, or with nanomolar affinity in the context of an extended WH2 domain containing the central region [12]. Given that WAVE1, 2 and 3 contain the only WH2 domains similar in sequence to GoLoco/GPR motifs, it must be considered highly unlikely that WAVE1 contains a bona fide GoLoco/GPR motif. A simple explanation is that WAVE1 contains a WH2 domain with fortuitous sequence similarity to GoLoco/GPR motifs. That is not to say that the WAVE1/Gαi interactions observed by Song et al. [8] should not be considered important. One could consider this intriguing dual usage of a WH2 domain as a possible case of convergent evolution. In terms of the known structural biology of WH2/actin and Gα/GoLoco complexes this is not out of the question. Both the WH2 domain and GoLoco motif are likely in a unstructured conformation when unbound to ligand [11, 15, 16]; thus it is possible that Gαi and actin could compete for WH2 domain binding. It is worth noting, however, that despite high levels of sequence identity between the three human WAVE isoform WH2 domains (Fig. 1C) only WAVE1 but not WAVE2 nor WAVE3 were found to have GDI activity on Gαi subunits (the signature biochemical activity of a GoLoco/GPR motif). This therefore argues against a conserved role for WH2 domains in heterotrimeric G-protein regulation, in general.
In summary, WH2 domains and GoLoco/GPR motifs are evolutionarily independent protein-binding sequences. Fortuitous sequence similarity between WH2 domains and GoLoco/GPR motifs should not be considered indicative of common ancestry and/or shared function.
Abbreviations
Arp2/3: Actin related protein 2/3; GPSM: G-protein signaling modulator; PCP-2: Purkinje cell protein-2; Rap1GAP: Rap1 GTPase activating protein; RBD: Ras binding domain; RGS: regulator of G-protein signaling; SCAR: suppressor of the Dictyostelium cAMP receptor; WASP: Wiskott-Aldrich syndrome protein; WAVE: WASP-family verprolin homologous protein.
Conflict of interests
Declared none.
References
1. Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987 ;56:615-649
2. Sato M, Blumer JB, Simon V, Lanier SM. Accessory proteins for G proteins: partners in signalling. Annu Rev Pharmacol Toxicol. 2006 ;46:151-187
3. Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005 ;1(2):51-66
4. Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem. 2004 ;73:925-951
5. Takesono A, Cismowski MJ, Ribas C, Bernard M, Chung P, Hazard S, Duzic E, Lanier SM. Receptor-independent activators of heterotrimeric G-protein signaling pathways. J Biol Chem. 1999 ;274(47):33202-33205
6. Siderovski DP, Diverse-Pierluissi M, De Vries L. The GoLoco motif: a Galphai/o binding motif and potential guanine-nucleotide exchange factor. Trends Biochem Sci. 1999 ;24(9):340-341
7. Ponting CP. Raf-like Ras/Rap-binding domains in RGS12- and still-life-like signalling proteins. J Mol Med. 1999 ;77(10):695-698
8. Song KS, Peterson YK, Freidman A, Blumer JB, Sato M, Lanier SM. Identification and characterization of a G-protein regulatory motif in WAVE1. FEBS Lett. 2006 ;580(8):1993-1998
9. Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998 ;95(11):5857-5864
10. Bompard G, Caron E. Regulation of WASP/WAVE proteins: making a long story short. J Cell Biol. 2004 ;166(7):957-962
11. Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005 ;102(46):16644-16649
12. Kelly AE, Kranitz H, Dotsch V, Mullins RD. Actin binding to the C domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. in press
13. Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003 ;10(8):591-598
14. Aspenstrom P. The verprolin family of proteins: regulators of cell morphogenesis and endocytosis. FEBS Lett. 2005 ;579(24):5253-5259
15. Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits. Nature. 2002 ;416(6883):878-881
16. Adhikari A, Sprang SR. Thermodynamic characterization of the binding of activator of G protein signaling 3 (AGS3) and peptides derived from AGS3 with G alpha i1. J Biol Chem. 2003 ;278(51):51825-51832
17. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 ;215(3):403-410
18. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003 ;31(13):3497-3500
Figures
The predicted domain architecture of all human GoLoco motif containing proteins. Domain boundaries were determined using BLAST [17], SMART [9], and ClustalW [18].
(A) Structure-based multiple sequence alignment of quintessential GoLoco motifs and the putative GoLoco motif of human WAVE1. Alpha symbols (α) and asterisks (*) denote alpha helical secondary structure and GoLoco motif contacts with Gα as determined in the X-ray crystallographic structure of the RGS14/Gαi1•GDP complex [15]. The alignment was constructed using ClustalW v1.83 [18] and BOXSHADE3.21 (http://www.ch.embnet.org/software/BOX_form.html). Accession numbers are PCP-2 (AAH38715), GPSM2 (CAG33067), Rap1GAP1b (BAA83674), RGS14 (O08773), and WAVE1 (Q8R5H6). (B) Overall domain architecture of WAVE1. The position of the putative GoLoco/GPR motif in WAVE1 is indicated by arrows and amino acid annotations. Domain nomenclature: EVH1 (Ena/VASP homology 1 domain; also known as the WASP homology 1 (WH1) domain), basic domain (B), proline-rich domain (POLY PRO), WH2 (WASP homology 2 domain), central domain (C), acidic domain (A). (C) Structure-based multiple sequence alignment of the VCA (verprolin homology (WH2)/central/acidic) region of WH2-domain containing proteins, adapted from Chereau et al. [11], and Kelly et al.[12]. Alpha symbols (α) and red asterisks (*) denote alpha helical secondary structure and actin monomer contacting residues as determined by NMR [12] and X-ray crystallography [11]. Blue circles (o) denote Arp2/3 interacting residues as determined by differential line broadening NMR [13]. Accession numbers are WAVE1 (Q8R5H6), WAVE2 (Q8BH43), WAVE3 (Q8VHI6), WASP (CAJ30264), N-WASP (BAA20128), WIP (NP_003378). The alignment was constructed as described in A.
Author contact
![]() Correspondence to: Dr. Francis S. Willard, Tel: 919-843-9364 Fax: 919-966-5640 Email: fwillardunc.edu
Correspondence to: Dr. Francis S. Willard, Tel: 919-843-9364 Fax: 919-966-5640 Email: fwillardunc.edu

 Global reach, higher impact
Global reach, higher impact